Abstract
An outbreak of human anthrax occurred in Sverdlovsk, Union of Soviet Socialists Republic (now Ekaterinburg, Russia) in April 1979. Officials attributed this to consumption of contaminated meat, but Western governments believed it resulted from inhalation of spores accidentally released from a nearby military research facility. Tissue samples from 11 victims were obtained and methods of efficiently extracting high-quality total DNA from these samples were developed. Extracted DNA was analyzed by using PCR to determine whether it contained Bacillus anthracis-specific sequences. Double PCR using “nested primers” increased sensitivity of the assay significantly. Tissue samples from 11 persons who died during the epidemic were examined. Results demonstrated that the entire complement of B. anthracis toxin and capsular antigen genes required for pathogenicity were present in tissues from each of these victims. Tissue from a vaccination site contained primarily nucleic acids from a live vaccine, although traces of genes from the infecting organisms were also present. PCR analysis using primers that detect the vrrA gene variable region on the B. anthracis chromosome demonstrated that at least four of the five known strain categories defined by this region were present in the tissue samples. Only one category is found in a single B. anthracis strain.
Keywords: vrrA gene, variable number tandem repeats
An unusual outbreak of human anthrax occurred in Sverdlovsk, Union of Soviet Socialists Republic (now Ekaterinburg, Russia) in April 1979. A 1980 publication attributed this to cutaneous and gastrointestinal infection resulting from consumption of Bacillus anthracis-contaminated meat (1). However, considerable interest in this outbreak was aroused by the suspicion that the epidemic was caused by release of spores from a military microbiology facility. This facility is located at the northern end of a narrow high-risk zone within which most of the victims lived or worked (2). Epidemiological studies strongly suggested that release of a B. anthracis spore aerosol from the facility on April 2, 1979, was responsible for the anthrax outbreak and subsequent death of the human victims (2). Patient records and autopsy reports were removed from the hospital by central government authorities. However, notes describing the autopsy details of 42 victims, microscope slides, paraffin blocks, and tissue samples from necropsies were kept in the possession of two of the authors. These samples were the basis for a series of articles published in the Russian literature. A report describing examination of these samples was also published in English (3). All results were consistent with B. anthracis being the causative agent.
The PCR can be used to sensitively identify pathogens and their remnants in medical samples. DNA oligomers that prime PCR to amplify pathogen sequences can be used to determine the presence of a particular organism if the primers are complementary to only pathogen-specific sequences. When combined with single-stranded conformational polymorphisms or DNA sequencing, such an analysis can detect differences in the DNA sequences of the same fragments from different samples. Herein we report DNA extraction and subsequent PCR analysis of 13 formalin-fixed paraffin-embedded samples from 11 Sverdlovsk victims. Sample DNAs were used as template in a PCR containing primers that amplify portions of the B. anthracis capA, capB, capC, cya, lef, and pag genes. These genes are located on the two large B. anthracis plasmids, pX01 and pX02, required for pathogenicity (4, 5). Samples were also analyzed by using primers that amplify chromosomal B. anthracis-specific DNA sequences. Results of this PCR-based study of the Sverdlovsk necropsies confirm that tissues from all victims contained B. anthracis. Further analysis with primers that distinguish among different B. anthracis strains (6) demonstrated that the Sverdlovsk victims were apparently infected by a mixture of different B. anthracis strains. The implications of these results are discussed.
MATERIALS AND METHODS
Tissue Samples.
Table 1 lists the formalin-fixed samples collected by two of the authors (F.A.A. and L.M.G.) and used for these studies. These were embedded in small (<0.5 cm3) paraffin blocks obtained for 11 of the Sverdlovsk victims. The first digit in the reference number (column 1) cross-references them to the samples listed in table 1 in Abramova et al. (3). The paraffin blocks were cut from the original samples kept by the pathologists (3). Paraffin blocks containing formalin-fixed tissues of anthrax-infected primates were a gift from Arthur Friedlander (U.S. Army Medical Research Institute for Infectious Diseases, Biomedical Research Laboratory, Ft. Detrick, MD). Paraffin slices containing formalin-fixed tissues from B. anthracis-infected bovine samples were provided by Darrell D. Johnson (Veterinary Science Department, South Dakota State University, Brookings, SD). Samples provided by U.S. Army Medical Research Institute of Infectious Diseases were cut from blocks by using individual sterile scalpels, and samples from South Dakota State University were sliced with a microtome.
Table 1.
