Abstract
In the guts of more than 25 species of arthropods we observed filaments containing refractile inclusions previously discovered and named “Arthromitus” in 1849 by Joseph Leidy [Leidy, J. (1849) Proc. Acad. Nat. Sci. Philadelphia 4, 225–233]. We cultivated these microbes from boiled intestines of 10 different species of surface-cleaned soil insects and isopod crustaceans. Literature review and these observations lead us to conclude that Arthromitus are spore-forming, variably motile, cultivable bacilli. As long rod-shaped bacteria, they lose their flagella, attach by fibers or fuzz to the intestinal epithelium, grow filamentously, and sporulate from their distal ends. When these organisms are incubated in culture, their life history stages are accelerated by light and inhibited by anoxia. Characterization of new Arthromitus isolates from digestive tracts of common sow bugs (Porcellio scaber), roaches (Gromphodorhina portentosa, Blaberus giganteus) and termites (Cryptotermes brevis, Kalotermes flavicollis) identifies these flagellated, spore-forming symbionts as a Bacillus sp. Complete sequencing of the 16S rRNA gene from four isolates (two sow bug, one hissing roach, one death’s head roach) confirms these as the low-G+C Gram-positive eubacterium Bacillus cereus. We suggest that B. cereus and its close relatives, easily isolated from soil and grown on nutrient agar, enjoy filamentous growth in moist nutrient-rich intestines of healthy arthropods and similar habitats.
Keywords: anthrax, Ftsz, light-sensitive bacilli, segmented filamentous bacteria, spore attachment fibers
Large, rod-shaped, aerobic, endospore-forming, Gram-positive eubacteria that are members of the genus Bacillus exist in laboratory cultures worldwide. Most are considered to be benign soil bacteria (1) except for a few confirmed pathogens (2). Most grow on rich media as uni- (single) and diplo- (paired) cells or as short filaments and stain Gram-positive or Gram-variable. Bacillus are used as research models to study bacterial nutrition, cell wall formation, sporogenesis, gene regulation and plasmid expression, production of insecticidal (3) and other toxins (e.g., germ-warfare agents in anthrax), immunogenesis in vaccine production, and resistance to extreme conditions (4). Arthromitus filaments from arthropods were accurately described by Philadelphia physician and naturalist Joseph Leidy (5–7). Leidy named several species of segmented filamentous Arthromitus “rooted” in the intestinal walls of the common subterranean termite from eastern North America, Termes [= Reticulitermes] flavipes. Leidy described these filamentous “plants” as invariably associated with motile wood-ingesting “animals” (parabasalid protoctists).
Leidy’s descriptions (Fig. 1) are augmented by our observations of over 25 species of healthy arthropods. Their symbiotic prokaryotes have been variously designated in termites as Arthromitus (8) or Coleonema (9), in ducks as Coleomitus (10), in rats, mice, and chickens as Anisomitus (11) or (provisionally) as Candidatus Arthromitus (12), and in mice as the family Arthromitaceae (arthromitids) (13). Mammals and other vertebrates harbor unclassified intestinal segmented filamentous bacteria, “SFBs” (14–16). Although Leidy labeled the refractile inclusions he observed “spores,” the filaments were first suggested to be bacteria by Duboscq and Grassé (9) in their description of Coleonema pruvoti “schizophytes” (old name for bacteria) from a Loyalty Island termite (Kalotermes sp.). No generic arthromitid name was validated in the Approved Lists of Bacterial Names (17).
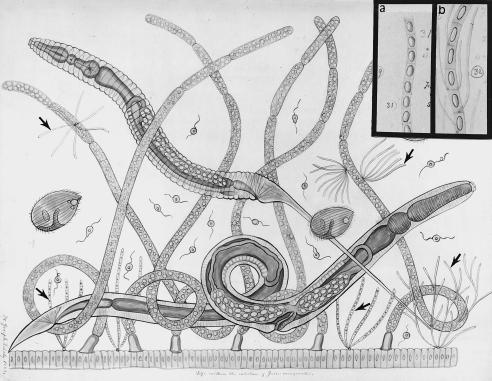
Figure 1.
Joseph Leidy’s teaching chart no. 60 courtesy of Philadelphia Academy of Natural Sciences. Intestinal microbial community of Julus marginatus, a juliform millipede (diplopod), Phylum Mandibulata (Uniramia). Arthromitus is attached both to the gut and to trichomycete fungal hyphae (arrows). Insets are from Leidy drawing no. 3 of “Termes,” his figures 31 (a) and 32 (b) (4).
