Abstract
Primitive neuroectodermal tumors (PNETs), including medulloblastoma (PNET/MB) and supratentorial PNET (sPNET), are the most common malignant brain tumors of childhood. The stabilization of telomere lengths by telomerase activation is an important step in carcinogenesis and cell immortalization. Epigallocatechin gallate (EGCG), the major polyphenol in green tea, is a telomerase inhibitor with antiproliferative and anticarcinogenic effects against different types of cancer. In this study, we used real-time reverse transcriptase-polymerase chain reaction to measure the mRNA expression of the human telomerase reverse transcriptase (hTERT) in 50 primary PNET samples (43 PNET/MB, 7 sPNET), 14 normal human brain samples, and 6 human PNET cell lines. Compared to normal human cerebellum, 38/50 (76%) primary PNET samples had ≥5-fold upregulated hTERT mRNA expression. We then examined PNET cell lines for telomerase activity using a quantitative telomeric repeat amplification protocol (TRAP), and for telomere length using terminal restriction fragment analysis. While a positive correlation between hTERT mRNA expression and telomerase activity was detected in PNET cell lines, no correlation was found between telomerase activity and telomere length. Treatment of PNET cell lines with EGCG resulted in a dose-dependent inhibition of telomerase activity at micromolar levels. Although EGCG displayed strong proliferation inhibitory effects against TRAP-positive PNET cell lines, it had no significant effect against TRAP-negative D425 cells. These results provide evidence for a possible role of telomerase in the pathogenesis of most PNETs and indicate that subsets of PNETs maintain telomere length by alternative mechanisms. Inhibition of telomerase function represents a novel experimental therapeutic strategy in childhood PNETs that warrants further investigation.
Primitive neuroectodermal tumors (PNETs)4 of the cerebellum, also termed medulloblastomas (PNET/ MBs), and supratentorial PNETs (sPNETs) are the most common malignant pediatric neoplasms of the central nervous system (Gurney et al., 1999). PNETs constitute more than 20% of all pediatric brain tumors and are characterized by their aggressive clinical behavior and a high risk of leptomeningeal dissemination. With current treatment strategies, nearly half of all patients will eventually die from progressive tumors. Accordingly, the identification of novel therapeutic strategies remains a major goal.
Unlimited replicative potential is an important acquired capacity of cancer (Hanahan and Weinberg, 2000). Telomere maintenance is evident in virtually all types of malignant cells; 85% to 90% of them show upregulated expression of the enzyme telomerase, which adds hexa-nucleotide repeats onto the ends of telomeric DNA (Bryan and Cech, 1999; Shay and Bacchetti, 1997), while most of the remainder use one or more different mechanisms known as alternative lengthening of telomeres (ALT) (Bryan et al., 1995; Henson et al., 2002; Neumann and Reddel, 2002).
Telomerase, or telomere terminal transferase, is a ribonucleoprotein that catalyzes the de novo synthesis and elongation of telomeric repeats at chromosomal ends by using the RNA segment within its molecule as a template (Blackburn, 1991; Bosoy et al., 2003; Feng et al., 1995). While functionally immortal germline cells express telomerase and maintain adequate telomeric repeats, most human somatic cells do not express telomerase. They fail to acquire telomerase activity in successive cultures and become senescent. In contrast, telomerase activity has been detected in numerous cancer cells and tissues (Kim et al., 1994; Mu and Wei, 2002).
Brain tumors in which telomerase activity has been detected include glioblastoma, anaplastic astrocytoma, oligodendroglioma, and meningioma (Hiraga et al., 1998; Langford et al., 1995; Sano et al., 1998). Increased expression of the telomerase RNA component was found to be associated with increased cell proliferation in astrocytomas and ependymomas (Grosso and Schiffer, 1998). Malignant brain tumors had a higher positive rate of telomerase activity than benign tumors (Sano et al., 1998). Moreover, telomerase activity in meningiomas was found to correlate with poor survival outcome, indicating that these apparently benign tumors may contain a population of immortal cells (Langford et al., 1997; Simon et al., 2000). Telomerase expression and activity in PNET have not yet been addressed specifically, and examination of the few existing reports yields controversial results. Falchetti et al. (2002) found no telomerase activity in two primary PNET/MB samples. Conversely, DeMasters et al. (1997) reported telomerase activity in 5/5 primary PNET samples, and Liu et al. (2000) reported telomerase activity in Med-3 cells.
In this study, we measured the mRNA expression of the human telomerase reverse transcriptase (hTERT) in 50 primary PNET samples, 14 normal brain samples, and 6 human PNET cell lines, using real-time reverse transcriptase (RT)-polymerase chain reaction (PCR). We then examined PNET cell lines for telomerase activity using a quantitative telomeric repeat amplification protocol (TRAP) and for telomere length using terminal restriction fragment (TRF) analysis. In addition, inhibition of telomerase in PNET cell lines was investigated for its potential as a novel therapeutic strategy in childhood PNET.
Materials and Methods
Tumor Cells
DAOY human PNET/MB cells were purchased from American Type Culture Collection (Manassas, Va.). D341 and D425 human PNET/MB cells, UW228-2 human PNET/MB cells, Med-1 human PNET/MB cells, and CHP707 human PNET cells were gifts from several universities and medical centers. DAOY, D341, and D425 cells were cultured in Richter’s Zinc Option medium/10% fetal bovine serum (FBS) (D341 and D425 additionally cultured with 1% nonessential amino acids); CHP707 cells were cultured in RPMI 1640/10% FBS; Med-1 and UW228-2 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco Invitrogen, Basel, Switzer-land)/10% FBS. All cell cultures were maintained at 37°C in a humidified atmosphere with 5% CO2.