PCR analysis of different tissues to detect B. anthracis genes
| Sample | Tissue | pX01 genes
|
pX02 genes
|
CS | VNTR(s) | ||||
|---|---|---|---|---|---|---|---|---|---|
| pag | lef | cya | capA | capB | capC | ||||
| 7.RA93.15.15 | Spleen | ++ | ++ | ++ | ++ | + | + | ++ | 4 |
| 31.RA93.39.3 | Spleen | ++ | ++ | ++ | ++ | +* | +* | + | 4 |
| 26.RA93.043 | Spleen | ++ | + | + | + | +* | + | +* | 2 |
| 40.RA93.40.5 | Spleen | + | + | ++ | +* | +* | + | +* | 2, 4, 5 |
| 27.RA93.30.3 | Spleen | +* | ++ | ++ | + | +* | ++ | +* | 2 |
| 37.RA93.35.4 | Vaccination site | ++ | + | + | +* | − | − | +* | 2, 4, 5 |
| 37.RA93.35.4 | Spleen | ++ | ++ | + | +* | +* | − | +* | 2, 4, 5 |
| 37.RA93.35.6 | Lung | + | + | + | + | +* | − | +* | 4 |
| 3.RA93.1.1 | Meninges | + | + | + | + | +* | +* | +* | 2, 5, 6 |
| 25.RA93.031 | Meninges | ++ | ++ | ++ | ++ | +* | ++ | +* | 4 |
| 1.RA93.42.1 | Meninges | + | ++ | + | ++ | + | + | +* | 4 |
| 33.RA93.20.5 | Meninges | ++ | ++ | + | ++ | ++ | ++ | + | 4 |
| 21.RA93.38.4 | Lymph node | ++ | ++ | + | +* | + | +* | ++ | 4 |
++, Strong PCR amplicon with first amplification; +, weak PCR amplicon with first amplification; +*, PCR amplicon only visible following nested PCR; −, no amplicon; CS, chromosomal sequence. Results are from analysis of at least three different amplifications.
DNA Extraction from Tissues.
All procedures were conducted in a Baker Biogard laminar flow hood (Baker, Sanford, ME). Each sample was handled independently, and a new sterile scalpel blade was used to cut shavings from each sample to minimize cross-contamination among samples. Single thin shavings from each sample were cut from blocks and placed into individual sterile 1.5-ml microcentrifuge tubes. One milliliter of xylene was added to each tube to extract the tissue from the paraffin. Tissues were incubated on a rocking shaker at room temperature until the paraffin was solubilized. Remaining tissue was pelleted by centrifugation at 12,000 × g for 2 min, and the solvent was removed. If paraffin was still present in the sample, the xylene extraction was repeated. Tissues were then washed with 0.5 ml of ice-cold 100% ethyl alcohol and collected by centrifugation as before, the ethyl alcohol removed, and the pellets suspended in 10 μl of acetone. Samples were air-dried at 55°C to remove the acetone. Dried pellets were suspended in 150 μl of a freshly prepared solution containing 50 mM Tris⋅HCl (pH 8.5), 1 mM EDTA, 0.1% Tween-20, and proteinase K (Boehringer Mannheim; 20 mg/ml). Samples were incubated overnight at 37°C. Resulting suspensions were centrifuged 15 sec at 12,000 × g, and the supernatant was collected and incubated at 95°C for 10 min to inactivate the proteinase. Samples were centrifuged as before, and the supernatant containing the DNA was transferred to a sterile microcentrifuge tube. Samples were stored at −20°C.
PCR Amplification of DNA.
Pigments present in the extracted DNA samples prevented direct photometric measurement of DNA concentrations. DNA samples were analyzed by gel electrophoresis and ethidium bromide staining to determine the concentration and average length of the DNA fragments. Dilutions of each sample were compared with known DNA standards to estimate the concentration. Purified DNA samples often contained sufficient impurities to inhibit PCRs. Therefore, undiluted and 1:10, 1:25, and 1:50 dilutions of DNA samples were used as template in a PCR containing bacterial-specific 16S ribosomal RNA primers to determine optimal concentrations for analysis. These primers were used as positive controls in all subsequent PCRs. DNA oligomers that functioned as PCR primers to amplify representative B. anthracis gene fragments are shown in Table 2. The initial PCR contained 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, all four dNTPs (each at 0.2 mM), 20 pmol of each primer, 2.5 units of Amplitaq DNA polymerase LD (Roche Molecular Systems, Branchburg, NJ), and 1–10 ng of template DNA in a 100-μl total reaction volume. Perkin–Elmer reagents were used for all reactions. Amplitaq DNA polymerase LD was used because the standard Amplitaq DNA polymerase preparations contained sufficient bacterial DNA contamination to support amplification of a bacterial 16S rDNA fragment in negative control reactions. Template DNA was initially denatured by heating at 94°C for 2 min. This was followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and primer extension at 72°C for 1 min. Incubation for 5 min at 72°C followed to complete extension. PCR was conducted in a Perkin–Elmer GeneAmp PCR System 9600 thermal cycler. PCR amplicons were purified by using QIAquick PCR purification kits (Qiagen, Chatworth, CA). Ten microliters of purified product from each of the first reactions was then used as DNA template in subsequent PCRs using “nested primers.” PCR conditions were as outlined above.
Table 2.