We compared intestinal filamentous bacteria in soil-dwellers (termite, beetle, roach, millipede, and terrestrial crustaceans) with those cultured from boiled whole guts of the same arthropods. Our work extends Leidy’s claim that these filaments grow in the hindguts of “healthy animals, as a natural condition” (5). Recent work from our laboratory (18) led us to hypothesize that, in nature, spore-forming aerobes in soil exist as one stage of the arthromitids. We further document that certain soil bacilli are arthropod gut symbionts.
METHODS AND MATERIALS
Microscopy and Cultivation.
The identification, geographical source, and maintenance conditions of the arthropods that harbor the filamentous bacteria are in Table 1. Intestinal contents were assessed by microscopy and photographed. Nestmates were surface cleaned by drowning the submerged arthropods in 95% ethanol. Intestines were removed under a Nikon dissecting microscope by using two sets of flamed forceps. The adult arthropod was held behind the head with coarse forceps while gentle pressure was applied to the intestine at the anal end with fine forceps. The termite intestines, first placed in insect Ringer’s or Trager’s solution (22), were punctured to allow the escape of the symbionts. Intestinal contents of other arthropods were removed in similar manner or by squeezing, and whole intestines were macerated with mortar and pestle for inspection of the microbial community with phase-contrast, darkfield, or Nomarski differential interference contrast (DIC) optics. Videographs of live microbes were taken with a Sony charge-coupled device (CCD)-Iris color video camera model DXC-107-A mounted on a microscope (Nikon Microphot, Fluorophot, or Optiphot) connected to a Sony ¾-inch VO-9600 videocassette recorder.
Table 1.
Sources of Arthromitus-like bacteria in nature
| KINGDOM* | |
|---|---|
| Phylum | |
| Order | |
| Genus species (common name) | Provider and identification |
| ANIMALIA | |
| Mandibulata (Uniramia, arthropods) | |
| Blattaria | |
| Blaberus giganteus (giant cockroach)† | J. Kunkel, UMA; R. Redfield, UBC |
| Cryptocercus punctulatus (wood-eating cockroach)‡ | Mountain Lake, VA; Florida |
| Gromphodorhina portentosa (hissing roach)† (20) | D. Winans, NESC, CBC |
| Isoptera | |
| Kalotermitids (dry wood-eating termites) | |
| Calotermes sp.§ | R. Scheffrahn, South Florida |
| Cryptotermes brevis† | R. Scheffrahn |
| Cryptotermes cavifrons† | M. Deyrup, Central Florida |
| Glyptotermes sp.† | Fixed slide preparations of H. Kirby (32) |
| Kalotermes approximatus‡ | Fig. 2e |
| Kalotermes flavicollis†‡ | M. Gajú, Andulusia, Spain |
| Kalotermes praecox‡ | |
| Kalotermes schwartzi,‡Incisitermes minor§ | Fig. 2g |
| Mastotermes darwiniensis‡ | |
| Pterotermes occidentis‡ | Fig. 2d |
| Hodotermitids (damp wood-eating termites) | |
| Zootermopsis angusticollis‡ | |
| Zootermopsis nevadensis‡ | |
| Rhinotermitids (subterranean termites) | |
| Coptotermes formosanus‡ | |
| Reticulitermes flavipes‡ | Airlie Conference Center, Airlie, VA; Fig. 2f |
| Reticulitermes hesperus‡ | |
| Reticulitermes tibialis‡ | Fig. 3 |
| Coleoptera | |
| Polydesmus (unicorn wood-beetle)‡ | |
| Odontotaenius disjunctus (patent-leather or bess beetle)† (= Passalus cornutus, Popilius disjunctus) |
NESC, CBC |
| Myriapoda | |
| Graphidostreptus? Fam. Julidae (Madagascar fire millipede)† | NESC, D. Winans |
| Spirobolus (millipede)‡ | |
| Julus marginatus (millipede)‡ | J. Leidy, 1849 (5) |
| Crustacea (isopods) | |
| Armadillidium sp. (pillbug)† | MBL |
| Porcellio scaber (sow bug)† | NESC (18), D. Winans; Fig. 2b |
| Chordata | |
| Vertebrates: amphibians, ducks | (13) |
| chickens, rats, mice, humans‡ | (12, 15) |
| PROTOCTISTA | |
| Gloeocystis major (green algal gel mass)† | Cider Mill Pond, Amherst, MA |
Abbreviations: MBL, Marine Biological Laboratory, Woods Hole, MA; NESC, New England Science Center, Worcester, MA; UMA, University of Massachusetts at Amherst; CBC, Carolina Biological Co.; UBC, University of British Columbia, Vancouver. Sow bugs were taken from the NESC exhibit housing Leontopithecus rosalia (golden lion tamarin; ref. 18). Several hundred animals were maintained on damp soil, wood chips, and dry leaves in plastic boxes. These and the roaches and termites were also housed at room temperature in boxes, well lit and covered with fine nylon mesh to prevent the entrance of fungal spores. The animal colonies were moistened when necessary by misting with distilled H2O; all but the termites were fed commercial dry dog food pellets dampened with distilled H2O. The termites were supplied with small pieces of wood of the same sort from which they were collected.
