Abstract
We identified a previously uncharacterized gene, spermatid maturation 1 (Spem1), encoding a protein exclusively expressed in the cytoplasm of steps 14–16 elongated spermatids in the mouse testis. This protein contains no known functional domains and is highly conserved across mammalian species. Male mice deficient in Spem1 were completely infertile because of deformed sperm characterized by a bent head wrapped around by the neck and the middle piece of the tail. We show that lack of Spem1 causes failure of the cytoplasm to become loose and detach from the head and the neck region of the developing spermatozoa. Retained cytoplasmic components mechanically obstruct the straightening of the sperm head and the stretching of the growing tail, leading to the bending of the head in the neck, followed by the wrapping of the head by the neck or the middle piece of the sperm tail. Our study reveals that proper cytoplasm removal is a genetically regulated process requiring the participation of Spem1 and that lack of Spem1 causes sperm deformation and male infertility.
Keywords: cytoplasmic droplets, gene knockout, spermatogenesis, spermiation, spermiogenesis
Mammalian male fertility depends on successful generation of motile spermatozoa carrying an intact paternal genome and capable of fertilizing the egg. Sperm is produced through a process called spermatogenesis, which can be divided into three phases: mitosis (self-renewal and multiplication of spermatogonia), meiosis (reduction of chromosomal number from diploid to haploid), and spermiogenesis (spermatid differentiation into spermatozoa). Unlike the first two steps of spermatogenesis, dramatic morphological changes occur during spermiogenesis, transforming round spermatids into elongated, tadpole-like spermatozoa with only one-fifth of their original sizes (1, 2). During spermiogenesis, spermatids undergo a complex restructuring program in which the acrosome and sperm tail are formed; DNA is tightly packed leading to a drastic reduction in the size of the nucleus; mitochondria are rearranged along the neck and middle piece of the tail; surface and transmembrane structures (e.g., receptors and ion channels) for zona pellucida binding and signaling are synthesized; and eventually most of the cytoplasm is removed to facilitate motility. These unique cellular reconstruction processes require spermatid-specific genes to execute their regulatory roles. The completion of the human and mouse genome projects has greatly facilitated the efforts of genomewide identification of germ cell-specific genes by using microarray analyses (3, 4) and in silico database mining strategies (5). It is estimated that ≈600–1,000 germ cell-specific genes participate in spermiogenesis (4). Over the past 15 years, gene knockout (KO) studies have identified >20 male germ cell-specific genes or gene isoforms that play essential roles in spermiogenesis (6). These genes are involved in the regulation of acrosome formation (Hrb, Gopc, and Csnk2a2) (7–9), tail formation (Tektin-t, Vdac3, Sepp1, Akap4, and Spag6) (10–14), chromosomal packaging (Prm1, Prm2, Tnp1, Tnp2, and H1t2) (15–18), surface molecules for zona binding and signaling (Adams1-3, Tenr, Apob, Clgn, Catsper1, and Catsper2) (19–25), and energy metabolism (Gapds and sAc) (26, 27). The ongoing efforts to define the function of all of the genes essential for spermiogenesis are of great significance because they allow for the identification of the causative genes for human infertility and thus will make genetic diagnosis available in the future. On the other hand, these genes can also serve as future nonhormonal contraceptive targets (28).
In our efforts to identify male germ cell-specific genes, we found a previously uncharacterized gene encoding a protein exclusively expressed in the cytoplasm of steps 14–16 spermatids (the last three steps of spermiogenesis, see Results). Because the expression of this protein is confined to the maturation phase during spermiogenesis, this gene has been named spermatid maturation 1 (Spem1) by the Mouse Genomic Nomenclature Committee (MGNC; www.informatics.jax.org/mgihome/nomen/#mgnc). To define the physiological role of Spem1, we generated KO mice lacking this gene. Here, we report that Spem1 is essential for proper cytoplasm removal, normal sperm morphology, and male fertility in mice.
Results and Discussion
SPEM1 Is Highly Conserved in Mammals.
In the UniGene collection Mm.159159 (www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Mm&CID=159159), there are 11 EST sequences derived from the same cDNA encoded by Spem1. Among these sequences, three contain an ORF encoding a putative protein with 310 aa. We performed rapid amplification of cDNA end (RACE) assays and obtained the sequences of the full-length cDNA for this gene, which had been deposited to the GenBank (accession no. EF120626). Spem1 has been predicted also in the Ensemble database (Ensemble gene ID ENSMUSG00000041165), where the Spem1 orthologous cDNAs and proteins derived from prediction or genomic/EST sequencing in five mammalian species including the rat, dog, cow, chimpanzee, and human are also available. Alignment analyses of the six SPEM1 orthologs revealed that they are highly conserved during evolution [see supporting information (SI) Fig. 6A]. The mouse SPEM1 shared 68% of its amino acids with the human SPEM1, and 97% amino acid identity was observed between chimpanzee and human SPEM1 proteins (SI Fig. 6B). The first 100 aa at the amino termini and the last 80 aa at the carboxyl termini in the six orthologous proteins are almost identical, suggesting important functional domains may exist in these regions. However, our search by using the InterPro Scan (www.ebi.ac.uk/InterProScan), an integrated search in the PROSITE, Pfam, PRINTS, and other protein family and domain databases failed to recognize any known functional domains in these proteins. We did not find orthologs for this gene in a search of other lower vertebrate genomic databases including those of the fly, zebrafish, and chicken, suggesting that this protein may belong to the mammalian kingdom. Highly conserved protein sequences across all mammalian species imply that this protein may play an important physiological role.
