Abstract
The accumulation of insoluble protein aggregates in intra and perinuclear inclusions is a hallmark of Huntington's disease (HD) and related glutamine-repeat disorders. A central question is whether protein aggregation plays a direct role in the pathogenesis of these neurodegenerative diseases. Here we show by using a filter retardation assay that the mAb 1C2, which specifically recognizes the elongated polyglutamine (polyQ) stretch in huntingtin, and the chemical compounds Congo red, thioflavine S, chrysamine G, and Direct fast yellow inhibit HD exon 1 protein aggregation in a dose-dependent manner. On the other hand, potential inhibitors of amyloid-β formation such as thioflavine T, gossypol, melatonin, and rifampicin had little or no inhibitory effect on huntingtin aggregation in vitro. The results obtained by the filtration assay were confirmed by electron microscopy, SDS/PAGE, and MS. Furthermore, cell culture studies revealed that the Congo red dye at micromolar concentrations reduced the extent of HD exon 1 aggregation in transiently transfected COS cells. Together, these findings contribute to a better understanding of the mechanism of huntingtin fibrillogenesis in vitro and provide the basis for the development of new huntingtin aggregation inhibitors that may be effective in treating HD.
Keywords: glutamine repeat, inhibition
Huntington's disease (HD) is a progressive neurodegenerative disorder with a generally midlife age of onset and a duration of 15–20 years. The disease is characterized by personality changes, motor impairment, and subcortical dementia (1). It is associated with selective neuronal cell death in the cortex and striatum (2). The mutation that causes HD is a CAG/polyglutamine (polyQ) repeat expansion in the first exon of the HD gene encoding the huntingtin protein. The molecular mechanisms responsible for delayed onset, selective pattern of neuropathology, and cell death observed in HD are unknown. However, insoluble huntingtin protein aggregates have been detected in an in vitro model system (3), as well as in transgenic animals (4), fly models (5), cell culture systems (6), and brains of HD patients (7). PolyQ-containing protein aggregates also have been found in related glutamine-repeat disorders such as dentatorubral pallidoluysian atrophy, spinal bulbar muscular atrophy, and the spinocerebellar ataxias type 1, 2, 3, and 7, suggesting that all of these neurodegenerative diseases are caused by deposition of toxic protein aggregates.
Although the causal relationship between aggregate formation and disease has not been proven, genetic, neuropathological, and biochemical evidence indicate that formation of insoluble protein aggregates plays an important role in the cellular distortions underlying HD and the related glutamine-repeat disorders. Recently, Ona et al. (8) have demonstrated that expression of a dominant-negative caspase-1 mutant slows down aggregate formation of the HD exon 1 protein as well as disease progression in transgenic mice. Furthermore, evidence has been presented that certain components of the proteasome, transcription factors, chaperons, and caspases, which normally are essential for cell viability, are recruited into polyQ-containing aggregates (9, 10). Accumulation of caspase-8 into insoluble protein aggregates, for example, is required for induction of cell death in primary rat neurons, whereas prevention of caspase-8 recruitment into aggregates blocks polyQ-induced cell death (11). Taken together these results suggest that formation of insoluble polyQ-containing protein aggregates is important both for the initiation and progression of these late-onset neurodegenerative disorders.
Here we report that the antibody 1C2, which selectively recognizes elongated polyQ chains, as well as the chemical compounds Congo red, thioflavine S, chrysamine G, and Direct fast yellow suppress the in vitro aggregation of HD exon 1 protein. We used a filter retardation assay, electron microscopy, SDS/PAGE, and MS to characterize the effect of the inhibitors of huntingtin fibrillogenesis.
Materials and Methods
Materials.
Thioflavine S, thioflavine T, Congo red, rifampicin, gossypol, melatonin, chrysamine G, N,N′-terephthalylidenebis-(4-aminosalicylic acid), Direct orange, Direct yellow 20, 4,4′-bis-(carboxyphenylamino)-3,3′-dimethoxybiphenyl, and DMSO were purchased from Sigma. Direct fast yellow was from TCI America (Portland, OR). All chemical compounds were dissolved in 100% DMSO at a conc. of 10 mM. Proteinase K was obtained from Quantum Appligene. Trypsin (modified version) was purchased from Boehringer Mannheim, and PreScission protease was purchased from Amersham Pharmacia Biotech.
Construction of Plasmids and Protein Purification.