*Classification follows ref. 19.
This paper.
See ref. 8 for details and earlier literature. If not otherwise mentioned, termites were collected and identified by the authors.
Calotermes [= Incisitermes (21) of the French literature] is spelled Kalotermes in English or German.
Macerated intestines of 1–10 animals, depending on size, were suspended in fresh salt solution in test tubes and placed for 8–12 min in a boiling water bath to kill non-spore-forming microbes. Upon cooling to lower than 60°C, samples were inoculated onto plates containing a nutrient agar medium (0.5% Difco yeast extract/0.5% Difco Bacto peptone/0.01% sodium acetate) at pH 7.5 ± 0.1 and incubated at room temperature, 31°C, or 38°C. To test anoxic growth some were incubated in Brewer jars to which gas packs were added (N2, CO2) or in a Coy glove box (model XOXX) with 7% H2/13% CO2/80% N2. Solutions of pH extremes were made up with hydrochloric acid and sodium hydroxide in distilled H2O. Entire guts or bacteria collected from culture plates were fixed in 1.5–2.5% glutaraldehyde, stained with osmium tetroxide, and thin-sectioned for transmission electron microscopy (TEM) (22).
16S rDNA Sequence Analysis.
Genomic DNA extraction, PCR-mediated amplification of the 16S rRNA gene (rDNA), and purification of PCR products were carried out as described by Rainey et al. (23). Purified PCR products were sequenced by using the Taq Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems) as directed in the manufacturer’s protocol. Sequence reaction products were electrophoresed in the Applied Biosystems 373A DNA Sequencer. The complete 16S rDNA sequences were aligned by the GenBank blast program against major low G+C bacilli: Bacillus anthracis, Bacillus cereus, Bacillus subtilis, and Bacillus mycoides. Sequences from the RDP (Ribosomal Database Project) were used for comparison.
RESULTS AND DISCUSSION
Long attached filaments of spore-forming Arthromitus sp. were conspicuous components of the intestinal community in all arthropods studied here except Porcellio scaber. Although long attached filaments were noted in dark-reared sow bugs, short filaments, uni- and diplocells were always more abundant in the typically dry intestines of sow bugs (18).
Sow bug, roach, and termite isolate phenotypes (ATCC no. 49589 “Arthromitus chasei”), (8) including the G+C mol% (24), lead us to conclude that arthromitids are stages of bacteria closely related to B. cereus (1, 25, 26).
The 16S rDNA sequence (1432 bp between Escherichia coli positions 31 and 1452) determined for strain JJ#1 (accession no. Y15466) (18) showed it to belong to the B. cereus/B. anthracis 16S rDNA phylogenetic cluster when compared with B. cereus sequences (accession nos. X55060 or D16266, Document ID BC16SRR1), B. anthracis (X55059), B. mycoides (X55061), and Bacillus thuringiensis (X55062). The bacterial isolate from Odontotaenius disjunctus exactly matched Leidy’s morphological description (6); its rDNA proved to be ≈99% equivalent to B. cereus 4 from the RDP 16S rDNA. Because strains of B. cereus and B. anthracis are indistinguishable by 16S rDNA sequence analysis (27), identification is based on physiology. Phenotypic data confirmed arthromitid JJ#1 as a strain of B. cereus (18).
On repeated occasions between 1988 and August 1997, isolations of spore-forming filaments were consistently successful from these animals: the bess or patent leather beetle Odontotaenius disjunctus (= Passalus cornutus), the death’s head or giant cockroach Blaberus giganteus, the hissing roach Gromphodorhina portentosa, the common sow bug Porcellio scaber, the pill bug Armadillidium sp., and the five drywood and subterranean termite species Cryptotermes brevis, C. cavifrons, Kalotermes flavicollis, K. schwartzi, and Reticulitermes flavipes. Our 100% success for new isolations led us to suspect the other animals listed (Table 1) would yield similar colonies were we to test them. Comparative morphology of the new isolates is depicted in Fig. 2.