SPEM1 Is Exclusively Expressed in Steps 14–16 Spermatids Right Before Spermiation.
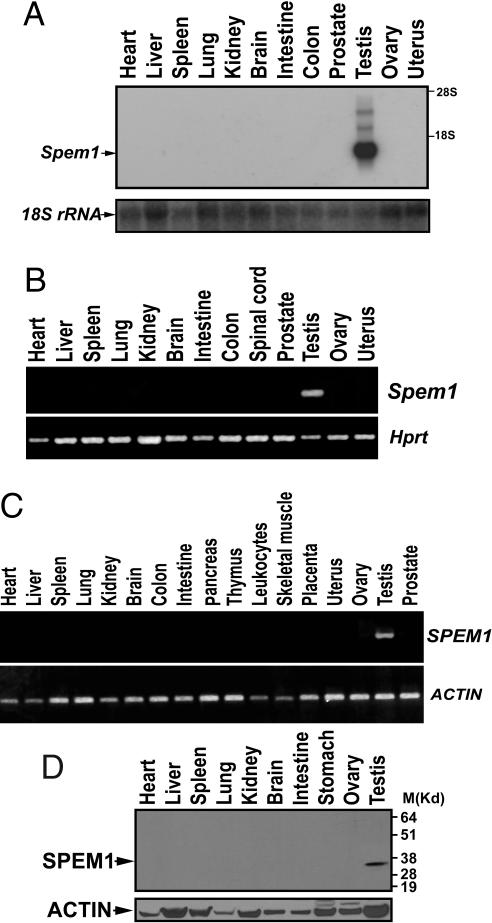
Nine of the 11 EST/cDNA sequences were from the testis in the UniGene database (one was predicted, the other one was from an uncharacterized tissue). Expression data from the Affymetrix GNF1M mouse genechip analyses (61 tissues and cells) also show that this cDNA is exclusively expressed in the testis (for database web site, see Methods). Consistent with these bioinformatic data, our multitissue Northern blot analyses showed that the Spem1 transcript with a size of ≈1.2 kb was detected exclusively in the testis (Fig. 1A). To exclude the possibility of false negativity due to the limited sensitivity of Northern blot analyses, we performed RT-PCR amplification (40 cycles) using cDNAs prepared from 13 mouse tissues, and Spem1 mRNA was detected only in the testis (Fig. 1B). Using human multitissue cDNA panels, we detected human SPEM1 cDNA exclusively in the testis in 17 human tissues tested (Fig. 1C). Consistent with the mRNA expression assays, SPEM1 protein was exclusively detected in the testis by using multitissue Western blot analyses (Fig. 1D). The bioinformatic data and our experimental data all demonstrate that Spem1 is a testis-specific gene.
Fig. 1.
Expression of Spem1 in multiple tissues of the mouse and human. (A) Northern blot analysis of Spem1 mRNA expression in 12 mouse organs. 18S rRNA was used as a loading control. (B) RT-PCR analysis of Spem1 mRNA expression in 13 mouse organs. Hprt was used as a loading control. (C) RT-PCR analysis of SPEM1 in 17 human organs. ACTIN was used a loading control. (D) Western blot analysis of SPEM1 protein expression in 10 mouse organs. ACTIN was used as a loading control.
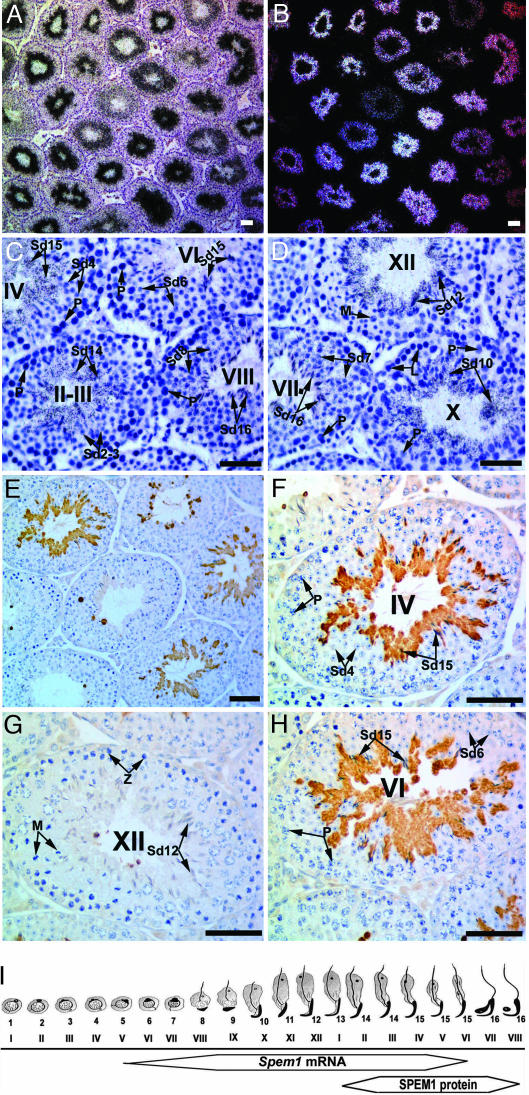
In situ hybridization analyses by using a Spem1-specific antisense riboprobe revealed that the hybridization signals were confined to the luminal compartment, where mainly haploid cells are located (Fig. 2). The expression appeared to be in a stage-specific manner (Fig. 2 A and B) with lower/absent intensity of hybridization signals at stages V–VII as compared with those in other stages (Fig. 2 C, D, and I). High-power microscopic examination revealed that the hybridization signals were overdeveloping spermatids at steps 6–15 (Fig. 2 C, D, and I), whereas no signals were detected in either the early male germ-cell types including spermatogonia, spermatocytes, or somatic cell types, including Sertoli cells and interstitial Leydig cells. Sense probe did not detect any signals above the background levels (data not shown). Interestingly, immunohistochemical detection of SPEM1 by using a polyclonal antibody raised against the full-length SPEM1 protein also unveiled a stage-specific localization pattern with stronger immunoreactivity at stages III–VII and weaker signals at stages I, II, and VIII. This stage-specific expression pattern results from confined expression of SPEM1 to the cytoplasm of steps 14–16 spermatids (Fig. 2 E–I). The majority of SPEM1 protein was removed into residual bodies after spermiation (stages IX–X), suggesting that this protein exerts its physiological role during late spermiogenesis rather than during posttesticular maturation of spermatozoa. The onset of the SPEM1 protein expression is later than that of Spem1 mRNA. The delayed protein expression is a phenomenon common to numerous genes that function during late spermiogenesis. This is because mRNAs encoding proteins that are required for late spermatid development (after step 9) have to be transcribed before transcription completely ceases when chromatin condensation and spermatid elongation start at step 9 and thereafter. The strictly confined expression of SPEM1 in elongated spermatids that are about to be released from the seminiferous epithelium imply that this protein may have a role in cytoplasm removal and/or sperm release (spermiation) because at steps 14–16 most of the key structures of future spermatozoa, including the acrosome, flagellum, and head (condensed and elongated nucleus), are mostly formed.
