Abstract
Survival periods vary considerably for patients with high-grade astrocytomas, and reliable prognostic markers are not currently available. We therefore investigated whether genetic losses from chromosomes 1p, 19q, 9p, or 10q were associated with survival in 89 high-grade astrocytomas using tissue microarrays (TMAs) derived from Radiation Therapy Oncology Group clinical trials. Cases included 15 anaplastic astrocytomas (AAs) and 74 glioblastomas (GBMs) selected on the basis of survival times significantly shorter or longer than the expected median. Genetic analysis was performed by TMA-fluorescence in situ hybridization (FISH) on array sections using 8 DNA probes, including those directed at 1p32, 19q13.4, 9p21 (p16/CDKN2A), and 10q (PTEN and DMBT1). Genetic status for each locus was correlated with patient survival group, and data were analyzed by using Fisher’s exact test of association (adjusted P = 0.025). Losses of chromosome 1p, either alone or in combination with 19q, were encountered in only 2 cases, both AAs. This contrasts with oligodendrogliomas, in which combined 1p and 19q losses are frequent and predictive of prolonged survival. Solitary 19q loss was noted in 3/15 AAs and in 7/70 GBMs and was more frequent in the long-term survival group (P = 0.041, AA and GBM combined). Chromosome 9p loss was seen in 5/8 AAs and 39/57 GBMs, whereas chromosome 10q loss was detected in 4/15 AAs and 48/68 GBMs. The 9p and 10q deletions were slightly more frequent in short-term survivors, though none of the comparisons achieved statistical significance. Long-term and short-term survival groups of high-grade astrocytomas appear to have dissimilar frequencies of 19q, 9p, and 10q deletions. TMA-FISH is a rapid and efficient way of evaluating genetic alterations in such tumors.
High-grade astrocytomas, which include anaplastic astrocytoma (AA3; WHO grade III) and glioblastoma (GBM; WHO grade IV), are the most common primary brain tumors (CBTRUS, 2002). These are fatal diseases, yet patient survival times vary substantially. Histological grade does not account for all the individual prognostic variability (Cavenee et al., 2000). Clinical variables including age, performance status, and mental health status are predictive of survival, especially when considered in a “grouped” fashion within a histologic category (Curran et al., 1993a). It is unclear whether differential prognosis is solely a function of the clinical parameters or whether clinical parameters reflect inherent biological diversity among neoplasms.
Interest has risen in defining genetic subtypes of infiltrating gliomas for enhanced prognostic accuracy and potentially for directing therapies (Brat et al., 2002; Hunter et al., 2003; Perry et al., 2003). Genetic studies have shown that 60% to 80% of oligodendrogliomas harbor deletions of chromosome 1p and 19q and that this “genetically favorable” subset has enhanced sensitivity to PCV (procarbazine, CCNU [lomustine], vincristine) chemotherapy (Cairncross et al., 1998; Smith et al., 2000). It is possible that other types of infiltrating gliomas with 1p and 19q losses, including high-grade astrocytomas, could be associated with prolonged survival or enhanced therapeutic responses. While astrocytomas have lower frequencies of 1p and 19q losses, the incidence of astrocytomas is 8- to 10-fold greater than that of oligodendrogliomas (CBTRUS, 2002). The prognostic significance of 1p and 19q losses in astrocytic neoplasms is not clear (Ino et al., 2002; Schmidt et al., 2002; Smith et al., 2000).
Likewise, well-defined markers of poor prognosis in high-grade astrocytomas would be valuable. Chromosome 10q losses are considered to be a late event in the progression of astrocytomas since they are more common in GBMs than in AAs and are tightly correlated with the GBM phenotype overall (Fujisawa et al., 1999; Kunwar et al., 2001). Chromosome 10 losses may also be an independent marker of aggressive behavior among high-grade astrocytomas (Balesaria et al., 1999; Perry et al., 1997a; Tada et al., 2001). Loss of chromosome 9p21, the site of the tumor suppressor gene p16/CDKN2A, has been associated with aggressive clinical behavior in oligodendrogliomas and astrocytomas (Cairncross et al., 1998; Kamiryo et al., 2002; Maruno et al., 1996; Perry et al., 1999).
We have investigated whether losses from chromosomes 1p, 19q, 9p, or 10q were associated with long versus short survival in 89 high-grade astrocytomas in order to determine their potential utility as genetic markers of prognosis. Fluorescence in situ hybridization (FISH) for these loci was performed on tissue microarrays (TMA-FISH), and the findings were compared among survival groups (Fuller and Perry, 2002; Fuller et al., 2002). The cases were retrieved from prior Radiation Therapy Oncology Group (RTOG) trials, providing a unique opportunity to study high-grade astrocytomas from patients with similar clinical enrollment criteria who had not received prior therapy and who had complete clinical follow-up.