Standard protocols for DNA manipulations were followed (12). Escherichia coli SURE (Stratagene) was used as host strain for plasmid construction and protein expression. Plasmids pCAG51, pCAG51ΔP, and pTL1-CAG51 have been described (3, 13, 14). pCAG51myc was generated by ligating a 0.3-kb EcoRI–SalI IT15 fragment, isolated from YEp105-CAG51 into pGEX-6P-1 (Amersham Pharmacia Biotech). For construction of YEp105-CAG51 a BamHI–SalI IT-15 fragment, isolated from pCAG51, was subcloned into YEp105. E. coli SURE carrying pCAG51, pCAG51ΔP, or pCAG51myc was used for expression of the glutathione S-transferase (GST)-HD51, GST-HD51ΔP, and GST-mycHD51 fusion proteins, respectively. Recombinant fusion proteins were purified under native conditions by affinity chromatography on glutathione agarose as described (14).
Antibodies and Immunoblotting.
The generation of the polyclonal huntingtin-specific antibodies HD1 and AG51 has been described (3, 15). The anti-huntingtin mAb 1C2 (16) was a kind gift of J.-L. Mandel (IGBMC, Strasbourg, France), and mouse IgG 2a (UPC-10) was purchased from Sigma. For Western blot analysis, proteins were separated by SDS/PAGE (10–12%) and transferred to nitrocellulose. Membranes were blocked with 3% nonfat dry milk in Tris-buffered saline containing 0.05% Tween-20 and incubated with the appropriate primary antibody. Secondary antibodies were peroxidase-conjugated anti-rabbit or anti-mouse IgG (Boehringer Mannheim). Immunoreactive protein was detected by using enhanced chemiluminescence reagent (Amersham Pharmacia).
Proteolytic Digestions and Filter Retardation Assay.
For in vitro aggregation studies in the presence of antibodies, 10 μl of a 5 μM solution of GST-mycHD51 fusion protein was treated for 2 h at 6°C with 0.5 units of PreScission protease under conditions as recommended by the supplier (Amersham Pharmacia Biotech). This resulted in >90% removal of the GST moiety from the fusion protein as estimated by SDS/PAGE and immunoblotting. Any aggregates formed during the cleavage reaction were pelleted by centrifugation at 25,000 × g for 15 min at 6°C. Then, 15 μl of either 1C2 antibody or mouse IgG 2a were added to the cleared cleavage reactions to give final IgG conc. of 1.5, 3, 6, and 9 μM, and incubation was continued for 16 h at 37°C to allow aggregate formation. The reaction was stopped by addition of 25 μl of 4% SDS/100 mM DTT followed by heating for 3 min at 98°C. Aliquots corresponding to 200 ng of GST-mycHD51 fusion protein were diluted into 0.2 ml of 2% SDS and filtered through a 0.2-μm cellulose acetate membrane. Captured aggregates were detected by incubation with HD1 antibody (1:5,000) followed by incubation with alkaline phosphatase-conjugated anti-rabbit secondary antibody (1:4,000) and the fluorescent substrate AttoPhos. The conditions for the proteolytic cleavage of the fusion proteins GST-HD51 and GST-HDΔP with trypsin have been described (3). The filter retardation assay for detection and quantification of SDS-insoluble HD exon 1 protein aggregates was performed as described (14, 15) by using aida 1.0 image analysis software (Raytest, Straubenhardt, Germany).
Mass Spectrometry.
On the target for matrix-assisted laser desorption/ionization–MS (MALDI-MS), 0.5 μl of sample solution was mixed with 0.5 μl of sinapic acid matrix solution (saturated in 35% acetonitrile/0.1% trifluoroacetic acid). After solvent evaporation, the samples were transferred into a Bruker Scout 384 Biflex III MALDI–time of flight (TOF) mass spectrometer and analyzed by using externally determined calibration constants. Exclusively positively charged ions were detected, and 100–150 single-shot spectra were accumulated for improved signal-to-noise ratio.
Microscopic Analysis.
For electron microscopic observation, the protease-digested GST-HD fusion proteins were adjusted to a final conc. of 50 μg/ml in 40 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl. Samples were negatively stained with 1% uranyl acetate and viewed in a Philips CM100 electron microscope.
Cell Lines and Cell Fractionation.
COS-1 cells were grown in DMEM (GIBCO/BRL) supplemented with 5% FCS and penicillin (5 units/ml) plus streptomycin (5 μg/ml). Transfection was performed as described (17). For expression of the HD51 protein, cells were plated to 30% confluence and cotransfected with 5 μg of pTL1-CAG51 along with 10 μg of carrier pBluescript DNA (10-cm dish). Cells were harvested 40 h after transfection, washed, scraped in ice-cold PBS, and pelleted. Cells were lysed on ice for 30 min in 50 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40, and protease inhibitors (13). Insoluble material was pelleted by centrifugation at 14,000 rpm for 10 min at 4°C, and portions (10 μg) of the supernatant were used for immunoblot analysis. The pellets containing the insoluble material were resuspended in 100 μl of 20 mM Tris⋅HCl (pH 8.0) and 15 mM MgCl2, and DNase I was added to a final conc. of 0.5 mg/ml followed by incubation at 37°C for 1 h. After DNase treatment the protein concentration was determined by the Dot Metric assay (Geno Technology, Maplewood, MO) and aliquots of 3 and 6 μl (corresponding to 6–15 and 10–25 μg of protein, respectively) were used for the cellulose acetate filter assay and immunoblotting.