Fig. 2.
Localization of Spem1 mRNA and protein in the mouse testis. (A–D) Localization of Spem1 mRNA by in situ hybridization. Bright (A, C, and D) and dark (B) field images are shown. Lower-magnification images (A and B) show that the hybridization signals are confined to the luminal compartment, and higher-magnification images (C and D) reveal that the hybridization signals are overelongating and elongated spermatids (steps 9–15). P, spermatocytes; Sd, spermatids. (E and F) Immunohistochemical localization of SPEM1 protein in the adult mouse testis. A lower-magnification image (E) shows that SPEM1 protein is expressed in a stage-specific fashion. Higher-magnification images (F–H) reveal that SPEM1 expression is confined to the cytoplasm of elongated spermatids at steps 14–16. Z, zygotene spermatocytes; M, meiotically dividing spermatocytes; P, pachytene spermatocytes. (I) Schematic summary of the localization of Spem1 mRNA and protein during spermiogenesis. Frames represent the expression windows of Spem1 mRNA and protein, and the width of the frames represents relative expression levels. Arabic numbers represent steps of spermatid development and Roman numerals indicate stages of the seminiferous epithelial cycles. (Scale bars: 50 μm.)
Generation of Spem1 KO Mice.
To define the function of Spem1, we generated a mouse line lacking the Spem1 gene. The Spem1 locus is on chromosome 11. We obtained a ≈16-kb genomic fragment containing the Spem1 gene from a mouse genomic library. A targeting construct was generated such that the entire Spem1 gene, including the 250-bp-long 5′ UTR, exons 1–3 (except for the last 88 bp of exon 3), and 2 introns in between would be deleted after homologous recombination in the R1 ES cells (SI Fig. 7A). Southern blot analysis was used to identify correctly targeted ES cell clones in which a 5′ external probe detected the KO allele as a 5.1-kb band and the WT allele as an 11.5-kb band (SI Fig. 7B). An internal 3′ probe was also used to further verify correct targeting (data not shown). A PCR-based genotyping protocol was developed to distinguish the KO and WT alleles once the germline transmission of the KO allele was confirmed by using Southern blot analyses. Genotyping analysis on a litter of six pups showed the Mendelian inheritance of the KO allele (SI Fig. 7C). Because the entire Spem1 gene (except for the last 88 bp of the last exon) was deleted, the KO allele should be functionally null. As expected, neither Spem1 mRNA nor protein was detected in Spem1−/− mouse testes (SI Fig. 7 D and E). Therefore, we generated Spem1-null mice with a complete inactivation of the Spem1gene.
Spem1-Null Males Are Infertile Because of Sperm Deformation.
Spem1−/−, Spem1+/−, and WT mice show no gross difference during development. Fertility tests by breeding WT, Spem1+/−, and Spem1−/− adult males (six for each genotype) with WT adult females over a period of 6 months revealed that Spem1+/− males display fertility comparable to WT males, whereas Spem1−/− males were completely infertile (SI Table 1). Spem1−/− females displayed normal fertility, which is consistent with the fact that Spem1 is a male germ cell-specific gene and is not expressed in females.