Primary Tumor and Normal Brain Samples
Frozen tumor tissue to perform RT-PCR was available from 50 PNET patients treated at the Children’s Hospital of Philadelphia, Pennsylvania, USA (n = 31), and the University Children’s Hospital of Vienna, Austria (n = 19). All diagnoses were confirmed by histological assessment of a tumor specimen obtained at surgery by experienced neuropathologists. Tumor samples were snap-frozen in liquid nitrogen in the operating room and then stored at –80°C until further analysis. The median age at diagnosis for these PNET patients was 6.4 years (range 0.3–32.3 years); 33 (66%) were male and 17 (34%) were female. Tumor location was cerebellar in 43 patients and supratentorial in 7 patients. RNA of normal brain samples was purchased from Stratagene (La Jolla, Calif.) and Clonetech (Palo Alto, Calif.); the samples included fetal brain samples (18 and 19 weeks of gestation), cerebellum, frontal cortex (50 years of age), and whole brain (15 years of age). Near-normal brain samples included cerebellum of a 4-year-old glioma patient, temporal cortex from a 4-year-old epilepsy surgery patient, temporal cortex from a 14-year-old epilepsy surgery patient, and occipital cortex from a 19-year-old epilepsy surgery patient. Normal, noncommercial brain samples were snap-frozen in liquid nitrogen in the operating room and then stored at –80°C until further analysis.
Real-Time Quantitative RT-PCR
Isolation of total RNA and cDNA synthesis was performed as previously described (Grotzer et al., 2001; Huber et al., 2001; Zuzak et al., 2002). Kinetic real-time PCR quantification of hTERT mRNA was performed by using an ABI Prism 7700 Sequence Detection System (Applied Biosystems, Rotkreuz, Switzerland), also as described previously (Bieche et al., 2000). Primers and probes for hTERT and the endogenous control 18S rRNA were purchased from Microsynth (Balgach, Switzerland). For each PCR run, a mix was prepared of 200 nM of each primer, 400 nM of probe, 30 ng of cDNA, and 1 × of PCR master mix (Applied Biosystems) in a final volume of 25 μ l. The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, and a 50-cycle countdown at 95°C for 15 s and 60°C for 1 min. Experiments were performed in triplicate for each data point. Each PCR run included the 5 points of the standard curve (serially diluted DAOY PNET cell line cDNAs), a no-template control, the calibrator cDNA, and patient cDNAs.
The precise amount of total RNA added to each reaction mix (based on absorbance) and its quality are difficult to assess. We therefore also quantified transcripts of the 18S rRNA housekeeping gene as endogenous controls, and each sample was normalized on the basis of its 18S rRNA content. Relative expression of hTERT mRNA was calculated by using the comparative threshold cycle (CT) method (Giulietti et al., 2001). The amount of hTERT, normalized to 18S rRNA and relative to the calibrator, is then given by 2−Δ Δ CT, where = Δ Δ CT Δ CT(sample) - Δ CT(calibrator), and Δ CT is the CT of the target gene (hTERT) subtracted from the CT of the housekeeping gene (18S rRNA). PCR efficiencies for hTERT and 18S rRNA were measured as previously described (Bustin, 2000; Heid et al., 1996) and were found to be >95%. Thus, the Δ Δ CT method, in which the efficiency of PCR amplification of the target gene must be approximately equal to that of the housekeeping gene, is applicable.
Telomerase Activity
Telomerase activity in PNET cell lines was measured by using the Telomerase PCR enzyme-linked immunosorbent assay (ELISA) kit (Roche Diagnostics, Basel, Switzerland). This assay combines the highly specific amplification of telomerase DNA products (TRAP assay [Kim et al., 1994]) with the nonradioactive detection of those PCR products by a photometric enzyme immunoassay (ELISA). For this procedure, 106 cells were lysed and homogenized in 200 μ l of lysis buffer (included in the telomerase detection kit). After a 30-min incubation on ice, the lysates were centrifuged at 12,000 rpm for 30 min at 4°C. Protein concentration was measured by using the DC protein reagent set (Bio-Rad, Reinach, Switzerland). In the elongation step, 2 μ l of cell extract (2 μ g of protein/μ l) was incubated in 50 μ l of reaction mixture at 25°C for 30 min to allow the telomerase to add telomeric repeats (TTAGGG) to the end of the biotin-labeled synthetic P1-TS primer. The products extended by telomerase were then amplified by PCR in the presence of the biotin-labeled P1-TS primer and another primer, P2. Amplification products from the PCR were next immobilized in the well of a streptavidin-coated microplate and hybridized to a digoxigenin (DIG)-labeled probe specific for the telomeric repeat sequence. Finally, the DIG-labeled hybrids were visualized with a peroxidase-conjugated anti-DIG antibody and a colorimetric peroxidase substrate and then quantified photometrically. To visualize the typical 6-nucleotide-ladder resulting from the TRAP assay, the amplification products from the PCR were separated by 12.5% polyacrylamide gel electrophoresis, blotted onto a positively charged membrane, and hybridized with a DIG-labeled probe specific for the telomeric repeat sequence. Telomerase activity in PNET samples was quantified (by using the Telomerase PCR ELISA kit) by measuring absorbance values at 450 nm reading against a blank (reference wavelength A690 nm). The level of telomerase activity was calculated by subtracting the mean of the absorbance readings of the negative controls from those of the samples. In this assay, samples are regarded as telomerase-positive if the difference in absorbance is higher than 0.2. The telomerase activity measured by this assay does not reflect hTERT enzyme activity in a rigorous biochemical sense, but may also depend on hTERT enzyme concentration.