Materials and Methods
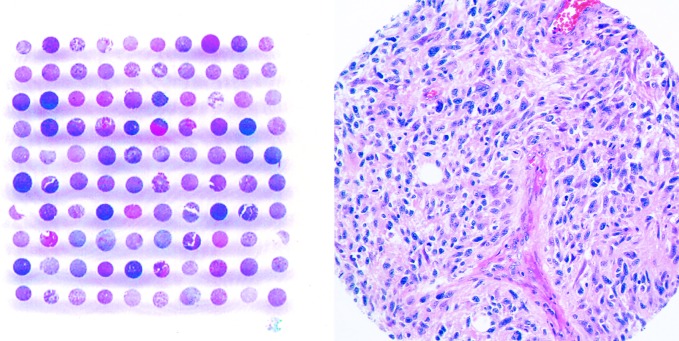
TMAs were constructed by using formalin-fixed, paraffin-embedded specimens from 18 AAs and 88 GBMs from RTOG protocols 7401, 7903, 7918, 8302, 9006, 9305, 9404, 9602, and 9806 (Table 1). Thus, the initial tissue array contained 106 tissue cores, each of which measured 0.7 mm in diameter (Fig. 1). All included neoplasms arose in adult patients and were primary high-grade astrocytomas that had not been previously treated. All were from patients who met clinical enrollment criteria for each protocol, available online (RTOG, 2003) or previously described (Curran, 1993b; Kurup, 1985; Nelson, 1986, 1988).
Table 1.
RTOG studies included in tissue microarray
| Study | Phase | Description | N = 89 | AA (n = 15) | GBM (n = 74) |
|---|---|---|---|---|---|
| 7401 | 3 | WBRT 1 (BCNU vs. MeCCNU+DTIC) | 32 (36%) | 6 | 26 |
| 7903 | 1/2 | Misonidazole and Neutrons | 1 (1%) | 1 | 0 |
| 7918 | 3 | WBRT 1 (BCNU vs. Misonidazole radiosensitizer 1 BCNU) | 3 (3%) | 0 | 3 |
| 8302 | 2 | Hyperfractionated RT 1 BCNU | 10 (11%) | 1 | 9 |
| 9006 | 3 | BCNU 1 (Hyperfractionated RT vs. RT) | 18 (20%) | 4 | 14 |
| 9305 | 3 | 6 SRS followed by RT 1 BCNU | 6 (7%) | 0 | 6 |
| 9404 | 3 | RT 1 PCV 6 BUDR | 3 (3%) | 3 | 0 |
| 9602 | 2 | RT 1 Taxol | 5 (6%) | 0 | 5 |
| 9806 | 2 | RT 1 Thalidomide | 11 (12%) | 0 | 11 |
Abbreviations: AA, anaplastic astrocytoma; BCNU, carmustine; BUDR, bromodeoxyuridine; DTIC, decarbazine; GBM, glioblastoma; MeCCNU, semustine; PCV, procarbazine; CCNU (lomustine), vincristine; RT, radiation therapy; RTOG, Radiation Therapy Oncology Group; SRS, stereotactic radiosurgery; WBRT, whole-brain radiation therapy.
Fig. 1.
Low (left panel) and high (right panel) magnification of H&E-stained tissue microarrays (TMAs) constructed from 106 high-grade astrocytomas from patients enrolled in RTOG clinical trials. Each tissue core on the TMA measures 0.7 mm in diameter.
Selection of cases from the RTOG Tissue Bank was based on the histology of AA or GBM and on survival periods that were defined as either short or long within each diagnostic category. Short and long survivals for AA were defined as <2 years and >4 years, respectively. For GBM, short and long survivals were defined as <6 months and >18 months, respectively. Survival time was calculated from the date of randomization or registration to the time of death for short-term survivors, and to the time of death or last follow-up for long-term survivors. Roughly one half of the tumors on the array were from long-term survivors (56), the remainder coming from short-term survivors. The mean age at presentation for GBM short-term survivors was 60.4 years (range, 50–79 years) compared to 49.0 years (range, 31–73 years) for the long-term survivors. Mean age for AA short-term survivors was 44.1 (range, 33 – 56) compared to 37.8 years (range, 23–50 years) for long-term survivors. The Karnofsky performance status (KPS) of 65% of GBM short-term survivors was greater than 70, whereas 85% of GBM long-term survivors had KPS scores greater than 70. The KPS of 88% of AA short-term survivors was greater than 70, whereas 100% of AA long-term survivors had a KPS greater than 70. Brief descriptions of protocols, the histologic classifications, and the number of cases successfully analyzed in this study are listed in Table 1.