Results
Inhibition of Huntingtin Fibrillogenesis by Antibodies.
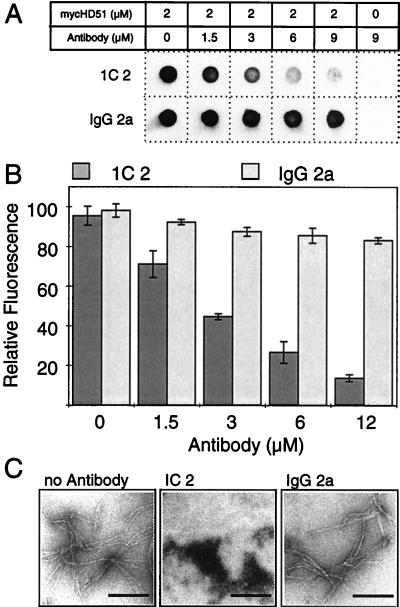
It has been proposed that small molecules that stabilize the native fold of an amyloidogenic protein should prevent its aggregation into ordered fibrillar structures (18). Trottier et al. (16) discovered that the mAb 1C2 specifically recognizes polyQ expansions in soluble proteins such as huntingtin. However, insoluble, high molecular weight polyQ-containing protein aggregates are not detected by this antibody (19). This suggests that 1C2 recognizes the conformation of an elongated polyQ tract in soluble proteins but not the array of glutamine residues in insoluble protein aggregates with a fibrillar morphology (3). Therefore, the addition of 1C2 to a preparation of soluble HD exon 1 protein with 51 glutamines (HD51) would be expected to stabilize the native conformation of the polyQ tract and prevent its aggregation into ordered fibrils. To test this hypothesis, purified GST-mycHD51 fusion protein was incubated with PreScission protease for 2 h at 6°C, resulting in almost complete (>90%) removal of the GST tag from the fusion protein. PreScission protease specifically cleaves the fusion protein between the GST tag and the mycHD51 portion of the protein. SDS/PAGE analysis revealed that 1C2 Ig is not attacked by this protease (data not shown). After cleavage, mAb 1C2 at various concentrations was added to the cleavage reaction and incubation was continued for 16 h at 37°C to permit aggregate formation. Proteins then were denaturated by boiling in 2% SDS and analyzed for the presence of insoluble aggregates by using a filter retardation assay (14). As shown in Fig. 1 A and B, the 1C2 antibody inhibited mycHD51 fibril formation in vitro in a dose-dependent manner. Thus, a 16 h incubation of mycHD51 in the presence of a 1.5-, 3-, and 4.5-fold molar excess of 1C2 resulted in an approximately 50%, 70%, and 85% reduction of the amount of SDS-insoluble mycHD51 aggregates retained on the filter. Under the same conditions the mouse IgG 2a, which does not recognize polyQ sequences, had little or no inhibitory effect. Results similar to those with 1C2 were obtained by using the affinity-purified polyclonal HD1 antibody (3), which also recognizes the polyQ tract in soluble GST-huntingtin fusion proteins. In contrast, an affinity purified polyclonal anti-GST antibody had no inhibitory effect (data not shown).
Figure 1.
Inhibition of HD exon 1 fibrillogenesis by antibodies. (A) Effect of increasing concentrations of 1C2 antibody or mouse IgG 2a on mycHD51 aggregation. Aggregate formation was detected by the filter retardation assay. (B) Quantitative analysis of dot-blot results shown in A. The relative amount of aggregate for each sample was quantified on a Fuji-Imager (LAS 2000). For each experiment, the signal intensity obtained from the samples without added antibodies was arbitrarily set as 100. Values shown are the average of triplicate determinations ± SE. (C) Electron micrographs of mycHD51 protein aggregates after antibody treatment. Proteolytically cleaved GST-mycHD51 fusion protein was incubated for 16 h at 37°C in the presence or absence of the indicated antibodies. Proteins were negatively stained with 1% uranyl acetate and viewed by electron microscopy. (Bar = 200 nm.)
When examined by electron microscopy, the protein mixture containing 1C2 antibody showed numerous amorphous protein aggregates but very little fibrillar material. In comparison, numerous clusters of large fibrillar structures were detected in the absence of 1C2 antibody or in the presence of the mouse IgG 2a control antibody (Fig. 1C).