DNA Isolation
Genomic DNA was isolated by using the QIAamp DNA Mini Kit (Qiagen AG, Basel, Switzerland) according to procedures recommended by the manufacturer. In brief, tumor cells were lysed with proteinase K and incubated with RNase A. Genomic DNA was then purified by a silica gel-based method. The yield of DNA from the various samples was calculated by spectrophotometry.
Telomere Length
Telomere length in PNET cells was determined by using the TeloTAGGG Telomere Length Assay (Roche Diagnostics, Basel, Switzerland) to measure the length of TRFs. In brief, 1 μ g of genomic DNA was digested overnight with the restriction enzymes HinfI and RsaI, which do not digest within telomeric repeats. Following DNA digestion, the DNA fragments were separated by 8% agarose gel electrophoresis and transferred to a nylon membrane by Southern blotting. The blotted DNA fragments were hybridized with a DIG-labeled probe that recognizes TRFs. Probe-TRF complexes were detected with alkaline phosphatase-conjugated anti-DIG antibody and CDP-Star (Applied Biosystems, Rotkreuz, Switzerland) chemiluminescent alkaline phosphatase substrate. The mean length for TRF was determined by comparing each fragment with a known molecular weight standard and by quantifying the intensity of the chemiluminescent signal from that fragment with the Lumi-Imager F1 workstation (Roche Diagnostics, Basel, Switzerland).
Cytotoxicity Assay
Cell viability was quantified by using a colorimetric 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS) assay (Promega, Wallisellen, Switzerland) as previously described (Grotzer et al., 2000). Briefly, 100 μ l of target cell suspension was added to each well of 96-well microtiter plates, and each plate was incubated at 37°C in a humidified 5% CO2 atmosphere. Following incubation, 10 μ l of MTS working solution was added to each culture well, and the cultures were incubated for 4 h at 37°C in a humidified 5% CO2 atmosphere. Each condition was performed in triplicate. The absorbance values of each well were measured with a microplate spectrophotometer (Molecular Devices, Sunnyvale, Calif.) at 490 nm.
Results
hTERT mRNA Expression
Using real-time quantitative RT-PCR, we determined hTERT mRNA expression in 43 primary PNET/MB samples, 7 sPNET samples, 14 normal brain samples, and 6 PNET cell lines. Compared to normal human cerebellum, 38/50 primary PNET samples (76%) had upregulated hTERT mRNA expression (defined as an hTERT expression level ≥ 5-fold normal human cerebellum; Fig. 1). hTERT mRNA expression levels in PNET/MB samples (median 24.65, range 0.01–183.55) and sPNET samples (median 22.37, range 0.02–55.46) were significantly higher than hTERT mRNA expression levels of normal brain samples (median 1.55, range 0.01– 3.5; Mann-Whitney U test, P < 0.0001, P = 0.006, respectively). No major differences were found between PNET/MB samples and sPNET samples (P = 0.13). However, the number of sPNET samples is too small to draw firm conclusions. No significant correlation was found between hTERT mRNA expression and metastatic stage, age, or gender. In PNET cell lines, the hTERT mRNA expression levels were 13 to 48 times higher than in normal human cerebellum. Med-1 and D341 PNET cells showed the highest hTERT mRNA expression levels, and DAOY and D425 PNET cells the lowest.
Fig 1.
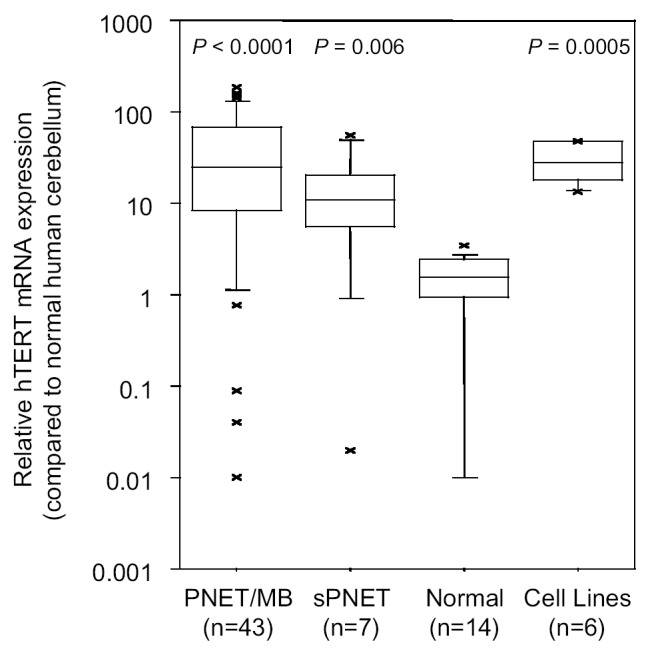
Human telomerase reverse transcriptase mRNA expression in 43 primary PNET/MB, 7 primary sPNET, and 14 normal brain samples, and in 6 PNET cell lines. hTERT mRNA expression was determined by quantitative real-time RT-PCR by using 18S rRNA as an endogenous control and normal human cerebellum as a calibrator. hTERT mRNA expression levels in PNET/MB, sPNET, and PNET cell lines were significantly higher than hTERT mRNA expression levels in normal brain samples. The floor and the ceiling of the box indicate 25th and 75th percentile values, respectively. The horizontal line inside the box represents the median (50th percentile). The top and bottom whiskers of the box identify the 90th and the 10th percentile values, respectively.