Dual-color FISH was performed on 5-mm sections from TMAs (Fuller and Perry, 2002; Fuller et al., 2002). Analysis was performed without knowledge of survival groups of the samples and was performed in duplicate on tumor tissue that was histologically representative of the resection specimen. For chromosome 1, paired DNA probes consisted of a fluorescein isothiocyanate (FITC)-labeled chromosome 1p32 probe (Human BAC RPCI-11 library clone 260I23, Research Genetics, Huntsville, Ala.) and a rhodamine-labeled 1q42 reference probe (CIT978SK-A clone 309C5, Research Genetics). For chromosome 19, a rhodamine-labeled chromosome 19q13.4 probe (RPCI-11 clone 426G3) was paired with an FITC-labeled 19p13 reference probe (RPCI-11 clone 575H1). A commercial probe cocktail for the p16/CDKN2A gene region on chromosome 9p included a SpectrumGreen-labeled centromere enumerating probe (CEP9) and SpectrumOrange-labeled 9p21 (p16/CDKN2A) (Vysis, Downers Grove, Ill.). Probes for 10q consisted of an FITC-labeled 10q23.3 (PTEN) and rhodamine-labeled 10q25.3-q26.1 (DMBT1) (DNA received as a gift). The pairing of PTEN and DBMT1 probes was chosen rather than including a chromosome 10 centromere probe because PTEN and DBMT1 are occasionally deleted independently in gliomas, and we wished to assess each marker individually (Steck et al., 1999). Unstained slides were deparaffinized, steam-cooked for DNA target retrieval with 10-mM citrate buffer, pH 6.0, and subjected to 30 min of pepsin digestion at 37°C. Paired FISH probes were diluted to 1:50 in DenHyb buffer (Insitus, Albuquerque, N.M.), applied to each slide, and co-denatured with the target DNA at 90°C for 13 min. Slides were incubated overnight at 37°C in a humidified oven and washed.
Fluorescent signals were visualized and quantitated under an Olympus BX60 fluorescence microscope (B & B Microscopes, Ltd., Warrendale, Penn.) with appropriate filters. A minimum of 100 nonoverlapping nuclei were assessed for each hybridization. A hemizygous deletion was defined as either >50% nuclei with only 1 fluorescent signal or a ratio of test to reference probe signals of <0.8. The latter criterion was used primarily in cases of extensive polyploidy/aneuploidy with a generalized state of chromosomal gains, wherein the region of interest remained deleted relative to the reference DNA copy numbers (e.g., nuclei with 4 copies of 1q, but 2 copies of 1p). Homozygous p16/CDKN2A deletions required >20% of tumor nuclei with CEP9 signals only. In cases where no tumor nuclei harbored p16/CDKN2A signals, the possibility of partial hybridization failure was excluded by the presence of p16/CDKN2A signals in nearby non-neoplastic cells (e.g., endothelial cells). For the purposes of this study, hemizygous and homozygous deletions of p16/CDKN2A were considered equivalent since previous studies have shown that homozygous deletions of p16/CDKN2A detected by quantitative polymerase chain reaction may appear as either homozygous or hemizygous deletions by FISH (Perry et al., 1997b). Tumors for which FISH signals were lacking or too weak were considered “non-informative.” Images were captured with a black and white, high-resolution COHU CCD camera, Z-stack motor, and CytoVision basic workstation (Applied Imaging, Santa Clara, Calif.) and were reconstituted in blue, green, and red pseudocolors with CytoVision software.
To evaluate the association of genetic status with survival, cases were dichotomized into short- or long-term survival groups as described previously. A distribution of survival groups as related to genetic status was generated, resulting in a 2-by-2 table for each marker. These contingency tables were analyzed by using Fisher’s exact test of association. AA and GBM groups were analyzed separately and together. Markers could potentially be evaluated for either retention or deletions at a given locus as it related to survival. Our analysis assessed deletions as they related to survival group. We analyzed 1p and 19q deletions, and 9p and 10q deletions, separately and together (either or both). With the use of an overall type 1 error (alpha) of 0.10 and by adjusting for multiple comparisons within each histologic group and for each marker combination, a P-value less than 0.025 was considered statistically significant.
A similar approach was taken to investigate any possible association between genetic status and patient age. To accomplish this, patients were dichotomized into groups that were ⩽60 years old (45 patients) and >60 years old (30 patients). This analysis was restricted to patients with GBM because there were no patients with AA who were >60 years old. A 2-by-2 table was generated that related age and genetic status for each marker, and associations were analyzed by using Fisher’s exact test.
Results
Among the 106 high-grade astrocytomas included in the TMAs, 6 could not be analyzed by FISH because of insufficient tissue, which was due either to tissue falling off the slide or to a lack of neoplastic tissue within the sample. Eleven cases were indeterminate by FISH analysis for all 4 markers tested. Tissue from the oldest clinical trial (RTOG 7401) had the highest number of indeterminate analyses, most likely because of the age-dependent decrease of DNA integrity within archived paraffin blocks. Thus, 89 tumors remained that yielded FISH data for at least 1 of the markers. We were able to analyze all 4 markers for 61 tumors; 3 of 4 markers for 13 tumors; 2 of 4 markers for 8 tumors; and 1 marker for 7 tumors.
The overall incidence of genetic deletions for high-grade astrocytomas (AA+GBM) was as follows: 3% (2/73) for 1p, 12% (10/85) for 19q, 68% (44/65) for 9p21, and 63% (52/83) for 10q. Chromosome 1p deletions were noted in 2/12 AAs (17%) and 0/61 GBMs, whereas 19q deletions were noted in 3/15 AAs (20%) and 7/70 GBMs (10%) (Fig. 2; Table 2). Combined 1p and 19q loss was detected in 2/12 AAs (17%), but was not seen in any of the 61 GBMs with both markers successfully analyzed. Chromosome 9p loss was noted in 5/8 AAs (63%) and 39/57 GBMs (68%) (Table 3). Chromosome 10q loss was detected in 4/15 AAs (27%) and 48/68 GBMs (71%).