Sequences of DNA oligodeoxynucleotides used as primers of PCR reactions to detect B. anthracis genes
| Primer set | P/N | Gene/accession no. | Location | Primer sequence | Amplicon size, bp |
|---|---|---|---|---|---|
| GPR-4 | P | ACAACTACCACCGATGGC | |||
| GPR-5 | P | vrrA/L48553 | C | TTATTTATCATATTAGTTGGATTCG | 377–425 |
| EWA-1 | N | TATGGTTGGTATTGCTG | |||
| EWA-2 | N | vrrA/L48553 | C | ATGGTTCCGCCTTATCG | 142–190 |
| PA-1F | P | CCAGACCGTGACAATGATG | |||
| PA-1R | P | pag/M22589 | pX01 | CAAGTTCTTTCCCCTGCTA | 508 |
| PA-2F | N | CGAAAAGGTTACAGGACGG | |||
| PA-1R | N | pag/M22589 | pX01 | CAAGTTCTTTCCCCTGCTA | 409 |
| LEF-1F | P | GGTGCGGATTTAGTTGATTC | |||
| LEF-1R | P | lef/M29081 | pX01 | CGCTTCATTTGTTCTCCC | 851 |
| LEF-2F | N | GAAACATCGGTCTGGAAAT | |||
| LEF-2R | N | lef/M29081 | pX01 | CCCTTTTGAATGAACTTGC | 403 |
| CYA-1F | P | GCGATGAAAACAACGAAGTA | |||
| CYA-2R | P | cya/M24074 | pX01 | TCGTCTTTGTCGCCACTATC | 720 |
| CYA-2F | N | CATTAGAAAAGCAAAAAGGTC | |||
| CYA-1R | N | cya/M24074 | pX01 | TCATTATCTTGCTCTGTGCC | 186 |
| CAPA-F | P | CAGAAGCAGTAGCACCAGTAA | |||
| CAPA-R | P | capA/M24150 | pX02 | ATTTTCACCAGCACCCAC | 397 |
| CAPA-Fnest | N | TGACGATGGTTGGTGACA | |||
| CAPA-Rnest | N | capA/M24150 | pX02 | CCTTATTGTATCTTTAGTTCCC | 302 |
| CAPB-F | P | CTGACCAATCTAAGCCTGC | |||
| CAPB-R | P | capB/M24150 | pX02 | TCGTTTCTCCAATCGCAAT | 221 |
| CAPC-F | P | GTACCTGGTTATTTAGCACTCG | |||
| CAPC-R | P | capC/M24150 | pX02 | ATCTCAAATGGCATAACAGG | 208 |
P, primary primer set; N, nested primer set; C, chromosomal; pX01, located on plasmid pX01; pX02, located on plasmid pX02.
Analysis of PCR Amplicons.
Large (>200 bp) PCR amplicons were analyzed by electrophoresis through 3% [2% (wt/vol) NuSieve GTG/1% SeaKem LE; FMC] agarose dissolved in 45 mM Tris borate (pH 8.3) and 1.0 mM EDTA. Smaller vrrA amplicons were separated by electrophoresis through 4.5% (wt/vol) FMC Metaphor agarose gels prepared in 90 mM Tris borate (pH 8.3) and 1.0 mM EDTA. Electrophoresis was for 1 hr at 200 V through 3% gels or for 5 hr at 70 V through 4.5% Metaphor gels. Gels were stained for 30 min to 2.5 hr with a solution containing ethidium bromide (1 μg/ml), destained in distilled water, and then visualized under UV. Images were captured electronically by using a Strategene Eagle Eye II still video system (Strategene).
DNA Sequencing.
GPR-4 and GPR-5 primers (Table 2) were used in the PCR to amplify 377- to 425-bp DNA fragments of the B. anthracis vrrA gene from the total DNA extracted from tissue samples. Resulting PCR amplicons were purified through QIAquick PCR purification columns and then used as template in subsequent sequencing reactions. EWA-1 and EWA-2 nested oligonucleotides (ref. 6 and Table 2) were used in Taq DyeDeoxy Terminator cycle sequencing reactions (Applied Biosystems) to sequence a 142- to 190-bp internal DNA fragment of the original amplicons containing the variable number tandem repeat sequence. Unincorporated dyes were removed from these reactions by using Centri-Sep spin columns (Princeton Separations, Adelphia, NJ), and sequencing was performed on an Applied Biosystems 373A Stretch DNA Sequencer. Both strands of each amplicon were sequenced twice. DNA sequence files were analyzed by using sequence navigator software (Applied Biosystems).