Telomerase Activity
To determine the enzymatic activity of telomerase in CHP707 (12.1 kpb), we performed a modified TRAP assay with the cell lysates of 6 human lines. After poly-acrylamide gel electrophoresis separation of the amplification products from the TRAP assay, we observed typical 6-nucleotide ladders in D341, DAOY, Med-1, and CHP707 PNET cells (Fig. 2A). To quantify telomerase activity, we then measured amplification products from the TRAP assay photometrically. D341, DAOY, Med-1, and CHP707 were TRAP-positive, UW228-2 had some telomerase activity, and D425 was TRAP-negative (Fig. 2B).
Fig. 2.
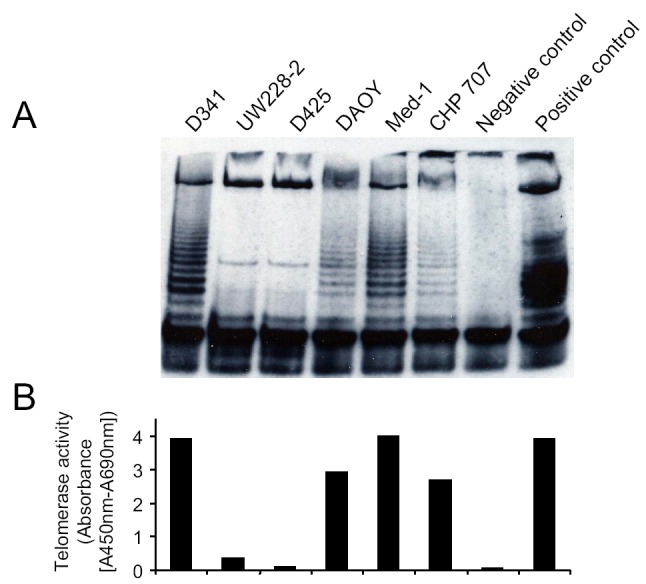
Telomerase activity in 6 PNET cell lines as determined by the TRAP assay (A) and the Telomerase PCR ELISA kit (B). Controls included cell extract prepared from immortalized telomerase-expressing human kidney cells (positive control) and DNase-free RNase-treated D341 cells (negative control). While telomerase activity was positive in D341, DAOY, Med-1, and CHP707 PNET cells, telomerase activity was weak in UW228-2 and negative in D425 PNET cells.
Telomere Length
Telomere lengths of PNET cells were analyzed by measuring the TRF length of TRF by using Southern blotting with a (TTAGGG) probe. D425 (11.9 kbp) and CHP707 (12.1 kbp) PNET cells had the longest TRF, and D341 (5.1 kbp) and UW228-2 (5 kbp) PNET cells the shortest (Fig. 3). This indicates greater telomere length in D425 and CHP707 than in D341 and UW228-2.
Fig. 3.
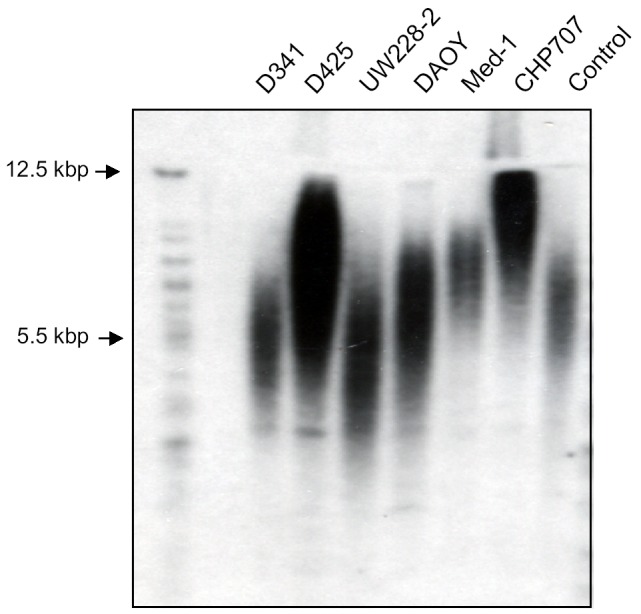
Telomere length in 6 PNET cell lines as determined by the TeloTAGGG Telomere Length Assay. The length of the TRF in each sample was determined by comparing each fragment with a known molecular weight standard. This length, together with the intensity of the chemiluminescent signal from each fragment, was used to calculate the mean TRF length for each cell line. CHP707 (12.1 kb) and D425 (11.9 kb) PNET cells had the longest TRF, and D341 (5.1 kb) and UW228-2 PNET cells (5 kb) PNET cells the shortest.