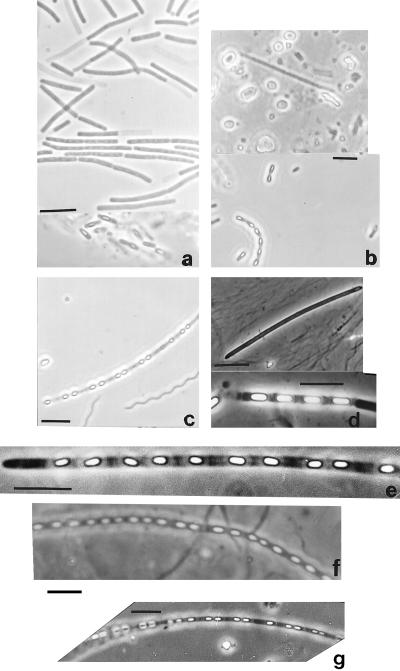
Figure 2.
Four Arthromitus types from roaches (a), sow bugs (b), and termites (c–g) compared. (a) Blaberus giganteus cultures. (b) Porcellio scaber (Upper, animal 44 days in the dark; Lower, culture). (c) Reticulitermes sp. intestine. (d) Pterotermes occidentis intestine. (e) Kalotermes approximatus intestine. (f) Reticulitermes flavipes intestine. (g) Incisitermes minor intestine. (Light micrographs of live bacteria; bars = 5 μm.)
In culture with growth and transfer, isolates grow as shorter filaments and diplocells but are found mainly as unicells. The extent of filament formation and the tendency to be motile decline as cultures mature and sporulate. Conditions favoring sporulation and retention of filament morphology (up to 180 cell lengths) were discerned for the sow bug strain JJ#1 (18). Sporulation was accelerated by light incubation but filaments were longer and more abundant in dark-incubated cultures of sow bug Arthromitus (Table 2). The presence in the medium of boiled gut material retarded sporulation relative to controls. Although cultures grown with boiled gut material favored fewer spores, this material enhanced the appearance of more filaments of greater length. Sporulation was inhibited for more than 3 months by anoxia in glove box cultures whether desiccated or amply wetted. Yet, upon removal from the glove box, spore formation always proceeded within 24 hr of exposure to any concentration of oxygen. Relative frequency of morphotype apparently depends on environmental contingencies.
Table 2.
Acceleration of development of Arthromitus-Bacillus* by light
| Time after inoculation | Colony morphology and light microscopic appearance
|
|
|---|---|---|
| Light-grown | Dark-grown | |
| Day 1 | ||
| 0–3 hr | cd 0.5–0.8 cm; unicells, diplocells | No apparent growth |
| 3–6 hr | cd 0.8–1.5 cm; diplocells, short filaments | cd <0.5 cm; unicells |
| 6–9 hr | Short and long filaments | cd 0.5 cm; unicells, diplocells |
| 9–24 hr | Filaments, some endospores | Short filaments |
| Day 2 | Unicells, diplocells, endospores, some mature spores | Unicells, diplocells, short and long filaments (5–180 cell lengths) |
| Day 3 | cd 2–3 cm; unicells, sporulated diplocells, no filaments | Short and long filaments, no spores; cd 0.5–1.0 cm |
| Day 4 | Mature spores | Long and short filaments, few spores |
Abbreviation: cd, colony diameter. “Endospores” if parent cell visible; “spores” refers to mature spores, no parental cells.
*Strain JJ#1 from Porcellio scaber (18).
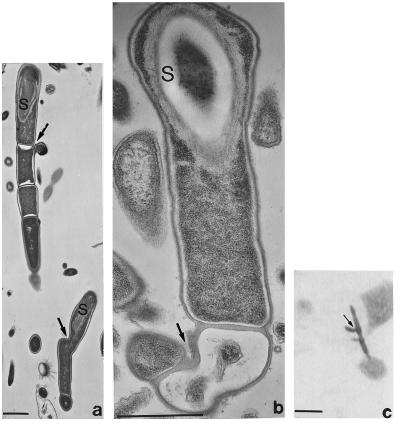
In insect guts a probable developmental cycle of these filamentous bacteria has been reconstructed from over 200 electron micrographs. Thin sections of arthromitids from reticultermitid intestines show that the endospores form attachment fibers and retain them on release from the parent cell (Figs. 3–5). The possibility that these attachment fibers are a form of pili or flagella was eliminated by comparison of our micrographs with D. DeRosier’s high-resolution computer-enhanced images. After germination into bacilli, the motile and filament growth stages ensue. New endospores form, generally from the distal unattached end of the filaments. We documented by videomicroscopy of live bacteria the swimming and “docking” of arthromitids at intestinal sites consistent with electron micrographs in Pterotermes occidentis, Cryptotermes brevis, and Blaberus giganteus (Fig. 6). Amorphous material attaches spore-forming filaments to the chitinous intestine of sow bugs; endospore fibers are lacking in this arthromitid.