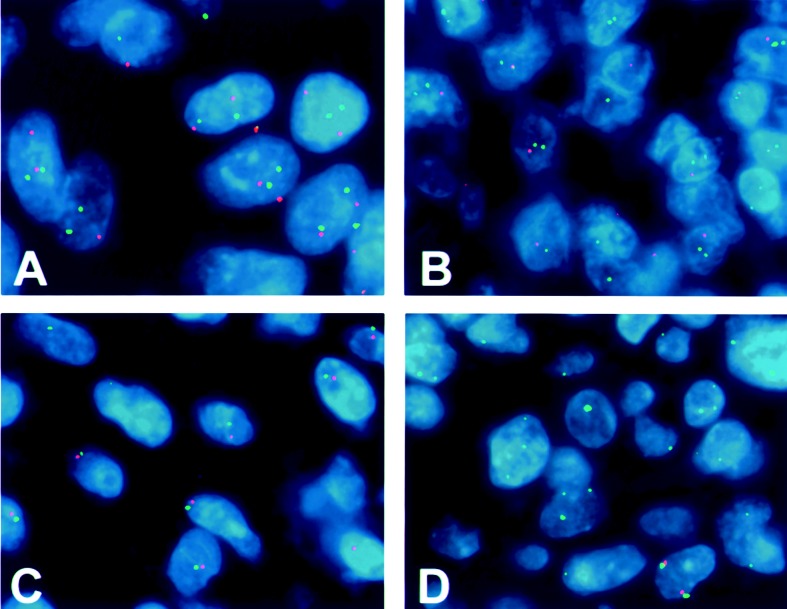
Fig. 2.
Representative FISH hybridizations performed on tissue microarrays. A. GBM with a normal disomic complement of 1p. Most cells have 2 green (1p32) and 2 red (1q42) signals. Some appear to have fewer than 2 copies because of the truncation artifact encountered in thin tissue sections (i.e., incomplete DNA complement in sectioned nuclei). Some signals are out of the plane of focus represented in the photograph. B. Pattern of 19q deletion in GBM showing 2 green (19p13) signals and 1 red (19q13.4) signal in most nuclei. C. Pattern of chromosome 10 deletion in a GBM. Only 1 green (PTEN) signal and 1 red (DMBT1) signal are detected in each nucleus, most likely representing either a large 10q deletion or the loss of an entire chromosome 10. D. Pattern of homozygous deletion of 9p21 (p16/CDKN2A) in a GBM. Only green signals from the centromeric probe are noted in the majority of nuclei. Signals from 9p21 (red) are almost entirely absent.
Table 2.
Association of 1p deletions and 19q deletions with survival group among patients with high-grade astrocytomas
| GBM | AA | AA+GBM | |||||
|---|---|---|---|---|---|---|---|
| Survival Groupa | Survival Groupb | Survival Group | |||||
| Marker | Short | Long | Short | Long | Short | Long | |
| 1p del | Yes | 0 | 0 | 0 | 2 (40%) | 0 | 2 (6%) |
| No | 30 (100%) | 31 (100%) | 7 (100%) | 3 (60%) | 37 (100%) | 4 (94%) | |
| 19q del | Yes | 2 (5%) | 5 (15%) | 0 | 3 (43%) | 2 (4%) | 8 (20%) |
| No | 35 (95%) | 28 (85%) | 8 (100%) | 4 (57%) | 43 (96%) | 32 (80%) | |
| Either | Yes | 2 (5%) | 5 (15%) | 0 | 3 (43%) | 2 (4%) | 8 (20%) |
| No | 38 (95%) | 29 (85%) | 8 (100%) | 4 (57%) | 46 (96%) | 33 (80%) | |
| Both | Yes | 0 | 0 | 0 | 2 (40%) | 0 | 2 (5%) |
| No | 30 (100%) | 31 (100%) | 7 (100%) | 3 (60%) | 37 (100%) | 34 (94%) | |
Abbreviations: AA, anaplastic astrocytoma; del, deletions; GBM, glioblastoma.
Short and long survivals for GBM were defined as <6 months and >18 months, respectively.
Short and long survivals for AA were defined as <2 years and >4 years, respectively.
Table 3.
Association of 9p deletions and 10q deletions with survival group among patients with high-grade astrocytomas
| GBM | AA | AA+GBM | |||||
|---|---|---|---|---|---|---|---|
| Survival Groupa | Survival Groupb | Survival Group | |||||
| Marker | Short | Long | Short | Long | Short | Long | |
| 9p del | Yes | 21 (72%) | 18 (64%) | 2 (67%) | 3 (60%) | 23 (72%) | 21 (64%) |
| No | 8 (28%) | 10 (36%) | 1 (33%) | 2 (40%) | 9 (28%) | 12 (36%) | |
| 10q del | Yes | 27 (75%) | 21 (66%) | 3 (38%) | 1 (14%) | 30 (68%) | 22 (56%) |
| No | 9 (25%) | 11 (34%) | 5 (63%) | 6 (86%) | 14 (32%) | 17 (44%) | |
| Either | Yes | 32 (80%) | 23 (68%) | 4 (50%) | 4 (57%) | 36 (75%) | 27 (66%) |
| No | 8 (20%) | 11 (32%) | 4 (50%) | 3 (43%) | 12 (25%) | 14 (34%) | |
| Both | Yes | 16 (59%) | 16 (59%) | 1 (33%) | 0 | 17 (57%) | 16 (50%) |
| No | 11 (41%) | 11 (41%) | 2 (67%) | 5 (100%) | 13 (43%) | 16 (50%) | |
Abbreviations: AA, anaplastic astrocytoma; del, deletions; GBM, glioblastoma.