Inhibition of Huntingtin Aggregation by Small Molecules.
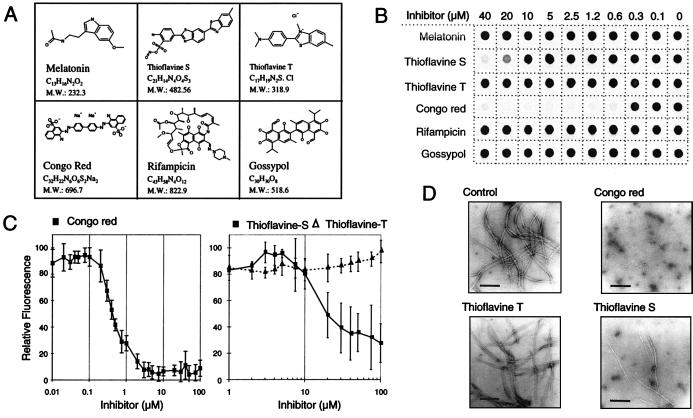
A number of potential inhibitors of amyloid-β (Aβ), scrapie isoform of prion protein (PrPsc), and microtubule fibril formation such as Congo red (20), rifampicin (21), melatonin (22), gossypol (23), thioflavine S (24), and thioflavine T (25) have been described (for structure see Fig. 2A). To test whether these structurally diverse chemical compounds are also effective in inhibiting huntingtin aggregation, they were added at various concentrations (0.15–40 μM) to a 1 μM solution of GST-HD51 fusion protein, which had been predigested with trypsin. Trypsin removes the GST-tag from the fusion protein plus an additional 15 aa from the N terminus of HD exon 1 protein (3). The mixtures then were incubated for 18 h at 37°C to enable the assembly of insoluble HD51 aggregates. Quantification of aggregate formation using the filter retardation assay (14) revealed that Congo red and thioflavine S inhibited HD51 aggregation in a dose-dependent manner, whereas rifampicin, melatonin, gossypol, and thioflavine T under the same conditions had no or only a minor inhibitory effect (Fig. 2B). Dose-response profiles for Congo red, thioflavine T, and thioflavine S are shown in Fig. 2C. Both Congo red and thioflavine S inhibited HD51 aggregation with IC50 values of ≈0.3 and 20 μM, respectively; whereas thioflavine T was ineffective. The inhibitory effect of Congo red and thioflavine S on HD51 aggregation was confirmed by electron microscopy (Fig. 2D). Treatment of trypsin-digested GST-HD51 fusion protein (final conc. 1 μM) with Congo red and thioflavine S at 2.5 and 40 μM, respectively, resulted in the appearance of only very few fibrils with a diameter of 6–7 nm. In strong contrast, after incubation of fusion protein without added compound or with thioflavine T, numerous clusters of high molecular weight fibrils with a ribbon-like morphology (diameter 20–40 nm) were detected by electron microscopy. These observations indicate that both Congo red and thioflavine S interfere with the assembly of ribbon-like structures, possibly by inhibiting the lateral aggregation of the 6- to 7-nm thick fibrils, which are produced by trypsin digestion of GST-HD51 protein (3).
Figure 2.
Inhibition of HD exon 1 aggregation by small molecules. (A) Structure of the chemical compounds examined for their ability to inhibit HD exon 1 fibrillogenesis. (B) Effect of various concentrations of the indicated chemical compounds on HD exon 1 aggregation as monitored by the filter retardation assay. GST-HD51 fusion protein at a conc. of 2.5 μM was predigested for 30 min at 37°C with trypsin at an enzyme/substrate ratio of 1:10 (wt/wt). Then chemical compounds were added to the cleavage reactions to give the indicated final concentration. Reaction mixtures were incubated for an additional 18 h at 37°C and insoluble polyQ-containing HD51 aggregates were detected by the filter retardation assay. (C) Quantification of the dot blot results obtained with Congo red, thioflavine S, and thioflavine T. The signal intensity obtained from the sample without added chemical compound was arbitrarily set as 100. Values shown are the average of triplicate incubations ± SE. (D) Electron micrographs of HD51 fibrils formed in the presence or absence of the indicated chemical compounds. Trypsin-digested GST-HD51 protein at 2.5 μM was incubated at 37°C for 24 h either without added chemical compound (control) or with Congo red (final conc. 2.5 μM), thioflavine T, and thioflavine S (final conc. 40 μM each). Representative samples of fibrillar structures are shown. (Bar = 200 nm.)