Comparison of Telomerase Activity, hTERT mRNA Expression, and TRF Length
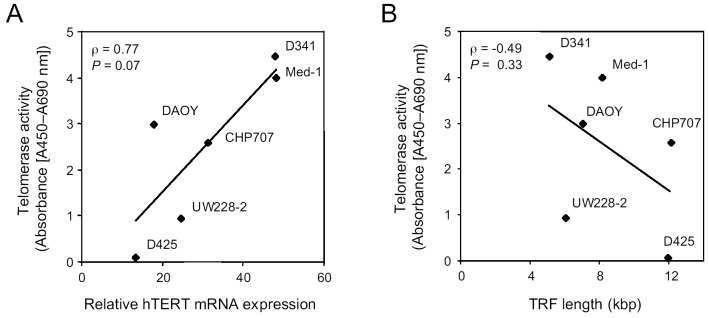
We then compared telomerase activity with hTERT mRNA expression and telomere length. While a marginally significant correlation between telomerase activity and hTERT mRNA expression was detected in PNET cell lines (Spearman’s correlation ρ = 0.77, P = 0.07; Fig. 4A), no significant correlation was found between telomerase activity and telomere length (ρ = –0.49, P = 0.33; Fig. 4B). TRAP-positive cell lines had a high hTERT mRNA expression, while TRAP-negative D425 PNET cells had the lowest hTERT mRNA expression. Interestingly, the TRAP-negative D425 PNET cells had a greater telomere length than most TRAP-positive PNET cells. This indicates that upregulated telomerase is functionally active in PNET cells and that telomere maintenance in PNET cells is not always the result of upregulated telomerase expression, but may also be the result of ALT.
Fig. 4.
Comparison of telomerase activity with hTERT mRNA expression (A) and telomere length (B) in 6 PNET cell lines. While telomerase activity correlated well with hTERT mRNA expression, it did not correlate with telomere length.
Suppression of PNET Cell Proliferation by EGCG
To test whether inhibition of telomerase alters proliferation of PNET cells, we treated TRAP-positive and TRAP-negative PNET cells with the telomerase inhibitor EGCG (Calbiochem, San Diego, Calif.) at various concentrations for 3 days (Fig. 5). Treatment of TRAP-positive DAOY, D341, and UW228-2 cells with EGCG resulted in dose-dependent cytotoxicity. However, treatment of TRAP-negative D425 cells resulted in significantly less growth inhibition. It remains to be evaluated whether inhibition of telomerase is the only mechanism by which EGCG inhibits proliferation in PNET cells.
Fig. 5.
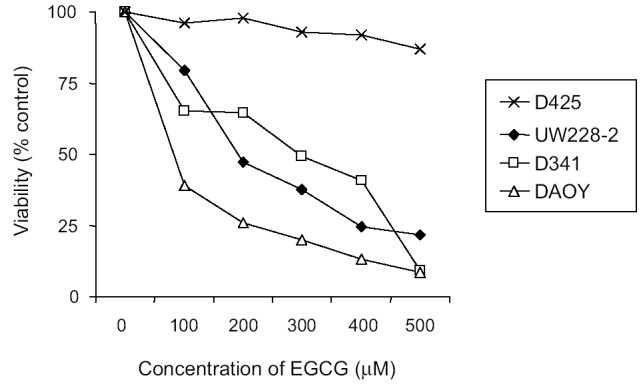
Dose-dependent cytotoxicity of EGCG to human PNET cells. PNET cells were cultured in 10% serum medium in 96-well plates and treated with EGCG at the concentrations indicated. Cell survival was determined after 72 h by the MTS assay. Values represent the mean percentage of survival compared to control cells (n = 3). Standard deviations were less than 10%.
In Vitro Telomerase Inhibition by EGCG
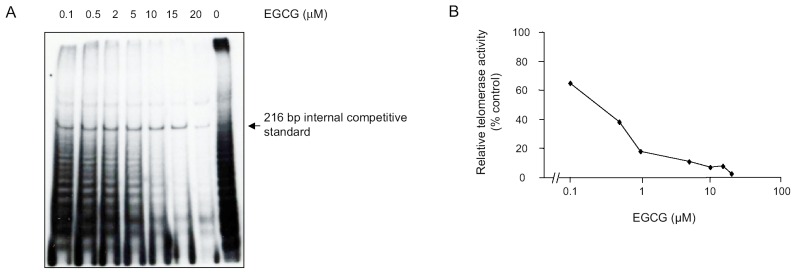
To identify the EGCG concentration that inhibits telomerase activity in PNET cells, TRAP assays were performed on cell lysates from DAOY cells treated with different concentrations of EGCG. Telomerase activity was found to be inhibited in a dose-dependent manner, with ≥ 50% inhibition at 0.5 μ M of EGCG (Fig. 6).
Fig. 6.
Dose-dependent inhibition of telomerase activity by EGCG in human DAOY PNET cells as determined by the TRAP assay (A) and the Telomerase PCR ELISA kit (B). Relative telomerase activity was inhibited in a dose-dependent manner, with ≥ 50% inhibition at 0.5 μ M EGCG.
Discussion
In this first comprehensive study of telomere maintenance in childhood PNET, we demonstrated that telomerase expression is upregulated in 76% of primary PNET samples. Since hTERT expression correlated well with telomerase activity in PNET cells, one can assume that upregulated telomerase is also functionally active in primary PNET. To confirm this, further studies in primary tumors are needed. The finding that the majority of childhood PNET samples express high levels of telomerase, whereas normal brain tissues do not, indicates that telomerase has an important role in the pathogenesis of PNET. This makes telomerase in PNET, as in other cancers, a potentially useful diagnostic marker as well as a therapeutic target (White et al., 2001).