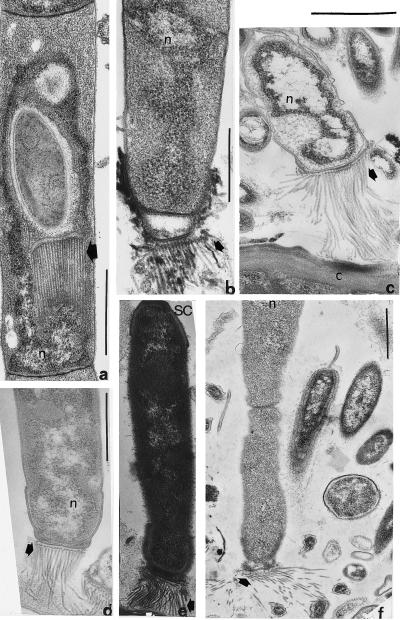
Figure 3.
Endospore development in an Arthromitus sp. from Reticulitermes tibialis; n = genetic material. (a) Early stage in spore formation. (b) Maturing spore and enigmatic tubular body (arrows) apposed to inner surface of membrane. (c) Growing filament still covered with spore coat (SC) material. The possibility that these tubular membrane connectors (arrowheads) are composed of filament temperature-sensitive Z protein (Ftsz) needs investigation. (d) Endospore in which nucleoid is apposed to the inner surface of the proteinaceous thickening of the membrane (arrowheads), transverse section comparable to b and c. (Bar = 1 μm.)
Figure 5.
Spore attachment fibers; n = genetic material. (a) Reticulitermes tibialis, attachment fibers produced inside spore parent cell (arrow). (b) R. tibialis, attachment fibers (arrow) emerge from an extra wall layer, here pulled away. (c) Reticulitermes hesperus, attachment fibers emerge from the thickened wall layer outside the membrane and attach to the chitinous gut wall, C. (d) R. hesperus, attachment fibers, arrow, connect spores to gut inhabitants. (e) R. tibialis, attachment fibers at arrow, spore coat not yet shed. (f) R. hesperus, the correct attachment of spore fibers (arrow) to chitin may precede the establishment of permanent holdfasts as in Fig. 6. (Bars = 1 μm.)
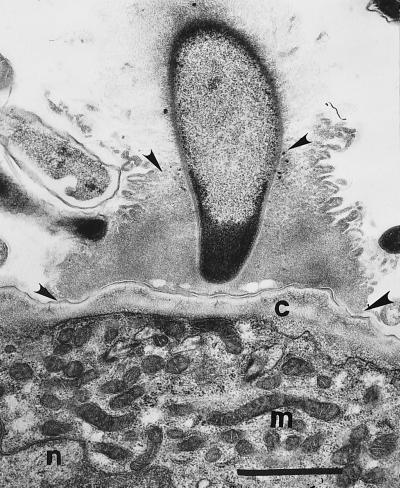
Figure 6.
Arthromitus holdfast thickening (arrowheads) at the insect’s chitinous wall, c; n = nucleus and m = mitochondria of termite gut cells. (Bar = 1 μm.)
We have documented Arthromitus inside animals and other nutrient-rich moist habitats, including vertebrate intestines. Promptly at dawn, active and immediate ingestion of Leontopithecus rosalia (golden lion tamarin monkey) feces by crowds of sow bugs was observed (Monkey House, New England Science Center, Worcester, MA.). That sow bugs walked over the surfaces of the tamarin food and that colonies indistinguishable from those of sow bug Arthromitus strain JJ#1 were isolated twice from sterile, boiled anal-swab cultures of a golden lion tamarin indicated the route by which spores are probably reingested. Filamentous spore-formers morphologically identical (central, oval, and nondistending of the parent cell) to some bacteria in roaches were found growing in Gloeocystis gel masses in Cider Mill Pond, Amherst, MA, where duck and other fecal material abounds. Isolated from cow dung and decaying plant material in ponds, “Lineola longa” (28) was later named “Bacillus macroides” (29, 30). These spore-forming filaments conform to Bacillus sphaericus (25). We suspect such “Lineola” observed in dung and algal gel masses is Arthromitus derived from cattle, sheep, duck, or other vertebrate feces.