RESULTS
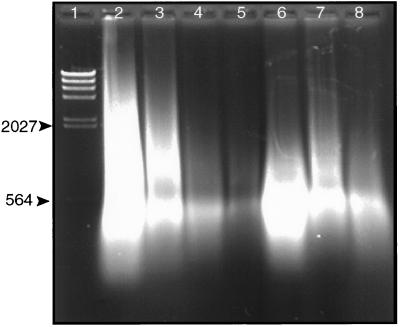
Fig. 1 shows DNA extracted from the tissues of different victims. The amount of DNA extracted varies considerably from one tissue to the next. However, the size of the DNA isolated is always between 1 kb and 300 bp, with the majority approximately 500 bp. This size dictated selection of primers and PCR strategy. The majority of the DNA isolated is of human origin. Moreover, there is no necessary correlation between the amount of total DNA present and the amount of B. anthracis DNA contained in each sample. PCR analysis and interpretation of the experimental results are influenced by the purity of the samples. Test extractions using other tissues demonstrated that use of the same tools to prepare multiple samples can lead to cross-contamination that is detectable by using PCR-based methods. Samples were therefore handled independently of one another. The possibility of cross-contamination among samples from different victims during prior handling and shipping exists. However, the only possible sources of B. anthracis DNA are the samples themselves. Therefore, possible cross-contamination among the different samples does not alter the conclusion that multiple strains were present in the samples regarded collectively.
Figure 1.
Agarose gel electrophoresis of DNA extracted from formalin-fixed paraffin-embedded Sverdlovsk victim tissue samples. Total DNA extracted from tissues was analyzed by electrophoresis through a 1% agarose gel. Electrophoresis was for 1.5 hr at 85 V. DNA was visualized under UV after staining with ethidium bromide. Lanes: l, λ DNA digested with HindIII; 2–8, DNA extracted from 7.RA93.15.15 spleen, 31.RA93.39.3 spleen, 27.RA93.30.3 spleen, 37.RA93.35.4 vaccination site, 37.RA93.35.4 spleen, 37.RA93.35.6 lung, and 26.RA93.043 spleen, respectively. Numbers on the left side of the gel refer to the size of DNA markers in bp.
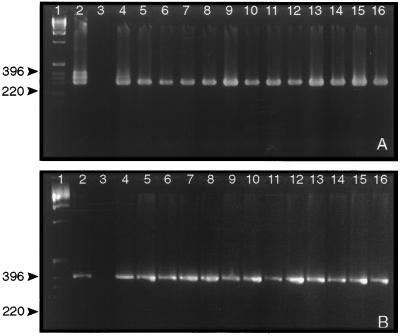
PCR analysis was conducted by using primers that amplified segments of each of the structural genes found on pX01 and pX02. Two sets of primer pairs were used to amplify fragments encoding portions of the pag, lef, cya, and capA genes. Primer pairs used in the initial PCR amplified a larger DNA fragment that contained the sequences complementary to the second set of primers. DNA products of the first amplification were used as template in a second reaction containing the second primer set. This use of “nested primers” significantly increased sensitivity and maintained the specificity of the reaction. B. anthracis-specific “nested primers” were not available for the capB and capC genes. Therefore, the second reaction contained the same primers as the first. Although this can lead to nonspecific amplification, the size of the amplicons produced was, in all cases, that expected. Fig. 2 shows the results of PCR amplification using double PCR to amplify portions of the capA (pX02), and lef (pX01) genes from the 13 samples (Fig. 2 A and B, respectively). These results demonstrate that the Sverdlovsk victims were infected by virulent B. anthracis containing both plasmids and not a vaccine strain. Samples from a vaccination site were also positive for capA in double PCR (Fig. 2A, lane 9). However, single PCR using primers for capA, capB, or capC did not produce detectable amplicons with DNA templates extracted from vaccination-site tissue (Table 1). This suggests that the capACB sequence and, therefore pX02, is underrepresented in this sample.
Figure 2.
Agarose gel electrophoresis of PCR amplicons after amplification of capA and lef gene-specific DNA fragments from tissue DNA. Total DNA extracted from tissues was used as template in PCR containing capA-specific (A) or lef-specific (B) primers to amplify a portion of these genes. Products of the first amplification were subsequently used in a second set of reactions containing “nested” PCR primers to further amplify a portion of the targeted DNA fragment. Samples were analyzed by electrophoresis through 3% (wt/vol) agarose gels. Gels were stained with ethidium bromide and DNA was visualized under UV. Lanes: 1, 1 kb DNA ladder marker (Life Technologies); 2–16 (both A and B), results of second PCRs containing B. anthracis (strain Vollum) control DNA, uninfected human DNA control, 7.RA93.15.15 spleen DNA, 31.RA93.39.3 spleen DNA, 26.RA93.043 spleen DNA, 40.RA93.40.5 spleen DNA, 27.RA93.30.3 spleen DNA, 37.RA93.35.4 vaccination site DNA, 37.RA93.35.4 spleen DNA, 37.RA93.35.6 lung DNA, 3.RA93.1.1 meninges DNA, 25.RA93.031 meninges DNA, 1.RA93.42.1 meninges DNA, 33.RA93.20.5 meninges DNA, and 21.RA93.38.4 lymph node DNA. The numbers to the left refer to the size (bp) of marker DNA fragments. The triplet bands in the capA control reaction (A, lane 2) result from incomplete removal of the first primer set prior to running the second “nested PCR.” This results in amplicons of 397, 350, 349, and 342 bp.