Reported telomerase inhibitors include antisense oligodeoxynucleotides (Feng et al., 1995), ribozymes (Folini et al., 2000; Yokoyama et al., 1998), RT inhibitors (Strahl and Blackburn, 1996), G-quadruplex interacting compounds (Harrison et al., 1999; Read et al., 1999), small molecule inhibitors (Naasani et al., 1999), isothiazolone derivatives (Hayakawa et al., 1999), rhodacyanine derivatives (Wadhwa et al., 2002), and natural compounds including EGCG, the major polyphenol in green tea (Fujiki et al., 2001; Kinjo et al., 2002; Lyu et al., 2002; Naasani et al., 1998, 2003). EGCG has been identified as a telomerase inhibitor with anticarcinogenic effects in various animal organs such as the esophagus, stomach, duodenum, colon, liver, pancreas, lung, breast, and skin (Naasani et al., 1998, 2003; Suganuma et al., 1999). In nude mice models bearing both telomerase-dependent and -independent xenograft tumors cloned from a single human cancer progeny, only the telomerase-dependent tumors responded to oral administration of EGCG (Naasani et al., 2003).
In the present study, EGCG was found to inhibit both telomerase activity and cellular proliferation in TRAP-positive cells. This indicates that EGCG also functions as a telomerase inhibitor in PNET cells. Further studies using PNET cells engineered to overexpress or under-express hTERT are therefore of interest. Interestingly, EGCG inhibited telomerase activity in PNET cells at a relatively low concentration (≥ 50% inhibition at 0.5 μ M EGCG). A single cup of green tea contains about 150 mg EGCG, and some tea lovers consume up to 10 cups a day (Yang and Wang, 1993). Normally, the blood level of EGCG after consuming the equivalent of 2 to 3 cups of tea is 0.1 to 0.6 μ M (Lee et al., 1995). In studies with mice and rats in which inhibition of skin, lung, and esophageal tumorigenesis was found, the blood level of EGCG was 0.1 to 0.3 μ M, and the tissue levels were 0.2 to 1 μ M (Yang et al., 1996). However, the EGCG level found in rat brain after a single administration of EGCG (500 mg/kg body weight) was only 0.5 nM (Nakagawa and Miyazawa, 1997). Therefore, it remains to be seen whether the telomerase inhibitory effect of EGCG can be exploited against brain tumors in vivo.
Mechanisms of EGCG anticancer activity other than inhibition of telomerase include inhibition of angiogenesis, antioxidant activity, and interaction with certain enzymes or proteins implicated in cancer (e.g., TNF-alpha; Cao and Cao, 1999; Fujiki et al., 1999; Hasaniya et al., 1997). Therefore, future studies must also evaluate whether inhibition of telomerase is the only mechanism by which EGCG inhibits proliferation in PNET cells.
Negative telomerase activity and increased mean telomere length in D425 PNET cells, as well as normal hTERT mRNA expression in 24% of primary PNET samples, indicate that subsets of PNET maintain telomere length by ALT. Successful telomere-targeted anti-cancer therapy for PNET might therefore require a combination of telomerase and ALT inhibitors.
Our results provide evidence for a possible role of telomerase in the pathogenesis of PNET and indicate the presence of ALT in subsets of PNET. Inhibition of telomerase function and ALT represents a novel experimental therapeutic strategy in childhood PNET that warrants further investigation.
Acknowledgments
D341 and D425 human PNET/MB cells were the gift of Dr. Henry Friedman (Duke University Medical Center, Durham, North Carolina, USA). UW228-2 human PNET/MB cells were given by Dr. John R. Silber (University of Washington, Seattle, Washington, USA), Med-1 human PNET/MB cells by Dr. Torsten Pietsch (University of Bonn, Germany), and CHP707 human PNET cells by Dr. David Pleasure (The Children’s Hospital of Philadelphia, Pennsylvania, USA). Tumor samples were the kind gifts of Dr. Peter Phillips (The Children’s Hospital of Philadelphia, USA) and Dr. Irene Slavc (University Children’s Hospital of Vienna, Austria). Near-normal brain samples were the kind gift of Dr. Peter Phillips (The Children’s Hospital of Philadelphia, Pennsylvania, USA).
Footnotes
Supported by the Schweizer Forschungsstiftung Kind und Krebs.
Abbreviations used are as follows: ALT, alternative lengthening of telomeres; DIG, digoxigenin; EGCG, epigallocatechin gallate; ELISA, enzyme-linked immunosorbent assay; FBS, fetal bovine serum; hTERT, human telomerase reverse transcriptase; MB, medulloblastoma; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt; PCR, polymerase chain reaction; PNET, primitive neuroectodermal tumor; RT, reverse transcriptase; sPNET, supratentorial PNET; TRAP, telomeric repeat amplification protocol; TRF, terminal restriction fragment.