These data suggest a tentative life history. Arthropods ingest feces or soil particles containing spores or cells that grow in the intestine. Newly germinated motile cells form short swimming filaments of 5–10 cell lengths. The cells reproduce rapidly to form filaments that attach to the epithelium, to protists, to trichomycete fungi, or to other intestinal substrata that prevent defecation by peristalsis (Fig. 1). The less-than-atmospheric concentration of oxygen in the arthropod intestine (31) reduces the rate of sporulation. Cells, including those without endospores, are released from the filaments and defecated to the soil by the arthropods. Increasing oxygen concentration, increased exposure to light, nutrient depletion, and desiccation enhance formation of single cells and spores. Apparently Bacillus spores are dormant stages poised to grow in decaying vegetation or nutrient-rich laboratory plates, or symbiotrophically in arthropod and mammal intestines. We predict the abundance and variation in the genus Bacillus will reflect arthropod and mammal diversity in and above the soil at any given location.
The arthromitids we isolated into axenic culture from arthropod intestines are symbiotrophs and not just “soil bacilli.” Because microbiologists seldom study motility of bacteria in soil-dwelling arthropods in situ, differentiation of spore attachment fibers and filamentous growth tend to be overlooked. Arthromitid genera were not officially reinstated by systematic bacteriologists due in part to the belief that they cannot be grown in culture (17). Arthromitids growing on rich-media plates are easily dismissed as laboratory contaminants.
Three growth points per bacillus cell leading to branched Arthromitus-like filaments were observed in electron micrographs of Reticulitermes hesperus intestine (Fig. 7 a and b) (8). Although they were not mentioned by Harold Kirby (as quoted in ref. 32), we saw these Arthromitus-like branched filaments attached by fibers to protists in his Glyptotermes sp. preparations (Fig. 7c). Study of calonymphid protists** in Kirby’s original preparations (≈1945) of the Glyptotermes hindgut revealed branched, spore-forming prokaryotes. Even if the branched arthromitids “tree” with Bacillus or Clostridium (33), they deserve their own new genus on ecological and morphological grounds.
Figure 7.
Branched Arthromitus-like spore formers. (a and b) Transmission electron micrographs; bars = 1 μm. (a) Arrows at branches, third growth region on cell. (b) Early development of the branch point (arrow; S = spore). (c) Branched bacterial filament on unidentified protist; light micrograph from Kirby’s stained slide of Glyptotermes sp. intestine, bar = 5 μm.
The relation of arthropod spore-formers to the Clostridium-like SFBs from mammals is unknown. Our isolates, which fail to grow under anoxic conditions, are not Clostridium. The SFBs of rodent intestines, as yet unculturable, are probably anaerobes closely related to Clostridium (12). Clostridium-like symbiotic SFBs in mammals increase the host immune response to pathogens (14, 34), and actin accumulates at their attachment sites in mouse intestinal epithelia (35). Like these vertebrate SFBs (12, 36), certain arthromitids in arthropods may be host specific. A single termite may contain more than four distinct arthromitid morphotypes. In situ DNA⋅DNA hybridization, immunostaining at the electron microscopic level, and sequence analysis of the 16S rRNA gene should be correlated with cell morphology to accurately define the taxonomic relationships of these bacilli (37). Other cultivated termite hindgut bacteria include the clearly different strictly anaerobic Gram-negative endospore-forming Sporomusa (38) and another short-filamentous anaerobic acetogen, Acetonema longum (39).
Leidy’s (5, 6) and later reports show that filamentous spore-forming bacteria are normal inhabitants of animal intestines, e.g., plasmid-bearing or aplasmidic Bacillus anthracis may be Arthromitus from sheep. Epibiotic methanogenic bacteria abundantly attach to arthromitids (40) (filamentous spore-forming prokaryotes) in the North American termite Reticulitermes flavipes in patterns described by Leidy (ref. 6, his figures 33 and 34). Of course, the identity of any arthropod Arthromitus with any Bacillus strain requires specific determination, yet in culture from arthropods, these kinds of bacteria (whether or not they retain their filamentous habit) precisely fit the description of the main cluster of the genus Bacillus to which B. cereus belongs. We conclude that Arthromitus is the normal intestinal stage of Bacillus cereus and its close relatives. Knowledge of the differentiation stages in the natural history of Arthromitus/Bacillus will permit more comprehensible interpretations of pathogenicity, gene expression, gene regulation, and prodigious insecticidal protein production. Whether or not the genus Arthromitus is formally resurrected for mammalian SFBs (34) or for Leidy’s plants (5), the symbiotic stages of these bacteria in animal intestines warrant far more scrutiny.