Table 1 shows results of PCR using primers that amplify portions of known B. anthracis genes found on pX01 and pX02. PCR results using a primer set that specifically amplifies a randomly selected portion of the B. anthracis chromosome are also shown. These results clearly demonstrate that B. anthracis was responsible for the 1979 epidemic.
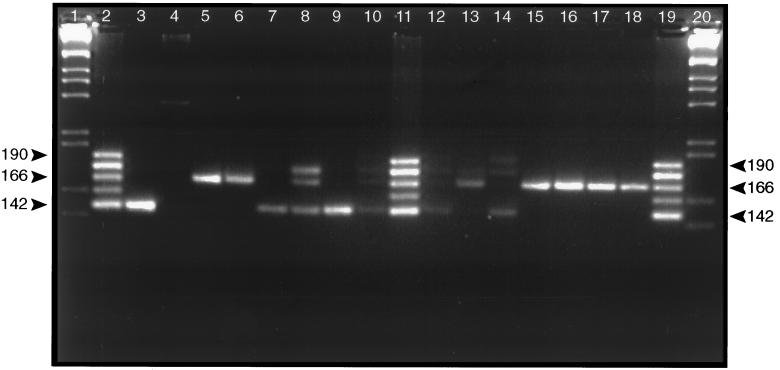
Single PCR analysis of tissue DNA using EWA-1 and EWA-2 primers to amplify the variable portion of the vrrA gene (6, 7) clearly demonstrated length polymorphisms among the samples (data not shown). However, some reactions produced only small amounts of amplicon. Therefore, amplicons from a first PCR using GPR-4 and GPR-5 primers were used as template in a second PCR containing EWA-1 and EWA-2 “nested primers.” Results produced much more pronounced patterns of amplicons consistent with the first PCR results. Separation on Metaphor agarose gels resolved amplicons containing variable number tandem repeats (VNTR) in the vrrA gene (Fig. 3). Four of the five different VNTR categories, (VNTR)2, (VNTR)4, (VNTR)5, and (VNTR)6, known to exist in different B. anthracis strains are present. The (VNTR)2 and (VNTR)4 categories predominate. Many tissues contain only one or a predominant VNTR category (lanes 5, 6, 7, 9, 13, and 15–18). However, several samples contain amplicons representing additional categories (lanes 8, 10, 12, and 14). Two samples (lanes 8 and 14) contain multiple VNTR amplicons that are approximately equally represented in the reaction. This suggests that, in at least some cases, tissues are infected with multiple B. anthracis strains representative of the different VNTR categories. DNA fragments from several of the reactions containing a single amplicon were sequenced to determine whether the visible amplicons contained the expected vrrA VNTR sequences or whether they were PCR artifacts. In all cases, sequences matched the DNA sequence appropriate for the VNTR category exactly (6). It may still be possible that variation in VNTR patterns may result from some PCR artifact associated with amplifying fragments from degraded DNA samples. However, 9 of 13 samples from the human victims contained only (VNTR)2 or (VNTR)4. Such errors would most likely generate all different combinations of the categories and not favor the two predominant categories present.
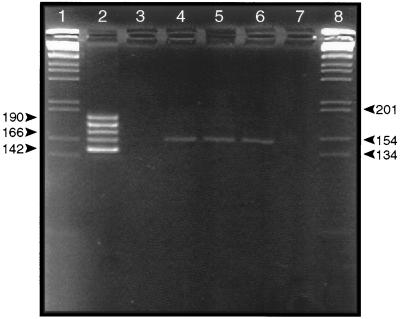
Figure 3.
VrrA VNTR amplicons produced by PCR of DNA extracted from 11 different Sverdlovsk anthrax victims. PCR amplicons were analyzed by electrophoresis through a 4.5% (wt/vol) Metaphor agarose gel. Electrophoresis was for 5 hr at 70 V. Gels were stained with ethidium bromide, and DNA was visualized under UV. Lanes: 1 and 20, 1 kb DNA ladder marker; 2, 11, and 19, VNTR ladder containing the five different VNTR categories found in B. anthracis (6); 3, results of PCR containing B. anthracis (strain Vollum) control DNA; 4, PCR containing uninfected human control DNA; 5–10, results of PCR containing 7.RA93.15.15 spleen DNA, 31.RA93.39.3 spleen DNA, 26.RA93.043 spleen DNA, 40.RA93.40.5 spleen DNA, 27.RA93.30.3 spleen DNA, and 37.RA93.35.4 vaccination site DNA, respectively; 12–18, results of PCR containing 37.RA93.35.4 spleen DNA, 37.RA93.35.6 lung DNA, 3.RA93.1.1 meninges DNA, 25.RA93.031 meninges DNA, 1.RA93.42.1 meninges DNA, 33.RA93.20.5 meninges DNA, and 21.RA93.38.4 lymph node DNA. Labels on the sides refer to the size of B. anthracis VNTR categories 2, 4, and 6 (142, 166, and 190 bp, respectively) (6).