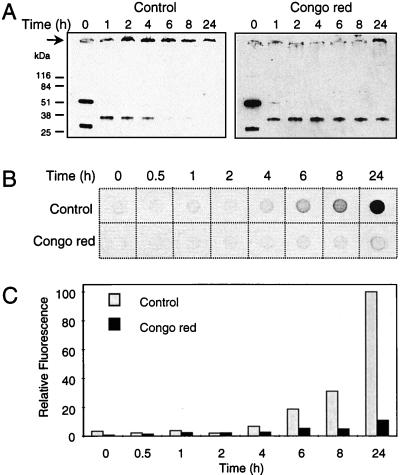
Recently, we have demonstrated that HD exon 1 fibrillogenesis is a nucleation-dependent polymerization, which critically depends on protein concentration and time (15). Thus, if a chemical compound interferes with nucleus formation, this should lead to a marked delay in the onset of HD51 aggregation. To test this hypothesis, a time-course experiment was performed in which the decrease of soluble HD51 protein in the presence or absence of Congo red was monitored over a period of 24 h by SDS/PAGE and immunoblotting using the AG51 antibody. Fig. 3A shows that incubation of trypsin-digested GST-HD51 fusion protein in the presence of Congo red resulted in the production of only a small amount of high molecular weight protein aggregates. Soluble, apparently monomeric HD51 protein migrating in a 12% SDS gel at ≈34 kDa was still detected even after 24 h of incubation in the presence of Congo red. In strong contrast, in the absence of Congo red HD51 aggregated after 6 h of incubation, resulting in a rapid decrease of the HD51 monomers concomitant with the appearance of high molecular weight protein aggregates at the top of the gel (Fig. 3B). As quantification of insoluble protein aggregates that do not enter the stacking gel by Western blot analysis is difficult, aliquots of the aggregation reaction mixtures also were analyzed by the filter retardation assay. Fig. 3C shows that the amount of aggregates formed after a 24-h incubation in the presence of Congo red was only ≈10% of that obtained in the absence of the dye.
Figure 3.
Time course of HD exon 1 aggregation in the presence or absence of Congo red. (A) Western blot analysis of aggregation reactions. GST-HD51 fusion protein at 2.5 μM was incubated at 37°C with trypsin. After 30 min of incubation, Congo red was added to a final conc. of 2.5 μM and incubation was continued for 24 h at 37°C. Samples corresponding to 200 ng of fusion protein were removed from the aggregation reactions at the indicated times and analyzed by SDS/PAGE and immunoblotting using the AG51 antibody. →, The origin of electrophoresis. (B) Analysis of aggregation by the filter retardation assay. Captured aggregates were detected by incubation with the HD1 antibody. (C) Quantitative analysis of the dot-blot results shown in B. The dot with the highest signal intensity was arbitrarily set as 100.
For confirmation of the above results, a separate time-course experiment using a truncated GST-HD exon 1 fusion protein with 51 glutamine residues was performed. This protein (GST-HD51ΔP) lacks the proline-rich region C terminal to the polyQ tract (26). Aggregation was initiated by addition of trypsin to the GST-HD51ΔP fusion protein, and after incubation for various times in the presence or absence of Congo red, aliquots were analyzed by MALDI-TOF-MS. After 24 h of incubation, monomeric HD51ΔP protein was only detected in the samples, which contained the Congo red dye (Fig. 4). In the absence of Congo red, trypsin-digested GST-HD51ΔP fusion protein escaped quantitatively from solution into high molecular weight protein aggregates not accessible to MALDI-TOF-MS analysis but detectable by the filter retardation assay (data not shown). The soluble HD51ΔP protein remaining after 24 h of incubation in the presence of Congo red was readily accessible to proteolytic degradation. Incubation for 30 min with proteinase K resulted in the complete degradation of monomeric HD51ΔP (Fig. 4), whereas the polyQ-containing high molecular weight protein aggregates were resistant to proteinase K treatment (data not shown).
Figure 4.
Effect of Congo red on HD exon 1 aggregation as monitored by MALDI-TOF-MS. GST-HD51ΔP fusion protein at 2.5 μM was incubated for 24 h at 37°C with trypsin (10 ng/μl) in the absence (control) or presence of Congo red (final conc. 2.5 μM). Reaction aliquots (0.5 μl) were taken at different time points and analyzed by MALDI-TOF-MS. (A) Undigested GST-HD51ΔP fusion protein. 2+, Doubly-charged molecular-ion signal of GST-HD51ΔP. (B) GST-HD51ΔP after 45 min incubation with trypsin resulting in release of a peptide, HD51ΔP, with the sequence SF[Q]51PPPPLERPHRD. *, Tryptic digestion products of GST. To confirm the identity of HD51ΔP, the isotopic pattern of HD51ΔP was resolved in high-resolution acquisition mode (Inset, right spectrum) enabling determination of the monoisotopic mass of 8,074.74 Da (expected: 8,074.78 Da). (C) GST-HD51ΔP after 24 h of incubation with trypsin. In the presence of Congo red, nonaggregated HD51ΔP is still detectable whereas in absence of the dye it is not. (D) Samples analyzed in C after 30 min of additional incubation with 10 ng/μl proteinase K (PK). The HD51ΔP monomers stabilized by Congo red are degraded by PK, demonstrating that these molecules are readily accessible for proteolytic digestion. a.i., arbitrary intensity.