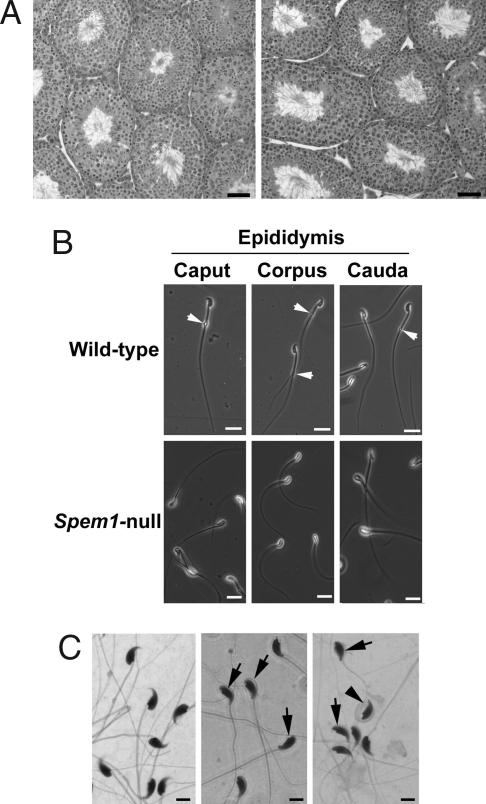
To unveil the reason for the infertility of Spem1−/− mice, we examined the testicular histology (Fig. 3A). WT and Spem1−/− testes showed robust spermatogenesis and no distinguishable differences were observed histologically. When we examined spermatozoa collected from the epididymis, however, we found that the Spem1-null spermatozoa were severely deformed (Fig. 3B). Under the phase contrast microscope, Spem1-null sperm lacked cytoplasmic droplets (Fig. 3B Lower), which are normally located between the neck and the middle piece of the sperm tail in WT mice (Fig. 3B Upper). An obvious defect was that the heads of Spem1-null sperm were all completely bent backward such that the tip of the head was pointing toward the tip of the tail (Fig. 3 B Lower and C). Approximately 85% of the Spem1-null sperm showed no motility and the remaining ≈15% were motile, but the motility appeared to be weaker than that of the WT sperm [see SI Table 1 and SI Movies 1 (for WT sperm) and 2 (for Spem1-null sperm)]. High-power microscopic examination of hematoxylin/eosin-stained sperm smears further confirmed that the bent head is the major defect for Spem1-null sperm and that the severity of head bending varied from a simple bending in the neck to a bent head wrapped around by the neck and the middle piece of the tail (Fig. 3C).
Fig. 3.
Morphological and histological analyses of Spem1-null testes and sperm. (A) Hematoxylin/eosin-stained sections of testes from WT (Left) and Spem1−/− (Right) mice. Both show robust spermatogenesis. (Scale bars: 50 μm.) (B) Phase-contrast microscopic analyses of WT and Spem1-null epididymal sperm. Note that CDs (arrows) are present in the middle piece or the junction between the middle and principal pieces of the tail in WT epididymal sperm, whereas Spem1-null epididymal sperm show no CDs. (Scale bars: 20 μm.) (C) Hematoxylin/eosin-stained sperm smear preparations. (Center and Right) Spem1-null sperm display deformation with heads bent backward pointing to tail tips (arrows) or bent heads wrapped around by necks and middle pieces of the tails (arrowhead). (Scale bars: 10 μm.)
Retention of Cytoplasmic Remnants in the Head/Neck Region Causes Sperm Deformation in Spem1-Null Mice.
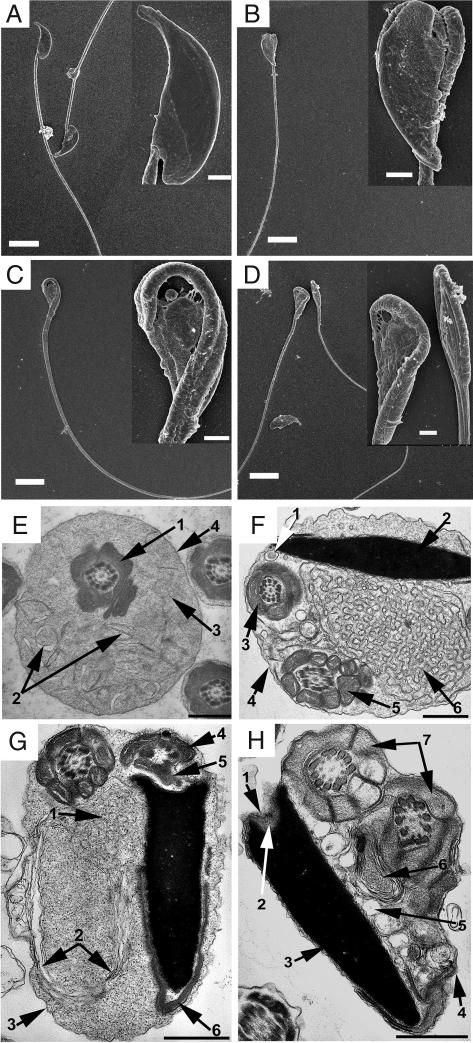
To further define the nature of the deformation of Spem1-null sperm, we examined the ultrastructure of Spem1-null sperm in comparison to WT sperm using scanning electron microscopy (SEM) and transmission electron microscopy (TEM). SEM analyses of epididymal spermatozoa revealed mainly three types of deformation in the head/neck region: First, the sperm head was bent at the neck region toward the tip of the tail at an angle of 180° and the head and the neck stuck together tightly (Fig. 4B); second, the head was bent gradually at the neck region, the neck appeared to be composed of two halves with a groove in the middle, and the head and the neck were connected loosely by membranous tissues (Fig. 4C); and third, the neck and the middle piece of the tail were wrapped several times around the head, forming a disk with the head in the center (Fig. 4D).
Fig. 4.