The possibility also exists that the VNTR region of the vrrA gene is modified during the course of the disease and multiple VNTR categories are generated in all animal infections by an unknown genetic mechanism. To test this possibility, DNA samples extracted from formalin-fixed paraffin-embedded tissues collected from three nonhuman primates infected with a single B. anthracis strain were analyzed with the same sets of primary and nested primers. Fig. 4 shows that B. anthracis-infected primate tissues contained only the single VNTR category consistent with the infecting strain regardless of the tissue tested. The same results were obtained for tissues prepared from all three primate samples. DNA extracted from brain tissue of the infected primate did not show any detectable VNTR sequence (Fig. 4, lane 7). However, histologic analysis of the same sample did not detect characteristics of B. anthracis tissue invasion (8). DNA extracted from formalin-fixed paraffin-embedded bovine samples collected from two different anthrax victims contained only one VNTR category (data not shown). Multiple single-colony isolates from a single bovine infection also contained only one VNTR category.
Figure 4.
Agarose gel electrophoresis of the B. anthracis vrrA VNTR amplicons in DNA extracted from formalin-fixed paraffin-embedded primate tissues infected with inhalational anthrax. DNA was extracted from primate tissue samples (8) and used as template in PCR containing primers that amplify the B. anthracis VNTR region. Amplification was as described for the results presented in Fig. 3 using nested primers. Lanes: 1 and 8, 1 kb DNA ladder marker; 2, VNTR ladder containing the five different VNTR categories found in B. anthracis (6); 3, results of amplification using tissue from an uninfected primate; 4–7, results of PCR amplification using template DNA from spleen, lung, liver, and brain of a single primate infected with a single strain of B. anthracis (8). Numbers on the left refer to the size of B. anthracis VNTR categories 2, 4, and 6 (6).
VNTRs have also been identified at other apparently unrelated genetic loci in the B. anthracis chromosome based on amplified fragment length polymorphism analysis of many B. anthracis isolates (9). PCR primers designed to amplify one such region also showed variability in the B. anthracis infecting the Sverdlovsk tissue samples (data to be presented elsewhere).
DISCUSSION
Formalin cross-links macromolecules when used to fix biological materials (10). Therefore, it is difficult to extract large DNA fragments from formalin-fixed tissues. It was necessary to optimize the yield and quality of the limited DNA available from these formalin-fixed paraffin-embedded tissues. The DNA extraction procedure removed the paraffin substrate and then relied on aqueous hydration and protease digestion in the presence of detergent to extract DNA from tissue. Although it is difficult to measure the amount of tissue extracted, results suggest that a large proportion of the total sample DNA is recovered by using this method. The isolated DNA is between 1 kb and 300 bp with the majority approximately 500 bp (Fig. 1). PCR primers were therefore designed to amplify relatively small DNA fragments. Only two primer sets were designed to produce amplicons larger than 500 bp. Both of these functioned to identify the appropriate sequences in DNA from the human tissues.
Most B. anthracis genes so far sequenced are located on one of two large plasmids that are required for pathogenicity. The larger pX01 (174 kb) plasmid contains the lef, pag, and cya genes encoding the lethal factor, protective antigen, and the edema factor proteins, respectively (11–13). The Atx A gene encoding the trans-acting positive regulator of toxin synthesis is also located on this plasmid (14). The smaller pX02 (95 kb) plasmid contains the capA, capB, and capC genes (15, 16). These encode information to produce a poly(d-glutamic acid) capsule. Anthrax virulence is due to production of this capsule in conjunction with the pX01-encoded proteins (13–15). A trans-acting regulatory protein is also encoded on pX02 (17). Two primer sets were used to amplify fragments encoding portions of the pag, lef, cya, and cap A genes and a chromosome-specific DNA fragment (Table 2). The “double PCR” amplification strategy optimized PCR sensitivity and selected against nonspecific fragment amplification. DNA products of the first amplification were used as template in a second reaction containing the “nested primers.” A single primer set was used in first and second reactions to amplify capB and capC.