Short and long survivals for GBM were defined as <6 months and >18 months, respectively.
Short and long survivals for AA were defined as <2 years and >4 years, respectively.
The frequencies of loss for each marker within long-term and short-term survival groups of AA, GBM, and AA+GBM patients are shown in Tables 2 and 3. Both of the 1p deletions in AAs were among the long-term survivors. Deletions of 19q were more frequent in long-term than short-term survivors for AA (43% vs. 0%), GBM (15% vs. 5%), and AA+GBM (20% vs. 4%; P = 0.041) (Table 2). The combination of 1p and 19q loss was seen in 2 AAs, both of which were in long-term survivors. Deletions of 9p were slightly more frequent in short-term than long-term survivors for AA (67% vs. 60%), GBM (72% vs. 64%), and AA+GBM (72% vs. 64%) (Table 3). Deletions of 10q were more frequent in short-term than long-term survivors for AA (38% vs. 14%), GBM (75% vs. 66%), and AA+GBM (68% vs. 56%). The combination of 9p and 10q losses was slightly more common in short-term than long-term survivors for AA (33% vs. 0%), GBM (59% vs. 47%), and AA+GBM (57% vs. 50%). None of the comparisons were statistically significant.
The frequency of 19q losses in GBMs from patients ⩽60 years old was equal to that in GBMs from patients >60 years old: 10% of GBMs in each age category had 19q losses. The frequency of 9p deletions in patients >60 years old (78%) was higher than that in patients ⩽60 years old (62%). The frequency of 10q deletions was also higher in patients >60 years old (80%) than in those ⩽60 years old (63%). No 1p losses were noted in GBMs from either age category. None of these comparisons reached statistical significance.
Discussion
Our analysis of genetic prognostic markers showed that combined losses of 1p and 19q are quite rare in high-grade astrocytomas. None of the GBMs and only 2 of the AAs in this study showed a combination of 1p and 19q loss. Interestingly, the 2 AAs with combined 1p and 19q loss were in the long-term survival group. Although the 1p and 19q findings are intriguing, the low number of high-grade astrocytomas with such losses makes it impossible to draw any meaningful conclusions regarding their clinical behavior. These findings for astrocytomas stand in stark contrast to those for oligodendrogliomas, in which the combined loss of 1p and 19q is both a frequent finding (60%–80%) and predictive of a favorable prognosis (Cairncross et al., 1998; Perry et al., 2003; Smith et al., 2000).
The significance of 1p and 19q losses in astrocytomas has been debated. In one of the first assessments, Smith et al. (2000) evaluated 1p and 19q status by FISH as it related to survival in a large series of oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. Combined loss of 1p and 19q was found to be predictive of prolonged overall survival only among oligodendrogliomas. As in our study, combined loss of 1p and 19q was quite uncommon in GBMs (8%), and the low numbers did not provide sufficient statistical power to adequately address prognostic significance.
In contrast, Schmidt et al. (2002) studied 97 GBMs and reported 33% with loss of heterozygosity (LOH) at 19q and 19% with LOH at 1p. The combination of 1p and 19q LOH occurred in 5 GBMs, and the mean survival of these patients was significantly longer than that of patients whose tumors retained 1p, 19q, or both. Others have suggested that 1p losses by themselves are associated with either unusually good responses to adjuvant therapy or unexpectedly long survivals in histologically diverse types of infiltrative gliomas (Ino et al., 2002).
Whereas 1p losses were rare in our study, we detected 19q losses in 12% of high-grade astrocytomas. Moreover, we found that long-term survivors had a higher frequency of such losses than short-term survivors in the AA, GBM, and AA+GBM groups (P = 0.041 for AA+GBM group). Other survival analyses of GBM have demonstrated a modest association between 19q loss and long survival (Smith et al., 2000). In a recent comparative genomic hybridization (CGH) analysis of long- and short-term survivors with GBM, solitary 19q losses were significantly more frequent in GBMs from long survivors, and no tumors from short-term survivors had 19q loss (Burton et al., 2002). Thus, while 1p losses are uncommon in astrocytic neoplasms, 19q losses occur in 12% to 33% and appear to be associated with longer survivals.