Ultrastructral analyses of Spem1-null sperm. (A–D) Scanning EM analyses of Spem1-null (B–D) and WT (A) epididymal sperm. Insets are the higher-magnification images of the head and neck region of the sperm. (Scale bars: 10 μm; Inset, 2 μm.) (E–H) TEM analyses of Spem1-null sperm head and neck region. (Scale bars: 1 μm.) (E) Cross-section of the CD of a WT spermatozoon. 1, Middle piece of the tail composed of mitochondrial sheath, outer dense fibers, and axoneme with typical “9 + 2” microtubule structure (9 pairs of peripheral and two central microtubules); 2, vacuoles within the droplet; 3, homogeneous-looking contents within the droplet; 4, outer membrane of the droplet. (F) A section through the bent head and neck region of a Spem1-null spermatozoon. 1, Vault of proximal centriole; 2, nucleus; 3, neck of the sperm composed of a sheath of two longitudinally aligned mitochondria, outer dense fibers, and axoneme with typical “9 + 2” microtubules; 4, outer membrane; 5, middle piece of the tail consisting of the sheath of circumscribing mitochondria and axoneme; 6, small membranous vacuoles with interconnections at areas of folding. (G) A section of a Spem1-null sperm head bent at the neck and wrapped by the middle piece of the tail. The head, neck, and the proximal portion of the middle piece of the tail are surrounded by membranous structures resembling cytoplasmic remnants. 1, Numerous small inter-connected membranous vacuoles; 2, single, large myelin-like vacuoles, which are likely to result from fusion and expansion of those small membranous vacuoles as seen in 1; 3, outer membrane; 4, sperm neck; 5, connecting piece; 6, acrosome. (H) A Spem1-null spermatozoon with the head bent at the neck and wrapped around by the middle piece of the tail. The middle piece and head are held together by cytoplasmic remnants with similar structures as seen in F and G. 1, Interspaces between the basal plate and connecting piece (absent here); 2, basal plate; 3, nucleus; 4, outer membrane of the cytoplasmic remnants; 5, large membranous vacuoles; 6, a large myelin-like vacuole; 7, middle pieces of the tail wrapping around the head.
One common feature of these various forms of deformation was that the head and the neck/middle piece of the tail were held together by tissues resembling remnants of the cytoplasm that should have been completely shed off during spermiation. It appears that the former cytoplasmic portion near the head and neck junction of future spermatozoon failed to become loose and to detach from the fully formed head and neck region, causing a mechanical obstruction that prevented the elongating/elongated sperm head and neck/middle piece from stretching straight. The SEM results strongly suggest that the mechanical obstruction occurs during late spermiogenesis when the cytoplasm begins to be removed before spermiation. We dissected seminiferous tubules at stage VIII and performed SEM analyses (SI Fig. 8). As expected, all forms of deformation observed in the epididymal spermatozoa had already occurred within the seminiferous epithelium at stage VIII, where spermatozoa are being released and cytoplasm has been/is being removed in the WT testes (SI Fig. 8). SEM analyses on spermatozoa in stage VIII tubules showed that the remnants of shed cytoplasm were the sources of mechanical obstruction, which was holding the head and the neck region together.
TEM analyses of epididymal sperm demonstrated that all of the structural components at the neck and middle piece of the tail are intact in Spem1-null sperm compared with WT sperm (SI Fig. 9). Consistent with light microscopic (LM) and SEM observation, TEM showed that the head bent at the neck/middle piece and the bent head and neck were held together by membranous tissues resembling the shed cytoplasm. The TEM characteristics of these cytoplasm remnant-like tissues include numerous interconnected membranous vacuoles (Fig. 4F), well bordered myelin-like discs (Fig. 4G), and single, large vacuoles (Fig. 4H). Given that all Spem1-null sperm do not have cytoplasmic droplets (CDs), we compared the ultrastructure of normal CDs (Fig. 4E) with the cytoplasmic remnants in the Spem1-null sperm head and neck region (Fig. 4 F–H). Although normal CDs are also membranous structures, the contents in the CDs at EM levels showed different features compared with those of cytoplasmic remnants on the Spem1-null sperm. The contents of CDs appeared to be homogenous and contain many slim leaf-shaped and evenly distributed vesicles. These findings imply that normal CD formation may serve as a hallmark for proper cytoplasmic removal. Alternatively, formation of normal CDs is accompanied by proper cytoplasmic removal. Therefore, it is likely that in the absence of Spem1, CDs fail to form because of impaired detaching and shedding of the cytoplasm from the nucleus and the neck region of the sperm during late spermiogenesis.
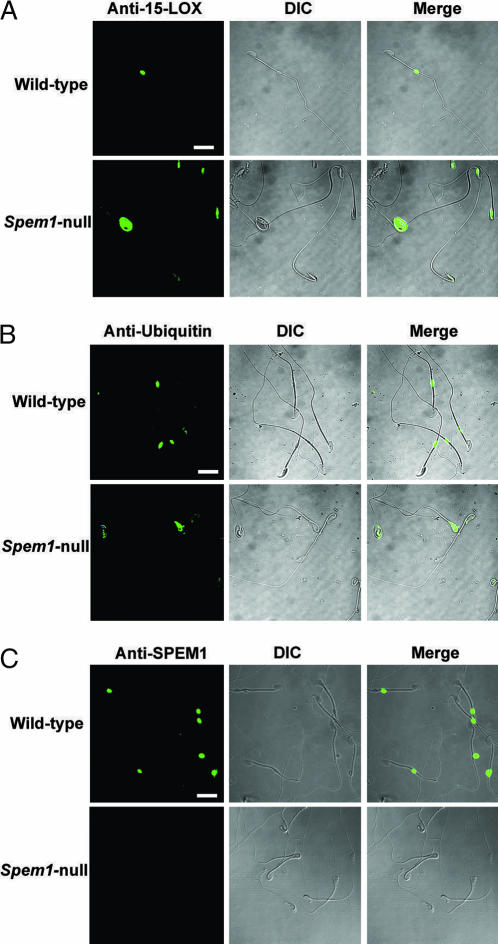
The Cytoplasmic Remnants in the Spem1-Null Sperm Head and Neck Region Contain Proteins Detectable in Normal Cytoplasmic Droplets.