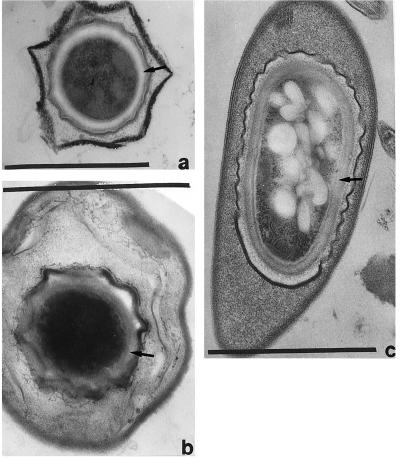
Figure 4.
Mature spores of three putative Arthromitus from intestinal thin sections of termites: Reticulitermes tibialis (a), Cryptotermes cavifrons (b), and Zootermopsis angusticollis (c). Spore coat at arrows. (Bars = 1 μm.)
Acknowledgments
The exquisite electron micrographs are by David Chase (1935–1986), to whose memory this work is dedicated. We are grateful to S. Ashcraft, J. B. Ashen, A. Becerra, T. Beveridge, B. Blunt-Harris, L. Brynes, D. J. DeRosier, M. Deyrup, S. Deyrup, M. Dolan, G. J. Domingue, S. Erlandsen, D. Forrest, S. Goodwin, A. G. Gristina, R. Guerrero, T. Guillemette, A. Haselton, J. Kunkel, S. C. Mohr, K. H. Nealson, L. Olendzenski, R. Rudner, K. Rusterholtz, R. Scheffrahn, P. Setlow, J. Snel, A. L. Sonenshein, T. H. Teal, C. B. Thorne, T. Wilkins, D. Winans, B. Wise, M. Yamin, and the New England Science Center for help with the work. We are especially grateful to G. E. Fox and J. Siefert, University of Houston, for the 16S rRNA sequence of the bess beetle and confirmation of the sequence in the sow bug isolate JJ#1 and D. Reppard for manuscript preparation. We thank the American Museum of Natural History and the Academy of Natural Sciences, Philadelphia, respectively, for use of Kirby’s slides and Leidy’s figures. NASA Space Sciences, the Richard Lounsbery Foundation, the University of Massachusetts Amherst Graduate School (Department of Geosciences, Graduate Program in Organismic and Evolutionary Biology), the Margaret E. and Howard E. Bigelow Awards of the Graduate Program in Organismic and Evolutionary Biology, and the Dean of the College of Natural Science and Mathematics provided financial support.
ABBREVIATIONS
- SFBs
segmented filamentous bacteria
- rDNA
rRNA gene
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y15466).
Dolan, M., Second European Congress of Protistology, July 24–28, 1995, Clermont-Ferrand, France, p. 42 (abstr.).
References
- 1.Slepecky R A, Hemphill H E. In: The Prokaryotes. 2nd Ed. Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. Vol. 2. New York: Springer; 1992. pp. 1663–1696. [Google Scholar]
- 2.Farrer W E, Reboli A C. In: The Prokaryotes. 2nd Ed. Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. Vol. 2. New York: Springer; 1992. pp. 1746–1768. [Google Scholar]
- 3.Agaisse H, Lereclus D. J Bacteriol. 1995;177:6027–6032. doi: 10.1128/jb.177.21.6027-6032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and Other Gram-positive Bacteria: Biochemistry, Physiology and Molecular Genetics. Washington, DC: Am. Soc. Microbiol.; 1993. [Google Scholar]
- 5.Leidy J. Proc Acad Nat Sci Philadelphia. 1849;4:225–233. [Google Scholar]
- 6.Leidy J. J Acad Nat Sci Philadelphia. 1881;8:425–447. [Google Scholar]
- 7.Glassman S, Bolt E A, Jr, Spamer E E. Proc Acad Nat Sci Philadelphia. 1993;144:1–19. [Google Scholar]
- 8.Margulis L, Olendzenski L, Afzelius B A. Symbiosis. 1990;8:95–116. , and ATCC strain description (1990) 8, 285. [PubMed] [Google Scholar]
- 9.Duboscq O, Grassé P. Arch Zool Expér Gén. 1929;68:8–15. [Google Scholar]
- 10.Duboscq O, Grassé P. Arch Zool Expér Gén. 1930;70:28. [Google Scholar]
- 11.Grassé P-P. Ann Parasitol Hum Comp. 1925;3:343–348. [Google Scholar]