Previously developed live vaccines use strains of B. anthracis that contain only pX01 and do not produce the capsule proteins (18–20). The Soviets used such a live vaccine to protect against this disease (18, 21). Therefore, analysis of samples using plasmid-specific primers definitively shows whether victims’ tissues contain vaccine or pathogenic strains. All samples supported amplification of the expected fragments in reactions containing primers for pX01-specific genes. They also supported amplification of a capA-specific sequence found on pX02. However, one sample (Table 1 and Fig. 2A, lane 9) contains DNA extracted from tissue collected from a vaccination site. The initial PCR using the first set of capA primers did not detect this sequence, and it was negative for capB- and capC-specific sequences. However, use of capA “nested primers” produced a positive result for this sample. The capA primers used consistently detect smaller numbers of template than capB or capC primers. It is possible that the samples were cross-contaminated during handling. The handling history of the human samples prior to their arrival in our laboratory is unknown, and it is unlikely that such precautions were taken during the initial sampling and histological analysis. However, it is also possible that the vaccination site contained a large quantity of the vaccine strain and a small amount of the bacteremic virulent strain because the patient died of systemic anthrax.
We have also designed and tested B. anthracis-specific PCR primers based on randomly selected B. anthracis chromosomal DNA sequences. Sample analysis with these primers demonstrated the presence of B. anthracis-specific chromosomal DNA in all the samples (Table 1). Thus, these results demonstrate that all victims of the Sverdlovsk anthrax outbreak represented in our samples were infected with virulent B. anthracis. Our results are in agreement with tissue analyses presented by Abramova et al. (3). Although Abramova et al. (3) report that they were unable to culture B. anthracis from tissues of victims 21 and 31, PCR results presented herein demonstrate that these victims too were infected with B. anthracis. It is possible that antibiotic treatment of these victims killed the bacteria or inhibited growth of B. anthracis in postmortem cultures.
Strain analysis using previously described sequencing of 16S rRNA and other variable regions is not sufficiently sensitive to differentiate among different B. anthracis strains because these sequences are identical among all B. anthracis strains tested (7). The B. anthracis vrrA VNTR region separates all known strains into five categories based on the presence of from two to six copies of a 12-bp tandem repeat (6). Results shown in Fig. 3 and summarized in Table 1 clearly demonstrate that more than one VNTR category is present in different victims. It is possible that these are PCR artifacts generated by an unknown mechanism. However, it is difficult to imagine such a mechanism that would produce category 2, 4, 5, and 6 but not category 3 amplicons. Several samples contain only category 2 or category 4 strains. DNA sequences of these different amplicons match those for the VNTR categories exactly (data not shown and ref. 6). No known B. anthracis strain contains more than one VNTR category (6). Although many tissue samples contain only one category, several (Fig. 4, lanes 8, 10, 12, and 14) contain more than one. It has been thought that only one strain will predominate in a mixed infection (22). However, this inference is based on measurement techniques far less sensitive than PCR. It may be possible that the remnants of multiple strains may be present in the samples but only one viable strain predominates. The PCR results presented are not quantitative so it is impossible to determine relative amounts of the different strains present in the samples.
In the analysis of 198 individual B. anthracis isolates from natural sources, each contained only one VNTR category (6). Primate samples infected with a single strain only show one VNTR category (Fig. 4). Only a single category was found in formalin-fixed tissues and cultures from two bovine victims of a natural anthrax outbreak in the United States (data not shown). Analysis of anthrax bovine and wood bison victims collected over 30 years in Canada showed no variability at the vrrA locus (6, 9). Collectively, this suggests that it is unlikely VNTR categories change frequently during the infection process or that more than one VNTR category is represented in a natural infection. Four of the five known VNTR categories are represented in the Sverdlovsk victims’ tissues. The simplest explanation for this is that the presence of multiple VNTR categories indicates that victims were exposed to a mixture of different B. anthracis strains. The strains could come from two possible sources, vaccination or inhalation. One of the authors present during the outbreak believes that only one of the victims analyzed in this study was vaccinated and several of the victims succumbed to the disease before the vaccination program was initiated. However, we have no written records that define the date that vaccinations were initiated. The (VNTR)2 and (VNTR)4 categories predominate in these samples. However, DNA extracted from a vaccination site from one victim contained at least three different VNTR categories. This is consistent with the presence of multiple B. anthracis VNTR categories in the vaccine. Two reports in the Russian literature state that live spore vaccines developed for immunization of humans contain two different B. anthracis strains (18, 21).
Approximately 60% of all known B. anthracis strains so far analyzed fall into the (VNTR)4 category including all Sterne and Ames isolates (6). Only 6% fall into category 2. However, three of five Vollum isolates analyzed are found in this category. Representation of four of five VNTR categories suggests that victims’ tissues contained a mixture of at least four strains. However, given the frequency of different VNTR categories in 198 different isolates (6), the probability that more than four strains were present is high. The function of the vrrA gene and the role of variability in this locus are unknown. Further analysis of the putative protein encoded by the vrrA ORF may provide insights into any role this protein might play in pathogenicity or virulence. In view of possible changes in the tandem repeats within the vrrA gene that might complicate interpretation of these results, it is probably worthwhile to continue analysis of these samples by sequencing genes encoding virulence factors, especially pag, from the different samples.