Although the ultrastructure of CDs is different from that of the cytoplasmic remnants on the Spem1-null sperm, they should share some left-over proteins from the removed cytoplasm if they have the same origin. Ubiquitin (29) and 15-lipoxygenase (15-LOX) (30) have been detected in CDs and are suggested to have a role in the cytoplasm removal. However, many proteins that are highly expressed in the cytoplasm of late spermatids (e.g., steps 13–16) can also be detected in CDs (H.Z., J.J., and W.Y., unpublished data), suggesting that these proteins may represent the residual proteins derived from the removed cytoplasm. In fact, ubiquitin, 15-LOX, and SPEM1 are all highly expressed in the cytoplasm of late spermatids and can all be detected in CDs (Fig. 5). Therefore, these proteins may not necessarily have a role in the posttesticular life of spermatozoa although they can be detected in CDs. Nevertheless, detection of these proteins does reflect that CDs structurally are derived from the cytoplasm of late spermatids. To examine whether the cytoplasmic remnants on the Spem1-null sperm are also derived from the components of former cytoplasm, we performed immunofluorescent detection of ubiquitin, 15-LOX, and SPEM1 (Fig. 5). As expected, both ubiquitin (Fig. 5A) and 15-LOX (Fig. 5B) are detected in CDs of WT epididymal sperm, whereas they were presenting the regions where the sperm head and neck were wrapped together on the Spem1-null sperm. These results further support our EM observations, suggesting that these membranous tissues are derived from the shed cytoplasm of late spermatids. The presence of SPEM1 on the WT sperm CDs and the lack of SPEM1 on the Spem1-null sperm further confirmed that SPEM1 had been completely inactivated in the KO mice (Fig. 5C).
Fig. 5.
Immunofluorescent detection of ubiquitin, 15-LOX, and SPEM1 in WT and Spem1-null epididymal sperm. (A) 15-LOX (green), which is normally detectable in the CD of the WT sperm, is located in the cytoplasmic remnants around the bent head and neck region of the Spem1-null sperm. (B) An anti-ubiquitin antibody immunoreactive to the CDs of the WT sperm show positive staining (green) in the region around the bent head and/or neck region. (C) Detection of SPEM1 (green) in the CDs in WT sperm. SPEM1 is absent in Spem1-null sperm. All panels are in the same magnification. (Scale bar: 20 μm.)
Intracytoplasmic Injection of Spem1-Null Sperm Heads into Eggs Produced Live-Born Pups.
Spem1-null sperms could not fertilize eggs by natural mating (SI Table 1) or in vitro fertilization (data not shown) because of significantly decreased motility and their bent heads and necks. The sperm with their necks wrapped around their heads failed to bind to the zona pellucida (ZP) because the acrosome and surface ZP-binding proteins were covered by the neck. Sperm with the head completely bent backward could bind ZP but failed to penetrate because the beating of the sperm flagellum generated forces that pulled the sperm head away from the eggs because of the wrong orientation. However, the defects in the sperm nucleus could not be excluded. We, therefore, injected the heads from the motile Spem1-null sperm collected from the epididymis into WT mouse eggs. No significant differences in the preimplantation development of injected eggs (two-cell to blastocyst stage; SI Table 2) and the number of live-born offsprings (SI Table 3) were observed between WT and Spem1-null sperm used, demonstrating that Spem1-null sperm heads contain normal paternal genome. This experiment confirms that a lack of SPEM1 does not impair the nuclear condensation and packaging process, which is consistent with our morphological analyses at the LM and EM levels showing no obvious structural defects in the Spem1-null sperm head.
Intriguingly, a subpopulation of spermatozoa in mice deficient of Tnp1 (31–34), Tnp2 (31–34), Prm1 (15, 35), Prm2 (15, 35), H1t2 (18), Camk4 (36), or Csnk2a2 (37) also shows failure of cytoplasmic removal resulting in the bending of the sperm head backward and the wrapping of the tail around the bent head. These genes encode nuclear proteins which participate in nuclear packaging and condensation during late spermiogenesis. The similar phenotype suggests that aberrant nuclear packaging and condensation may affect the same pathway that is affected by the absence of SPEM1. Because SPEM1 is cytoplasmic and Spem1-null spermatozoa uniformly display the above-mentioned deformation, it is likely that SPEM1 is involved in a specific pathway responsible for the proper removal of cytoplasm in late spermiogenesis.
Cytoplasmic removal from late-elongated spermatids is important to the generation of functional gametes. Although the structural and morphological aspects of cytoplasm removal during spermiation have been studied (38–42), the genetic control of this important process remains poorly understood. The apoptosis regulators caspases and cytochrome c have been shown to be involved in cytoplasmic removal and spermatid individualization in Drosophila (43, 44). However, no genes have been directly linked to the regulation of cytoplasm removal from late elongated spermatids in mammals. Our data support the notion that proper cytoplasmic removal is a genetically regulated process. Lack of Spem1 impairs the process of cytoplasm removal probably by preventing the cytoplasm from detaching from the spermatid nucleus and the neck region of the developing flagellum. Retained cytoplasmic components obstruct the straightening of the head and/or the stretching of the growing tail especially in the neck region, thus resulting in the bending of the head in the neck region or the wrapping of the neck around the head. Interestingly, the aberrant removal of spermatid cytoplasm is associated with a lack of CDs, suggesting that the presence of CDs is a reflection of normal cytoplasm removal. Although their function remains debatable (45), CDs appear to be a functional apparatus of sperm that may be important for sperm maturation through the epididymal transition.