- 12.Snel J, Heinen P P, Blok H J, Carman R J, Duncan A J, Allen P C, Collins M D. Int J Syst Bacteriol. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- 13.Chase D G, Erlandsen S L. J Bacteriol. 1976;127:572–583. doi: 10.1128/jb.127.1.572-583.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klaasen H L B M, Koopman J P, Poelma F G J, Beynen A C. FEMS Microbiol Rev. 1992;88:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 15.Klaasen H L B M, Koopman J P, Van Den Brink M E, Bakker M H, Poelma F G J, Beynen A C. Lab Anim. 1993;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- 16.Klaasen H L B M, Van der Heijden P J, Stok W, Poelma F G J, Koopman J P, Van den Brink M E, Bakker M H, Eling W M C, Beynen A C. Infect Immun. 1993;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skerman V B D, McGowan V, Sneath P H A. Int J Syst Bacteriol. 1980;30:225–420. [Google Scholar]
- 18.Jorgensen J, Dolan S, Haselton A, Kolchinsky R. Can J Microbiol. 1997;43:129–135. [Google Scholar]
- 19.Margulis L, Schwartz K V. Five Kingdoms. 3rd. Ed. New York: Freeman; 1998. [Google Scholar]
- 20.Dailey P J, Graves R C. Ann Ent Soc Amer. 1976;69:589–602. [Google Scholar]
- 21.Krishna K. Bull Amer Mus Nat Hist. 1961;122:303–408. [Google Scholar]
- 22.To L P, Margulis L, Nutting W L, Chase D. BioSystems. 1980;13:109–137. doi: 10.1016/0303-2647(80)90007-6. [DOI] [PubMed] [Google Scholar]
- 23.Rainey F A, Ward-Rainey N, Kroppenstedt R M, Stackebrandt E. Int J Syst Bacteriol. 1996;46:1088–1092. doi: 10.1099/00207713-46-4-1088. [DOI] [PubMed] [Google Scholar]
- 24.Hinkle G. Ph.D. thesis. Boston: Boston Univ.; 1992. [Google Scholar]
- 25.Sneath P H A. In: Bergey’s Manual of Systematic Bacteriology. Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Vol. 2. Baltimore: Williams & Wilkins; 1986. pp. 1104–1139. [Google Scholar]
- 26.Gordon R E, Haynes W C, Pang C H N. The Genus Bacillus. Washington, DC: U.S. Dept. Agric.; 1973. , Handbook no. 427. [Google Scholar]
- 27.Ash C, Farrow J A E, Dorsch M, Stackebrandt E, Collins M D. Int J Syst Bacteriol. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- 28.Pringsheim E G. J Gen Microbiol. 1950;4:198–209. doi: 10.1099/00221287-4-2-198. [DOI] [PubMed] [Google Scholar]
- 29.Bennett J F, Canale-Parola E. Arch Mikrobiol. 1965;52:197–205. [Google Scholar]
- 30.Buchanan R E, Gibbons N E, editors. Bergey’s Manual of Determinative Bacteriology. 8th Ed. Baltimore: Williams & Wilkins; 1974. [Google Scholar]
- 31.Brune A, Emerson D, Breznak J A. Appl Environ Microbiol. 1995;61:2681–2687. doi: 10.1128/aem.61.7.2681-2687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirby, H. (c. 1945) annotated by Margulis, L. (1994) Symbiosis 16, 7–63. [PubMed]
- 33.Snel J, Blok H J, Kengen H M P, Ludwig W, Poelma F G J, Koopman J P, Akkermans A D L. Syst Appl Microbiol. 1994;17:172–179. [Google Scholar]
- 34.Snel J. Ph.D. thesis. Nijmegen, The Netherlands: Univ. of Nijmegen; 1997. [Google Scholar]
- 35.Jepson M A, Clark M A, Simmons N L, Hirst B H. Infect Immun. 1993;61:4001–4004. doi: 10.1128/iai.61.9.4001-4004.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannock G W, Miller J R, Savage D C. Appl Environ Microbiol. 1984;47:441–442. doi: 10.1128/aem.47.2.441-442.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox G E, Wisotzkey J D, Jurtshuk P., Jr Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 38.Breznak J A, Switzer J M, Seitz H-J. Arch Microbiol. 1988;150:282–288. [Google Scholar]
- 39.Kane M D, Breznak J A. Arch Microbiol. 1991;156:91–98. doi: 10.1007/BF00290979. [DOI] [PubMed] [Google Scholar]
- 40.Leadbetter J R, Breznak J A. Appl Environ Microbiol. 1996;62:3620–3631. doi: 10.1128/aem.62.10.3620-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]