Results presented herein demonstrate the value of PCR and other molecular techniques to genetically analyze archived forensic samples for pathogen content. Future development of appropriate PCR primers and the associated technology should provide a thorough rapid characterization of forensic, environmental, veterinary, and medical samples for their pathogenic content and provide a wealth of information about the specific genetic character of each pathogen. This should be valuable for designing treatments to combat a specific disease and for understanding factors influencing outbreaks and spread of diseases.
Acknowledgments
We are indebted to Drs. Gary Andersen, David H. Walker, Philip Hanna, Theresa Koehler, Joshua Lederberg, Matthew Meselson, and Kenneth Wilson for helpful suggestions and for providing expertise and review of this work. This research was performed under the auspices of the U.S. Department of Energy.
ABBREVIATION
- VNTR
variable number tandem repeats
References
- 1.Bezdenezhnykh I S, Nikiforov V N. Zh Mikrobiol Epidemiol Immunobiol. 1980;5:111–113. [PubMed] [Google Scholar]
- 2.Meselson M, Guillemin J, Hugh-Jones M, Langmuir A, Popova I, Shelokov A, Yampolskaya O. Science. 1994;266:1202–1208. doi: 10.1126/science.7973702. [DOI] [PubMed] [Google Scholar]
- 3.Abramova F A, Grinberg L M, Yampolskaya O V, Walker D H. Proc Natl Acad Sci USA. 1993;90:2291–2294. doi: 10.1073/pnas.90.6.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green B D, Battisti L, Koehler T M, Thorne C B, Ivins B E. Infect Immun. 1985;49:291–297. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uchida I, Hashimoto K, Terakado N. J Gen Microbiol. 1986;132:557–559. doi: 10.1099/00221287-132-2-557. [DOI] [PubMed] [Google Scholar]
- 6.Jackson P J, Walthers E A, Kalif A S, Richmond K L, Adair D M, Hill K K, Kuske C R, Andersen G L, Wilson K H, Hugh-Jones M E, Keim P. Appl Environ Microbiol. 1996;63:1400–1405. doi: 10.1128/aem.63.4.1400-1405.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen G L, Simchock J M, Wilson K H. J Bacteriol. 1996;178:377–384. doi: 10.1128/jb.178.2.377-384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz D L, Jaax N K, Lawrence W B, Davis K J, Pitt M L M, Ezzell J W, Friedlander A M. Lab Invest. 1995;73:691–702. [PubMed] [Google Scholar]
- 9.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Jackson P. J Bacteriol. 1996;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overton W R, McCoy J P., Jr Cytometry. 1994;16:351–356. doi: 10.1002/cyto.990160410. [DOI] [PubMed] [Google Scholar]
- 11.Bragg T S, Robertson D L. Gene. 1989;81:45–54. doi: 10.1016/0378-1119(89)90335-1. [DOI] [PubMed] [Google Scholar]
- 12.Robertson D L, Tippetts M T, Leppla S H. Gene. 1988;73:363–371. doi: 10.1016/0378-1119(88)90501-x. [DOI] [PubMed] [Google Scholar]
- 13.Welkos S L, Lowe J R, Eden-McCutchan F, Vodkin M, Leppla S H, Schmidt J J. Gene. 1988;69:287–300. doi: 10.1016/0378-1119(88)90439-8. [DOI] [PubMed] [Google Scholar]
- 14.Uchida I, Hornung J M, Thorne C B, Klimpel K R, Leppla S H. J Bacteriol. 1993;175:5329–5338. doi: 10.1128/jb.175.17.5329-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green B D, Battisti L, Koehler T M, Thorne C B, Ivins B E. Infect Immun. 1985;49:291–297. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uchida I, Sekizaki T, Hashimoto K, Terakado N. J Gen Microbiol. 1985;131:363–367. doi: 10.1099/00221287-131-2-363. [DOI] [PubMed] [Google Scholar]
- 17.Vietri N J, Marrero R, Hoover T A, Welkos S L. Gene. 1995;152:1–9. doi: 10.1016/0378-1119(94)00662-c. [DOI] [PubMed] [Google Scholar]
- 18.Lesnyak O T, Saltykov R A. Zh Mikrobiol Epidemiol Immunobiol. 1970;47:32–35. [PubMed] [Google Scholar]
- 19.Little S F, Knudson G B. Infect Immun. 1986;52:509–512. doi: 10.1128/iai.52.2.509-512.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivins B E, Ezzell J W, Jr, Jemski J, Hedlund K W, Ristroph J D, Leppla S H. Infect Immun. 1986;52:454–458. doi: 10.1128/iai.52.2.454-458.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuliak V P. Zh Mikrobiol Epidemiol Immunobiol. 1970;47:117–120. [PubMed] [Google Scholar]
- 22.Zelle M R, Lincoln R E, Young G A., Jr J Infect Dis. 1946;79:247–253. doi: 10.1093/infdis/79.3.247. [DOI] [PubMed] [Google Scholar]