Taken together, our data demonstrate that the lack of Spem1 causes retention of cytoplasmic remnants on the head and neck region, and the retained cytoplasmic remnants obstruct the straightening and stretching of the sperm head and neck, leading to sperm deformation and male infertility. Because the SPEM1 protein is reasonably conserved (≈68%) between mice and humans, mutations in human SPEM1 gene may lead to similar sperm deformation and male infertility. However, no human cases with this specific type of sperm deformation have been reported. More careful sperm morphological examination by high-power phase-contrast microscopy may be necessary to identify this type of defect. Further studies on the molecular action of SPEM1 will help us gain more insight into the genetic control of cytoplasm removal during spermiogenesis.
Materials and methods
RNA Analyses.
RT-PCR and Northern blot analyses were performed as described in refs. 46 and 47.
Protein Analyses.
Immunohistochemical and immunofluorescent staining were performed as described in refs. 46 and 47.
Generation of Spem1 KO Mice.
Targeting vector construction, electroporation, selection of targeted ES cells, blastocyst injection, and chimeric mouse breeding were performed as described in ref. 48.
Intracytoplasmic Sperm Injection.
Intracytoplasmic sperm injection was performed as previously described in ref. 49.
For an extensive description of the materials and methods, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Vicki Madden and Dr. Deborah A. O'Brien (University of North Carolina, Chapel Hill, NC) for sharing protocols for electron microscopy analyses, David Young for editing the text, Dr. Sean Ward for help with confocal imaging, and the Nevada Transgenic Center (University of Nevada) for blastocyst injection. The Zeiss LSM 510 confocal microscope was obtained with support from National Institutes of Health Grant 1 S10 RR16871. This work was supported by a start-up fund from the University of Nevada (Reno) and also in part by National Institutes of Health Grants HD048855 and HD050281 (to W.Y.).
Abbreviations
- Spem1
spermatid maturation 1
- KO
knockout
- TEM
transmission electron microscopy
- CD
cytoplasmic droplet
- 15-LOX
15-lipoxygenase.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. EF120626).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701669104/DC1.
References
- 1.Oakberg EF. Am J Anat. 1956;99:391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- 2.Fawcett DW. Dev Biol. 1975;44:394–436. doi: 10.1016/0012-1606(75)90411-x. [DOI] [PubMed] [Google Scholar]
- 3.Shima JE, McLean DJ, McCarrey JR, Griswold MD. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 4.Schultz N, Hamra FK, Garbers DL. Proc Natl Acad Sci USA. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin YN, Matzuk MM. Semin Reprod Med. 2005;23:201–212. doi: 10.1055/s-2005-872448. [DOI] [PubMed] [Google Scholar]
- 6.Matzuk MM, Lamb DJ. Nat Cell Biol. 2002;4(Suppl):s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- 7.Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM. Science. 2001;294:1531–1533. doi: 10.1126/science.1063665. [DOI] [PubMed] [Google Scholar]
- 8.Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, Yanagida K, Sato A, Toshimori K, Noda T. Proc Natl Acad Sci USA. 2002;99:11211–11216. doi: 10.1073/pnas.162027899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escalier D, Silvius D, Xu X. Mol Reprod Dev. 2003;66:190–201. doi: 10.1002/mrd.10346. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, Nishimune Y. Mol Cell Biol. 2004;24:7958–7964. doi: 10.1128/MCB.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampson MJ, Decker WK, Beaudet AL, Ruitenbeek W, Armstrong D, Hicks MJ, Craigen WJ. J Biol Chem. 2001;276:39206–39212. doi: 10.1074/jbc.M104724200. [DOI] [PubMed] [Google Scholar]
- 12.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Biol Reprod. 2005;73:201–211. doi: 10.1095/biolreprod.105.040360. [DOI] [PubMed] [Google Scholar]
- 13.Miki K, Willis WD, Brown PR, Goulding EH, Fulcher KD, Eddy EM. Dev Biol. 2002;248:331–342. doi: 10.1006/dbio.2002.0728. [DOI] [PubMed] [Google Scholar]
- 14.Sapiro R, Kostetskii I, Olds-Clarke P, Gerton GL, Radice GL, Strauss IJ. Mol Cell Biol. 2002;22:6298–6305. doi: 10.1128/MCB.22.17.6298-6305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Nat Genet. 2001;28:82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- 16.Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, Weil MM, Behringer RR, Meistrich ML. Proc Natl Acad Sci USA. 2000;97:4683–4688. doi: 10.1073/pnas.97.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Proc Natl Acad Sci USA. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, et al. Mol Cell Biol. 2005;25:7107–7119. doi: 10.1128/MCB.25.16.7107-7119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuasnicu PS, Ellerman DA, Cohen DJ, Busso D, Morgenfeld MM, Da Ros VG. Arch Med Res. 2001;32:614–618. doi: 10.1016/s0188-4409(01)00321-6. [DOI] [PubMed] [Google Scholar]