Genetic losses on chromosome 10 occur in 70% to 90% of GBMs and can manifest as losses of the whole chromosome, as losses of only portions of the long or short arms, or as smaller genetic losses detected as LOH (Balesaria et al., 1999; Perry et al., 1997a; Tada et al., 2001). Three regions of genetic loss have received the most attention: 10p14-pter, 10q23-q24, and 10q25-qter (Fujisawa et al., 1999). Our studies focused on the 10q arm, since it is the region most commonly implicated in high-grade progression and aggressive clinical behavior. It is also the location of PTEN, the first tumor suppressor gene identified in this region. Mutations of PTEN occur in 20% to 40% of GBMs, are more common in high-grade astrocytomas than in grade II lesions, and generally occur in the setting of chromosome 10 allelic loss (Cavenee et al., 2000; Smith et al., 2001; Zhou et al., 1999). Both chromosome 10 losses and PTEN mutations are thought to be late events in the progression to GBM, since both are more frequent in GBMs than in AAs or grade II astrocytomas (Kunwar et al., 2001; Smith et al., 2001; Tada et al., 2001; Zhou et al., 1999). In our study, 10q losses were detected in 27% of AAs and 71% of GBMs. The vast majority of these included both the PTEN and DMBT1 loci, consistent with either large 10q deletions, loss of the entire long arm, or monsomy 10.
The high frequency of chromosome 10 losses in GBMs and the tight correlation of its loss with the glioblastoma phenotype might suggest that it would not be a useful prognostic marker across tumor grades. In previous studies that included both AAs and GBMs, LOH of chromosome 10 (either 10p or 10q) was found to be an independent predictor of poor prognosis, in large part because LOH was highly associated with AAs in patients who had short survivals (Balesaria et al., 1999; Tada et al., 2001). Some investigators have therefore suggested that AAs with LOH of 10q may behave clinically as GBMs. Supporting such a contention, Ganju et al. (1994) found that chromosome 10 loss was associated with short survival because of its association with high-grade histology, rather than as an independent variable. However, a recent investigation that included only GBMs revealed that LOH of 10q was associated with short survival (mean survival, 8.8 months) as compared to survival in GBM cases with no LOH of 10q (mean survival, 18 months) (Schmidt et al., 2002). PTEN mutations were also associated with a slightly shorter survival in this study, albeit not in a statistically significant way.
In our study, we found that 10q deletions were slightly more frequent among short-term than long-term survivors, which suggests that other alterations may also be relevant to poor prognosis. These findings complement prior CGH studies of GBM survival groups, which have shown that both 10q and 10p losses are more frequent among short-term survivors, with the association achieving statistical significance for 10q loss (Burton et al., 2002). The collective evidence suggests that chromosome 10 loss and PTEN mutations are late events in the evolution to GBM, which makes them potentially more powerful prognostic markers for AA than for GBM.
Losses of genetic material from the short arm of chromosome 9 have been documented in 50% to 70% of GBMs (Cavenee et al., 2000). The p16/CDKN2A tumor suppressor gene found at 9p21 is one of the critical cell cycle regulatory genes lost in this region during the high-grade progression of astrocytomas. Both LOH of 9p21 and homozygous deletion of p16/CDKN2A are more common in high-grade astrocytomas than in grade II tumors (Maruno et al., 1996; Perry et al., 1999). The frequency of 9p deletions in our study was similar among patients with AAs and patients with GBMs (63% and 68%, respectively), which suggests that 9p loss is likely to be an earlier event than 10q loss. There were only slightly more frequent 9p losses among short-term than long-term survivors, in keeping with the findings of previous CGH and molecular analysis (Burton et al., 2002; Kraus et al., 2000). Cohort studies have not proven the utility of 9p losses as an independent marker of overall survival in astrocytomas (Perry et al., 1999).
Our FISH analysis of high-grade astrocytomas from RTOG clinical trials suggests that 19q losses are more frequent in long-term survivors and may be a useful marker of improved survival. Deletions of 9p and 10q are slightly more frequent in short-term survivors. The present investigation was an exploratory evaluation of prognostic markers in short- and long-term survivors rather than a multivariate analysis of survival. The statistical power of our study design was inherently low because of the limited number of cases, the inclusion of multiple markers, and the low frequency of 1p and 19q losses. In addition, the interpretation of our results should take into account the low statistical power, dichotomized survival, and the lack of data from patients who were not short- or long-term survivors. Nonetheless, TMAs of RTOG material allowed us to study genetic markers of prognosis among patients who met entry criteria for RTOG clinical trials, who had not been previously treated, and who had carefully monitored clinical follow-up. In this regard, the findings of dissimilar frequencies of 19q, 9p, and 10q losses in long- and short-term survivors are intriguing and warrant further investigation of overall survival, which could define the independent prognostic significance of these genetic markers more definitively.
Acknowledgment
DNA for the 10q FISH analysis was generously donated by Dr. Robert Jenkins of the Mayo Clinic in Rochester, Minn.
Footnotes
Supported in part by U.S. Public Health Service and the National Institutes of Health, grants CA21661, CA37422, CA32115 (RTOG), and NS42934 (D.J.B.).
Abbreviations used are as follows: AA, anaplastic astrocytoma; CGH, comparative genomic hybridization; FISH, fluorescence in situ hybridization; FITC, fluorescein isothiocyanate; GBM, glioblastoma; KPS, Karnofsky performance status; LOH, loss of heterozygosity; RTOG, Radiation Therapy Oncology Group; TMA, tissue microarray.