References
- Bieche I, Nogues C, Paradis V, Olivi M, Bedossa P, Lidereau R, Vidaud M. Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000;6:452–459. [PubMed] [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Bosoy D, Peng Y, Mian IS, Lue NF. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J Biol Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. Embo J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Cao Y, Cao R. Angiogenesis inhibited by drinking tea. Nature. 1999;398:381. doi: 10.1038/18793. [DOI] [PubMed] [Google Scholar]
- DeMasters BK, Markham N, Lillehei KO, Shroyer KR. Differential telomerase expression in human primary intracranial tumors. Am J Clin Pathol. 1997;107:548–554. doi: 10.1093/ajcp/107.5.548. [DOI] [PubMed] [Google Scholar]
- Falchetti ML, Larocca LM, Pallini R. Telomerase in brain tumors. Childs Nerv Syst. 2002;18:112–117. doi: 10.1007/s00381-002-0562-7. [DOI] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang S.-S, Weinrich SL, Avilion AA, Chiu C.-P, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Folini M, Colella G, Villa R, Lualdi S, Daidone MG, Zaffaroni N. Inhibition of telomerase activity by a hammerhead ribozyme targeting the RNA component of telomerase in human melanoma cells. J Invest Dermatol. 2000;114:259–267. doi: 10.1046/j.1523-1747.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- Fujiki H, Suganuma M, Okabe S, Sueoka E, Sueoka N, Fujimoto N, Goto Y, Matsuyama S, Imai K, Nakachi K. Cancer prevention with green tea and monitoring by a new biomarker, hnRNP B1. Mutat Res. 2001;480–481:299–304. doi: 10.1016/s0027-5107(01)00189-0. [DOI] [PubMed] [Google Scholar]
- Fujiki H, Suganuma M, Okabe S, Sueoka E, Suga K, Imai K, Nakachi K, Kimura S. Mechanistic findings of green tea as cancer preventive for humans. Proc Soc Exp Biol Med. 1999;220:225–228. doi: 10.1046/j.1525-1373.1999.d01-38.x. [DOI] [PubMed] [Google Scholar]
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- Grosso R, Schiffer D. Prognostic significance of telomerase in brain tumors. Crit Rev Neurosurg. 1998;8:244–247. doi: 10.1007/s003290050083. [DOI] [PubMed] [Google Scholar]
- Grotzer MA, Eggert A, Zuzak TJ, Janss AJ, Marwaha S, Wiewrodt BR, Ikegaki N, Brodeur GM, Phillips PC. Resistance to TRAIL-induced apoptosis in primitive neuroectodermal brain tumor cells correlates with a loss of caspase-8 expression. Oncogene. 2000;19:4604–4610. doi: 10.1038/sj.onc.1203816. [DOI] [PubMed] [Google Scholar]
- Grotzer MA, Hogarty MD, Janss AJ, Liu X, Zhao H, Eggert A, Sutton LN, Rorke LB, Brodeur GM, Phillips PC. MYC messenger RNA expression predicts survival outcome in childhood primitive neuroectodermal tumor/medulloblastoma. Clin Cancer Res. 2001;7:2425–2433. [PubMed] [Google Scholar]
- Gurney, J.G., Smith, M.A., and Bunin, G.R. (1999) CNS and miscellaneous intracranial and intraspinal neoplasms. In: Ries, L.A.G., Smith, M.A., Gurney, J.G., Linet, M., Tamra, T., Young, J.L. and Bunin, G.R. (Eds.), Cancer Incidence and Survival Among Children and Adolescents: United States SEER Program 1975–1995, National Cancer Institute, SEER Program. NIH Pub. No. 99-4649. Bethesda, Md.: National Institutes of Health.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harrison RJ, Gowan SM, Kelland LR, Neidle S. Human telomerase inhibition by substituted acridine derivatives. Bioorg Med Chem Lett. 1999;9:2463–2468. doi: 10.1016/s0960-894x(99)00394-7. [DOI] [PubMed] [Google Scholar]
- Hasaniya N, Youn K, Xu M, Hernaez J, Dashwood R. Inhibitory activity of green and black tea in a free radical-generating system using 2-amino-3-methylimidazo[4,5-f]quinoline as substrate. Jpn J Cancer Res. 1997;88:553–558. doi: 10.1111/j.1349-7006.1997.tb00418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa N, Nozawa K, Ogawa A, Kato N, Yoshida K, Akamatsu K, Tsuchiya M, Nagasaka A, Yoshida S. Isothiazolone derivatives selectively inhibit telomerase from human and rat cancer cells in vitro. Biochemistry. 1999;38:11501–11507. doi: 10.1021/bi982829k. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Ohnishi T, Izumoto S, Miyahara E, Kanemura Y, Matsumura H, Arita N. Telomerase activity and alterations in telomere length in human brain tumors. Cancer Res. 1998;58:2117–2125. [PubMed] [Google Scholar]
- Huber H, Eggert A, Janss AJ, Wiewrodt R, Zhao H, Sutton LN, Rorke LB, Phillips PC, Grotzer MA. Angiogenic profile of childhood primitive neuroectodermal brain tumours/medulloblastomas. Eur J Cancer. 2001;37:2064–2072. doi: 10.1016/s0959-8049(01)00225-8. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kinjo J, Nagao T, Tanaka T, Nonaka G, Okawa M, Nohara T, Okabe H. Activity-guided fractionation of green tea extract with antiproliferative activity against human stomach cancer cells. Biol Pharm Bull. 2002;25:1238–1240. doi: 10.1248/bpb.25.1238. [DOI] [PubMed] [Google Scholar]