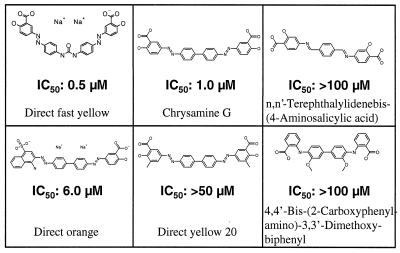
Because the therapeutic potential of Congo red is diminished by the fact that it does not cross the blood–brain barrier well, several structural analogs of Congo red such as chrysamine G (27), Direct yellow 20, Direct orange, Direct fast yellow, 4,4′-bis-(carboxyphenylamino)-3,3′-dimethoxybiphenyl, and N,N′-terephthalylidenebis-(4-aminosalicylic acid) were tested for their ability to inhibit HD51 aggregation in vitro (Fig. 5). Dose-response curves with trypsin-digested GST-HD51 fusion protein similar to those shown in Fig. 2C were produced by using the filter retardation assay. For each compound, IC50 values obtained with this assay are shown in Fig. 5. The more lipophilic Congo red derivatives, Direct fast yellow, and chrysamine G exhibited apparent IC50 values of ≈0.5 and 1 μM, respectively. In comparison, Direct orange inhibited HD51 aggregation with an IC50 of ≈6 μM and the compounds Direct yellow 20, 4,4′-bis-(carboxyphenylamino)-3,3′-dimethoxybiphenyl, and N,N′-terephthalylidenebis-(4-aminosalicylic acid) exhibited IC50 values of >50 μM. These results indicate that the highly acidic naphthalenesulphonic acid moieties of Congo red can be replaced by the more lipophilic salicylic acid moieties without significantly changing the ability of the chemical compounds to block HD51 aggregation. Furthermore, they suggest that a spacing of ≈19–20 Å between the acidic groups is important for inhibiting polyglutamine aggregation. For the most potent huntingtin aggregation inhibitors Congo red, chrysamine G, and Direct fast yellow the distance between the carboxylic groups is between 19 and 20 Å, whereas in the ≈100-fold less effective compound N,N′-terephthalylidenebis-(4-aminosalicylic acid) the distance between the carboxylic groups is only 14 Å. This suggests that a repetitive structural feature with a spacing of ≈19–20 Å is present in HD exon 1 proteins with a long polyQ sequence. However, besides the spacing between the acidic groups other structural features also appear to be critical for efficient inhibition of huntingtin aggregate formation. The presence of a methyl group at position C-3 in the salicylic acid moiety of Direct yellow 20 reduced the inhibitory activity of the compound ≈50-fold compared with chrysamine G. Thus, we conclude that the salicylic acid structure in chrysamine G and Direct fast yellow is important for efficient inhibition of huntingtin aggregation.
Figure 5.
Structure and IC50 values of lipid-soluble Congo red analogs. IC50 values were determined as described in Fig. 2 by using the filter retardation assay. GST-HD51 fusion protein at 2.5 μM was predigested for 30 min at 37°C with 10 ng/μl trypsin and chemical compounds at various concentrations (0.15–40 μM) were added to the cleavage reactions. After incubation for 18 h at 37°C formation of aggregates was quantified by using the filter retardation assay. Dose-response profiles for each chemical compound were generated, and IC50 values were calculated from data sets of four independent experiments.
Inhibition of Huntingtin Aggregation in Cell Culture Systems.