References
- Balesaria S, Brock C, Bower M, Clark J, Nicholson SK, Lewis P, de Sanctis S, Evans H, Peterson D, Mendoza N, Glaser MG, Newlands ES, Fisher RA. Loss of chromosome 10 is an independent prognostic factor in high-grade gliomas. Br J Cancer. 1999;81:1371–1377. doi: 10.1038/sj.bjc.6693403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat DJ, Castellano-Sanchez A, Kaur B, Van Meir EG. Genetic and biologic progression in astrocytomas and their relation to angiogenic dysregulation. Adv Anat Pathol. 2002;9:24–36. doi: 10.1097/00125480-200201000-00004. [DOI] [PubMed] [Google Scholar]
- Burton EC, Lamborn KR, Feuerstein BG, Prados M, Scott J, Forsyth P, Passe S, Jenkins RB, Aldape KD. Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res. 2002;62:6205–6210. [PubMed] [Google Scholar]
- CBTRUS. Central Brain Tumor Registry of the United States (2002) Statistical Report: Primary Brain Tumors in the United States, 1995 –1999.Chicago, Ill.: Central Brain Tumor Registry of the United States.
- Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- Cavenee, W.K., Furnari, F.B., Nagane, M., Huang, H.-J.S., Newcombe, E.W., Bigner, D.D., Weller, M., Berens, M.E., Plate, K.H., Israel, M.A., Noble, M.D., and Kleihues, P. (2000) Diffusely infiltrating astrocytomas. In Kleihues, P., and Cavenee, W.K., eds. Pathology and Genetics of Tumours of the Nervous System 2nd ed. (Kleihues, P., and Sobin, L.H., series eds. World Health Organization Classification of Tumors), pp. 10–21. Lyon: IARC Press.
- Curran WJ, Jr, Scott CB, Horton J, Nelson JS, Weinstein AS, Fischbach AJ, Chang CH, Rotman M, Asbell SO, Krisch RE. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst. 1993a;85:704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- Curran WJ, Jr, Scott CB, Weinstein AS, Martin LA, Nelson JS, Phillips TL, Murray K, Fischbach AJ, Yakar D, Schwade JG. Survival comparison of radiosurgery-eligible and -ineligible malignant glioma patients treated with hyperfractionated radiation therapy and carmustine: A report of Radiation Therapy Oncology Group 83-02. J Clin Oncol. 1993b;11:857–862. doi: 10.1200/JCO.1993.11.5.857. [DOI] [PubMed] [Google Scholar]
- Fujisawa H, Kurrer M, Reis RM, Yonekawa Y, Kleihues P, Ohgaki H. Acquisition of the glioblastoma phenotype during astrocytoma progression is associated with loss of heterozygosity on 10q25-qter. Am J Pathol. 1999;155:387–394. doi: 10.1016/S0002-9440(10)65135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller CE, Perry A. Fluorescence in situ hybridization (FISH) in diagnostic and investigative neuropathology. Brain Pathol. 2002;12:67–86. doi: 10.1111/j.1750-3639.2002.tb00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller CE, Wang H, Zhang W, Fuller GN, Perry A. High-throughput molecular profiling of high-grade astrocytomas: The utility of fluorescence in situ hybridization on tissue microarrays (TMA-FISH) J Neuropathol Exp Neurol. 2002;61:1078–1084. doi: 10.1093/jnen/61.12.1078. [DOI] [PubMed] [Google Scholar]
- Ganju V, Jenkins RB, O’Fallon JR, Scheithauer BW, Ransom DT, Katzmann JA, Kimmel DW. Prognostic factors in gliomas. A multivariate analysis of clinical, pathologic, flow cytometric, cytogenetic, and molecular markers. Cancer. 1994;74:920–927. doi: 10.1002/1097-0142(19940801)74:3<920::aid-cncr2820740320>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Hunter SB, Brat DJ, Olson JJ, von Deimling A, Zhou W, Van Meir EG. Alterations in molecular pathways of diffusely infiltrating glial neoplasms: Application to tumor classification and anti-tumor therapy. Int J Oncol. 2003;23:857–869. [PubMed] [Google Scholar]
- Ino Y, Zlatescu MC, Sasaki H, Macdonald DR, Stemmer-Rachamimov AO, Jhung S, Ramsay DA, von Deimling A, Louis DN, Cairncross JG. Long survival and therapeutic responses in patients with histologically disparate high-grade gliomas demonstrating chromosome 1p loss. J Neurosurg. 2002;92:983–990. doi: 10.3171/jns.2000.92.6.0983. [DOI] [PubMed] [Google Scholar]
- Kamiryo T, Tada K, Shiraishi S, Shinojima N, Nakamura H, Kochi M, Kuratsu J, Saya H, Ushio Y. Analysis of homozygous deletion of the p16 gene and correlation with survival in patients with glioblastoma multiforme. J Neurosurg. 2002;96:815–822. doi: 10.3171/jns.2002.96.5.0815. [DOI] [PubMed] [Google Scholar]