- 20.Kaji K, Kudo A. Reproduction. 2004;127:423–429. doi: 10.1530/rep.1.00163. [DOI] [PubMed] [Google Scholar]
- 21.Connolly CM, Dearth AT, Braun RE. Dev Biol. 2005;278:13–21. doi: 10.1016/j.ydbio.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Huang LS, Voyiaziakis E, Markenson DF, Sokol KA, Hayek T, Breslow JL. J Clin Invest. 1995;96:2152–2161. doi: 10.1172/JCI118269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikawa M, Nakanishi T, Yamada S, Wada I, Kominami K, Tanaka H, Nozaki M, Nishimune Y, Okabe M. Dev Biol. 2001;240:254–261. doi: 10.1006/dbio.2001.0462. [DOI] [PubMed] [Google Scholar]
- 24.Ren D, Navarro B, Perez G, Jackson AC, Hsu S, Shi Q, Tilly JL, Clapham DE. Nature. 2001;413:603–609. doi: 10.1038/35098027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quill TA, Sugden SA, Rossi KL, Doolittle LK, Hammer RE, Garbers DL. Proc Natl Acad Sci USA. 2003;100:14869–14874. doi: 10.1073/pnas.2136654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O'Brien DA. Proc Natl Acad Sci USA. 2004;101:16501–16506. doi: 10.1073/pnas.0407708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esposito G, Jaiswal BS, Xie F, Krajnc-Franken MA, Robben TJ, Strik AM, Kuil C, Philipsen RL, van Duin M, Conti M, et al. Proc Natl Acad Sci USA. 2004;101:2993–2998. doi: 10.1073/pnas.0400050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nass SJ, Strauss JF., III Science. 2004;303:1769–1771. doi: 10.1126/science.303.5665.1769b. [DOI] [PubMed] [Google Scholar]
- 29.Kuster CE, Hess RA, Althouse GC. J Androl. 2004;25:340–347. doi: 10.1002/j.1939-4640.2004.tb02798.x. [DOI] [PubMed] [Google Scholar]
- 30.Fischer KA, Van Leyen K, Lovercamp KW, Manandhar G, Sutovsky M, Feng D, Safranski T, Sutovsky P. Reproduction. 2005;130:213–222. doi: 10.1530/rep.1.00646. [DOI] [PubMed] [Google Scholar]
- 31.Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- 32.Zhao M, Shirley CR, Yu YE, Mohapatra B, Zhang Y, Unni E, Deng JM, Arango NA, Terry NH, Weil MM, et al. Mol Cell Biol. 2001;21:7243–7255. doi: 10.1128/MCB.21.21.7243-7255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao M, Shirley CR, Mounsey S, Meistrich ML. Biol Reprod. 2004;71:1016–1025. doi: 10.1095/biolreprod.104.028191. [DOI] [PubMed] [Google Scholar]
- 34.Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Biol Reprod. 2004;71:1220–1229. doi: 10.1095/biolreprod.104.029363. [DOI] [PubMed] [Google Scholar]
- 35.Cho C, Jung-Ha H, Willis WD, Goulding EH, Stein P, Xu Z, Schultz RM, Hecht NB, Eddy EM. Biol Reprod. 2003;69:211–217. doi: 10.1095/biolreprod.102.015115. [DOI] [PubMed] [Google Scholar]
- 36.Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. Nat Genet. 2000;25:448–452. doi: 10.1038/78153. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Toselli PA, Russell LD, Seldin DC. Nat Genet. 1999;23:118–121. doi: 10.1038/12729. [DOI] [PubMed] [Google Scholar]
- 38.Russell LD, Saxena NK, Turner TT. Tissue Cell. 1989;21:361–379. doi: 10.1016/0040-8166(89)90051-7. [DOI] [PubMed] [Google Scholar]
- 39.Russell LD, Goh JC, Rashed RM, Vogl AW. Biol Reprod. 1988;39:105–118. doi: 10.1095/biolreprod39.1.105. [DOI] [PubMed] [Google Scholar]
- 40.Weber JE, Russell LD. Am J Anat. 1987;180:1–24. doi: 10.1002/aja.1001800102. [DOI] [PubMed] [Google Scholar]
- 41.Sprando RL, Russell LD. Tissue Cell. 1987;19:479–493. doi: 10.1016/0040-8166(87)90042-5. [DOI] [PubMed] [Google Scholar]
- 42.Russell L, Clermont Y. Anat Rec. 1976;185:259–278. doi: 10.1002/ar.1091850302. [DOI] [PubMed] [Google Scholar]
- 43.Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, Guo M, Baehrecke EH, Hay BA. Development (Cambridge, UK) 2006;133:3305–3315. doi: 10.1242/dev.02495. [DOI] [PubMed] [Google Scholar]
- 44.Arama E, Agapite J, Steller H. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 45.Cooper TG. Hum Reprod. 2005;20:9–11. doi: 10.1093/humrep/deh555. [DOI] [PubMed] [Google Scholar]
- 46.Wang S, Zheng H, Esaki Y, Kelly F, Yan W. Biol Reprod. 2006;74:102–108. doi: 10.1095/biolreprod.105.045484. [DOI] [PubMed] [Google Scholar]
- 47.Jin JL, O'Doherty AM, Wang S, Zheng H, Sanders KM, Yan W. Biol Reprod. 2005;73:1235–1242. doi: 10.1095/biolreprod.105.045468. [DOI] [PubMed] [Google Scholar]
- 48.Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 49.Yanagimachi R, Wakayama T, Kishikawa H, Fimia GM, Monaco L, Sassone-Corsi P. Proc Natl Acad Sci USA. 2004;101:1691–1695. doi: 10.1073/pnas.0307832100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.