- Langford LA, Piatyszek MA, Xu R, Schold SC, Jr, Shay JW. Telomerase activity in human brain tumours. Lancet. 1995;346:1267–1268. doi: 10.1016/s0140-6736(95)91865-5. [DOI] [PubMed] [Google Scholar]
- Langford LA, Piatyszek MA, Xu R, Schold SC, Jr, Wright WE, Shay JW. Telomerase activity in ordinary meningiomas predicts poor outcome. Human Pathol. 1997;28:416–420. doi: 10.1016/s0046-8177(97)90029-0. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Wang ZY, Li H, Chen L, Sun Y, Gobbo S, Balentine DA, Yang CS. Analysis of plasma and urinary tea polyphenols in human subjects. Cancer Epidemiol Biomarkers Prev. 1995;4:393–399. [PubMed] [Google Scholar]
- Liu J, Guo L, Luo Y, Li JW, Li H. All transretinoic acid suppresses in vitro growth and down-regulates LIF gene expression as well as telomerase activity of human medulloblastoma cells. Anticancer Res. 2000;20:2659–2664. [PubMed] [Google Scholar]
- Lyu SY, Choi SH, Park WB. Korean mistletoe lectin-induced apoptosis in hepatocarcinoma cells is associated with inhibition of telomerase via mitochondrial controlled pathway independent of p53. Arch Pharm Res. 2002;25:93–101. doi: 10.1007/BF02975269. [DOI] [PubMed] [Google Scholar]
- Mu J, Wei LX. Telomere and telomerase in oncology. Cell Res. 2002;12:1–7. doi: 10.1038/sj.cr.7290104. [DOI] [PubMed] [Google Scholar]
- Naasani I, Seimiya H, Tsuruo T. Telomerase inhibition, telomere shortening, and senescence of cancer cells by tea catechins. Biochem Biophys Res Commun. 1998;249:391–396. doi: 10.1006/bbrc.1998.9075. [DOI] [PubMed] [Google Scholar]
- Naasani I, Seimiya H, Yamori T, Tsuruo T. FJ5002: A potent telomerase inhibitor identified by exploiting the disease-oriented screening program with COMPARE analysis. Cancer Res. 1999;59:4004–4011. [PubMed] [Google Scholar]
- Naasani I, Oh-Hashi F, Oh-Hara T, Feng WY, Johnston J, Chan K, Tsuruo T. Blocking telomerase by dietary polyphenols is a major mechanism for limiting the growth of human cancer cells in vitro and in vivo. Cancer Res. 2003;63:824–830. [PubMed] [Google Scholar]
- Nakagawa K, Miyazawa T. Absorption and distribution of tea catechin, (-)-epigallocatechin-3-gallate, in the rat. J Nutr Sci Vitaminol (Tokyo) 1997;43:679–684. doi: 10.3177/jnsv.43.679. [DOI] [PubMed] [Google Scholar]
- Neumann AA, Reddel RR. Telomere maintenance and cancer—look, no telomerase. Nature Rev Cancer. 2002;2:879–884. doi: 10.1038/nrc929. [DOI] [PubMed] [Google Scholar]
- Read MA, Wood AA, Harrison JR, Gowan SM, Kelland LR, Dosanjh HS, Neidle S. Molecular modeling studies on G-quadruplex complexes of telomerase inhibitors: Structure-activity relationships. J Med Chem. 1999;42:4538–4546. doi: 10.1021/jm990287e. [DOI] [PubMed] [Google Scholar]
- Sano T, Asai A, Mishima K, Fujimaki T, Kirino T. Telomerase activity in 144 brain tumours. Br J Cancer. 1998;77:1633–1637. doi: 10.1038/bjc.1998.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Simon M, Park TW, Leuenroth S, Hans VH, Loning T, Schramm J. Telomerase activity and expression of the telomerase catalytic subunit, hTERT, in meningioma progression. J Neurosurg. 2000;92:832–840. doi: 10.3171/jns.2000.92.5.0832. [DOI] [PubMed] [Google Scholar]
- Strahl C, Blackburn EH. Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalized human cell lines. Mol Cell Biol. 1996;16:53–65. doi: 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma M, Okabe S, Sueoka N, Sueoka E, Matsuyama S, Imai K, Nakachi K, Fujiki H. Green tea and cancer chemoprevention. Mutat Res. 1999;428:339–344. doi: 10.1016/s1383-5742(99)00059-9. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Colgin L, Yaguchi T, Taira K, Reddel RR, Kaul SC. Rhodacyanine dye MKT-077 inhibits in vitro telomerase assay but has no detectable effects on telomerase activity in vivo. Cancer Res. 2002;62:4434–4438. [PubMed] [Google Scholar]
- White LK, Wright WE, Shay JW. Telomerase inhibitors. Trends Biotechnol. 2001;19:114–120. doi: 10.1016/s0167-7799(00)01541-9. [DOI] [PubMed] [Google Scholar]
- Yang CS, Chen L, Lee MJ, Landau JM. Effects of tea on carcinogenesis in animal models and humans. Adv Exp Med Biol. 1996;401:51–61. doi: 10.1007/978-1-4613-0399-2_5. [DOI] [PubMed] [Google Scholar]
- Yang CS, Wang ZY. Tea and cancer. J Natl Cancer Inst. 1993;85:1038–1049. doi: 10.1093/jnci/85.13.1038. [DOI] [PubMed] [Google Scholar]
- Yokoyama Y, Takahashi Y, Shinohara A, Lian Z, Wan X, Niwa K, Tamaya T. Attenuation of telomerase activity by a hammerhead ribozyme targeting the template region of telomerase RNA in endometrial carcinoma cells. Cancer Res. 1998;58:5406–5410. [PubMed] [Google Scholar]
- Zuzak TJ, Steinhoff DF, Sutton LN, Phillips PC, Eggert A, Grotzer MA. Loss of caspase-8 mRNA expression is common in childhood primitive neuroectodermal brain tumour/medulloblastoma. Eur J Cancer. 2002;38:83–91. doi: 10.1016/s0959-8049(01)00355-0. [DOI] [PubMed] [Google Scholar]