- Kraus JA, Glesmann N, Beck M, Krex D, Klockgether T, Schackert G, Schlegel U. Molecular analysis of the PTEN, TP53 and CDKN2A tumor suppressor genes in long-term survivors of glioblastoma multiforme. J Neurooncol. 2000;48:89–94. doi: 10.1023/a:1006402614838. [DOI] [PubMed] [Google Scholar]
- Kunwar S, Mohapatra G, Bollen A, Lamborn KR, Prados M, Feuerstein BG. Genetic subgroups of anaplastic astrocytomas correlate with patient age and survival. Cancer Res. 2001;61:7683–7688. [PubMed] [Google Scholar]
- Kurup PD, Pajak TF, Hendrickson FR, Nelson JS, Mansell J, Cohen L, Awschalom M, Rosenberg I, Ten Haken RK. Fast neutrons and misonidazole for malignant astrocytomas. Int J Radiat Oncol Biol Phys. 1985;11:679–686. doi: 10.1016/0360-3016(85)90298-6. [DOI] [PubMed] [Google Scholar]
- Maruno M, Yoshimine T, Muhammad AK, Tokiyoshi K, Hayakawa T. Loss of heterozygosity of microsatellite loci on chromosome 9p in astrocytic tumors and its prognostic implications. J Neurooncol. 1996;30:19–24. doi: 10.1007/BF00177439. [DOI] [PubMed] [Google Scholar]
- Nelson DF, Diener-West M, Weinstein AS, Schoenfeld D, Nelson JS, Sause WT, Chang CH, Goodman R, Carabell S. A randomized comparison of misonidazole sensitized radiotherapy plus BCNU and radiotherapy plus BCNU for treatment of malignant glioma after surgery: Final report of an RTOG study. Int J Radiat Oncol Biol Phys. 1986;12:1793–1800. doi: 10.1016/0360-3016(86)90321-4. [DOI] [PubMed] [Google Scholar]
- Nelson DF, Diener-West M, Horton J, Chang CH, Schoenfeld D, Nelson JS. Combined modality approach to treatment of malignant gliomas—re-evaluation of RTOG 7401/ECOG 1374 with long-term follow-up: A joint study of the Radiation Therapy Oncology Group and the Eastern Cooperative Oncology Group. NCI Monogr. 1988;6:279–284. [PubMed] [Google Scholar]
- Perry A, Tonk V, McIntire DD, White CL., 3rd Interphase cytogenetic (in situ hybridization) analysis of astrocytomas using archival, formalin-fixed, paraffin-embedded tissue and nonfluorescent light microscopy. Am J Clin Pathol. 1997a;108:166–174. doi: 10.1093/ajcp/108.2.166. [DOI] [PubMed] [Google Scholar]
- Perry A, Nobori T, Ru N, Anderl K, Borell TJ, Mohapatra G, Feuerstein BG, Jenkins RB, Carson DA. Detection of p16 gene deletions in gliomas: A comparison of fluorescence in situ hybridization (FISH) versus quantitative PCR. J Neuropathol Exp Neurol. 1997b;56:999–1008. doi: 10.1097/00005072-199709000-00005. [DOI] [PubMed] [Google Scholar]
- Perry A, Anderl K, Borell TJ, Kimmel DW, Wang CH, O’Fallon JR, Feuerstein BG, Scheithauer BW, Jenkins RB. Detection of p16, RB, CDK4, and p53 gene deletion and amplification by fluorescence in situ hybridization in 96 gliomas. Am J Clin Pathol. 1999;112:801–809. doi: 10.1093/ajcp/112.6.801. [DOI] [PubMed] [Google Scholar]
- Perry A, Fuller CE, Banerjee R, Brat DJ, Scheithauer BW. Ancillary FISH analysis for 1p and 19q status: Preliminary observations in 287 gliomas and oligodendroglioma mimics. Front Biosci. 2003;8:a1–a9. doi: 10.2741/896. [DOI] [PubMed] [Google Scholar]
- RTOG. Radiation Therapy Oncology Group (2003) Protocols available at http://www.rtog.org
- Schmidt MC, Antweiler S, Urban N, Mueller W, Kuklik A, Meyer-Puttlitz B, Wiestler OD, Louis DN, Fimmers R, von Deimling A. Impact of genotype and morphology on the prognosis of glioblastoma. J Neuropathol Exp Neurol. 2002;61:321–328. doi: 10.1093/jnen/61.4.321. [DOI] [PubMed] [Google Scholar]
- Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- Steck PA, Lin H, Langford LA, Jasser SA, Koul D, Yung WK, Pershouse MA. Functional and molecular analyses of 10q deletions in human gliomas. Genes Chromosomes Cancer. 1999;24:135–143. doi: 10.1002/(sici)1098-2264(199902)24:2<135::aid-gcc6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Tada K, Shiraishi S, Kamiryo T, Nakamura H, Hirano H, Kuratsu J, Kochi M, Saya H, Ushio Y. Analysis of loss of heterozygosity on chromosome 10 in patients with malignant astrocytic tumors: Correlation with patient age and survival. J Neurosurg. 2001;95:651–659. doi: 10.3171/jns.2001.95.4.0651. [DOI] [PubMed] [Google Scholar]
- Zhou XP, Li YJ, Hoang-Xuan K, Laurent-Puig P, Mokhtari K, Longy M, Sanson M, Delattre JY, Thomas G, Hamelin R. Mutational analysis of the PTEN gene in gliomas: Molecular and pathological correlations. Int J Cancer. 1999;84:150–154. doi: 10.1002/(sici)1097-0215(19990420)84:2<150::aid-ijc10>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]