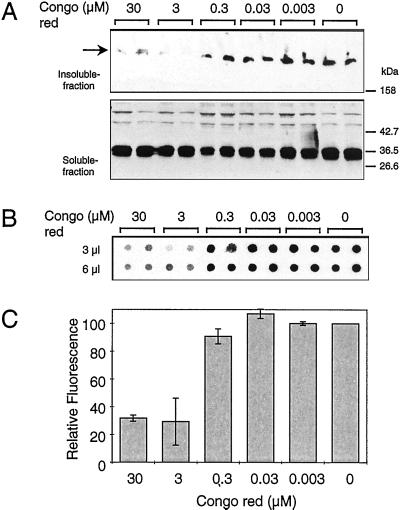
COS cells were grown for 1 wk in the presence or absence of various concentrations of Congo red (0.003–30 μM) and then transfected with an expression construct encoding the HD51 protein (13). Cells were lysed 42 h after transfection, and the soluble proteins were separated from the insoluble material by centrifugation. Both supernatant and insoluble material were analyzed by SDS/PAGE and Western blotting using the HD1 antibody. In the supernatant fractions of both the untreated and Congo red-treated cells, a band migrating on SDS/PAGE at ≈34 kDa corresponding to the soluble HD51 protein was detected, indicating that treatment of cells with Congo red had no dramatic effect on HD51 expression (Fig. 6A). HD51 aggregates that remained near the top of the stacking gel were detected only in the insoluble fraction. For quantification of these insoluble aggregates, the filter retardation assay was used (Fig. 6 B and C). Treatment of COS cells with different concentrations of Congo red inhibited HD51 aggregation in a dose-dependent manner. Growth of cells in the presence of 3 and 30 μM Congo red resulted in an ≈60% and 70% reduction, respectively, of the amount of HD51 aggregates retained on the filter. Similar results of dose-dependent inhibition of HD51 aggregation in transiently transfected COS cells also were obtained with the more lipophilic compound Chrysamine G (data not shown).
Figure 6.
Inhibition of HD exon 1 aggregation in COS cells. (A) Western blot analysis of soluble and insoluble fractions of transfected COS-1 cells after Congo red treatment. COS-1 cells grown for 1 wk in the presence or absence of various concentrations of Congo red (0.003–30 μM) were transfected with the pTL-CAG51 construct. Cell extracts were prepared and fractionated into soluble and insoluble fractions as described in Materials and Methods. Proteins were separated by SDS/PAGE and analyzed by immunoblotting using the HD1 antibody. (B) Filter retardation assay performed on the insoluble fraction of the transfected cell extracts. The SDS-insoluble protein aggregates retained on the filter were detected with the HD1 antibody. (C) Quantitative analysis of the dot blot results shown in B. The dot corresponding to the control experiment without added Congo red was arbitrarily set as 100.
Discussion
Our study demonstrates that the mAb 1C2 as well as the chemical compounds Congo red, thioflavine S, Direct fast yellow, and chrysamine G are capable of preventing huntingtin aggregation in vitro, at least partially. Although the molecular mechanism by which the 1C2 antibody and the various compounds inhibit huntingtin fibrillogenesis is unknown, we suggest that the antibody has a chaperone-like activity and blocks aggregation by stabilizing the native conformation of the elongated polyQ tract, whereas Congo red and its derivatives, which are known to selectively bind to β-sheet structures (27) may slow down huntingtin aggregation by interfering with nucleus formation and/or growth of the fibrils. Perutz (28) proposed that elongated polyQ chains beyond a critical length (>40 glutamines) may lead to a phase change from random coils to hydrogen-bonded hairpins that assemble into β-sheet structures. The finding that the 1C2 antibody, which does recognize elongated polyQ chains in soluble proteins but not in SDS-insoluble aggregates (16, 19), inhibits huntingtin aggregation supports this hypothesis and suggests that other molecules that are capable of stabilizing the native conformation of an elongated polyQ tract should be effective in inhibiting huntingtin aggregation. However, experiments to study antigen/antibody binding and its effect on the self-assembly of polyQ aggregates in more detail are necessary. A major drawback of the in vivo use of antibodies or peptides is that they may be degraded by proteases and that they have a poor blood–brain permeability. In addition, they may not enter the neuronal cells, where the aggregates are formed. Therefore, in future studies small molecules or chemically modified peptides should be screened that are protease resistant, have a reasonable brain permeability, and stabilize the conformation of the polyQ tract in the mutant huntingtin protein. Our results suggests that lipophilic derivatives of Congo red such as chrysamine G and Direct fast yellow that specifically bind to amyloid-like β-sheet structures have a considerable therapeutic potential. Recently, we proposed that inhibition of nucleus formation is a feasible therapeutic strategy against HD and the related glutamine-repeat disorders (15). Our findings support this hypothesis because treatment of transiently transfected COS cells with Congo red significantly reduced the formation of insoluble huntingtin aggregates in vivo. Furthermore, after overnight incubation of trypsin-treated GST-HD51 fusion protein soluble HD51 protein was detected only by immunoblotting or MS when Congo red was present during incubation (Figs. 3 and 4). These results suggest that Congo red interferes with nucleus formation and thereby significantly elongates the lag time of the aggregation process. However, more extensive research, including toxicological studies will be necessary to determine whether the more lipophilic chemical compounds chrysamine G and Direct fast yellow can be used as huntingtin aggregation inhibitors in vivo. In addition, a large-scale screening for the identification of new huntingtin aggregation inhibitors by using an automated membrane filter retardation assay is in progress.
Although it has not been formally proven whether the formation of ordered huntingtin aggregates is the cause of HD, we propose that inhibition of huntingtin fibrillogenesis by small molecules is a very attractive therapeutic strategy. Drugs that selectively bind to elongated polyQ sequences should delay the onset and progression of HD and related glutamine-repeat disorders. Moreover, they also may be useful for treatment of other neurodegenerative disorders caused by aberrant protein folding such as Alzheimer's disease, Parkinson's disease, or the prion diseases.
Acknowledgments
We thank A. Sittler and J.-L. Mandel for providing the 1C2 antibody and S. Schnögl for reading the manuscript. We also thank I. Dunkel, W. Broeker, and R. Hasenbank for excellent technical assistance. This work has been supported by grants from the Huntington's Disease Society of America, Deutsche Forschungsgemeinschaft (Wa1151/2–1), and the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BioFuture project: 0311853).
Abbreviations
- HD
Huntington's disease
- polyQ
polyglutamine
- GST
glutathione S-transferase
- MALDI-TOF
matrix-assisted laser desorption/ionization–time of flight
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110138997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110138997
References
- 1.Harper P S. Huntington's Disease. Philadelphia: Saunders; 1991. [Google Scholar]
- 2.Vonsattel J-P, Meyers R H, Stevens T J, Ferrante R J, Bird E D, Richardson E P. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 4.Davies S W, Trumaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 5.Warrick J M, Paulson H L, Gray-Board G L, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 6.Lunkes A, Mandel J L. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- 7.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Ona V O, Li M, Vonsattel J P, Andrews L J, Khan S Q, Chung W M, Frey A S, Menon A S, Li X J, Stieg P E, et al. Nature (London) 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 9.Perez M K, Paulson H L, Pendse S J, Saionz S J, Bonini N M, Pittman R N. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings C J, Mancini M A, Antalffy B, DeFranco D B, Orr H T, Zoghbi H Y. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez I, Xu C-J, Juo P, Kakizuka A, Blenis J, Yuan J. Neuron. 1999;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 12.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 13.Sittler A, Walter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates G P, Lehrach H, Wanker E E. Mol Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 14.Wanker E E, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 15.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, et al. Nature (London) 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 17.Sittler A, Devys D, Weber C, Mandel J L. Hum Mol Gen. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 18.Miroy G J, Lai Z, Lashuel H A, Peterson S A, Strang C, Kelly J W. Proc Natl Acad Sci USA. 1996;93:15051–15056. doi: 10.1073/pnas.93.26.15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merry D E, Kobayashi Y, Bailey C K, Taye A A, Fischbeck K H. Hum Mol Genet. 1998;7:693–701. doi: 10.1093/hmg/7.4.693. [DOI] [PubMed] [Google Scholar]
- 20.Caughey B, Race R E. J Neurochem. 1992;59:768–771. doi: 10.1111/j.1471-4159.1992.tb09437.x. [DOI] [PubMed] [Google Scholar]
- 21.Tomiyama T, Shoji A, Kataoka K, Suwa Y, Asano S, Kaneko H, Endo N. J Biol Chem. 1996;271:6839–6844. doi: 10.1074/jbc.271.12.6839. [DOI] [PubMed] [Google Scholar]
- 22.Pappolla M, Bozner P, Soto C, Shao H, Robakis N K, Zagorski M, Frangione B, Ghiso J. J Biol Chem. 1998;273:7185–7188. doi: 10.1074/jbc.273.13.7185. [DOI] [PubMed] [Google Scholar]
- 23.Medrano F J, Andreu J M. Eur J Biochem. 1986;158:63–69. doi: 10.1111/j.1432-1033.1986.tb09721.x. [DOI] [PubMed] [Google Scholar]
- 24.Guntern R, Bouras C, Hof P R, Vallet P G. Experientia. 1992;48:8–10. doi: 10.1007/BF01923594. [DOI] [PubMed] [Google Scholar]
- 25.LeVine H., III Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollenbach B, Scherzinger E, Schweiger K, Lurz R, Lehrach H, Wanker E E. Philos Trans R Soc London B. 1999;354:991–994. doi: 10.1098/rstb.1999.0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klunk W E, Debnath M L, Koros A M, Pettegrew J W. Life Sci. 1998;63:1807–1814. doi: 10.1016/s0024-3205(98)00454-8. [DOI] [PubMed] [Google Scholar]
- 28.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]