Abstract
Calcineurin is a conserved Ca2+-calmodulin-activated, serine/threonine-specific protein phosphatase that regulates a variety of physiological processes, e.g., cell cycle progression, polarized growth, and adaptation to salt and alkaline pH stresses. In the pathogenic yeast Cryptococcus neoformans, calcineurin is also essential for growth at 37°C and virulence. To investigate whether calcineurin plays a role in the virulence of Candida albicans, the major fungal pathogen of humans, we constructed C. albicans mutants in which both alleles of the CMP1 gene, encoding the calcineurin catalytic subunit, were deleted. The C. albicans Δcmp1 mutants displayed hypersensitivity to elevated Na+, Li+, and Mn2+ concentrations and to alkaline pH, phenotypes that have been described after calcineurin inactivation in the related yeast Saccharomyces cerevisiae. Unlike S. cerevisiae calcineurin mutants, which exhibit reduced susceptibility to high Ca2+ concentrations, growth of C. albicans was inhibited in the presence of 300 mM CaCl2 after the deletion of CMP1, demonstrating that there are also differences in calcineurin-mediated cellular responses between these two yeast species. In contrast to C. neoformans, inactivation of calcineurin did not cause temperature sensitivity in C. albicans. In addition, hyphal growth, an important virulence attribute of C. albicans, was not impaired in the Δcmp1 mutants under a variety of inducing conditions. Nevertheless, the virulence of the mutants was strongly attenuated in a mouse model of systemic candidiasis, demonstrating that calcineurin signaling is essential for virulence in C. albicans.
Calcineurin is a widely conserved Ca2+-calmodulin-activated, serine/threonine-specific protein phosphatase that is required for signal transduction, e.g., during the activation of T cells (45). Calcineurin signaling is inhibited by the immunosuppressive drugs cyclosporine A (CsA) and FK506, which diffuse into the cells and bind to the immunophilins cyclophilin and FKBP12, respectively. The resulting cyclophilin-CsA and FKBP12-FK506 complexes then inhibit calcineurin (28). CsA and FK506 also exhibit antifungal activity by inhibiting calcineurin through the same mechanism as in T cells (3, 5).
Calcineurin consists of a catalytic A subunit and a Ca2+-binding regulatory B subunit, both of which are required for enzymatic function. In the yeast Saccharomyces cerevisiae, the catalytic subunit is encoded by the two redundant genes CNA1 (CMP1) and CNA2 (CMP2), and the regulatory subunit is encoded by the CNB1 gene (10, 11, 23, 29). Mutant analyses have demonstrated that calcineurin is not essential for viability of wild-type cells, but it is required for recovery from pheromone-induced growth arrest, cell wall biosynthesis, and adaptation to high salt concentrations and alkaline conditions (10, 28, 34, 36, 40, 41). These responses are mediated by the calcineurin-dependent dephosphorylation and nuclear localization of the transcription factor Crz1p/Tcn1p, which regulates the expression of genes involved in cell wall biosynthesis and cation homeostasis (33, 35, 48, 49).
In the pathogenic yeast Cryptococcus neoformans, calcineurin is essential for growth at 37°C but not at ambient temperature (43). Consequently, C. neoformans cna1 and cnb1 mutants, in which the gene encoding the catalytic or regulatory subunit of calcineurin is inactivated, are avirulent in animal models of cryptococcosis (8, 16, 43). In addition, calcineurin is also required for hyphal elongation during mating and haploid fruiting in C. neoformans (6), similar to its role in the filamentous fungus Neurospora crassa, in which calcineurin is essential for hyphal growth and morphology (44).
In contrast to C. neoformans, which is found in the environment and accidentally infects humans by inhalation, the opportunistic fungal pathogen Candida albicans is a member of the microflora in healthy humans. Therefore, growth at 37°C is intrinsic to the normal lifestyle of C. albicans. In immunocompromised patients, C. albicans can develop from a commensal into an infectious organism, and a variety of virulence-associated characteristics contribute to its pathogenic potential (42). C. albicans can switch between yeast and filamentous growth forms, and the ability to form hyphae during an infection is believed to be an important virulence factor (30).
Because of its essentiality at body temperature and its role in hyphal formation and virulence in C. neoformans, we were interested in whether calcineurin plays a similar role in C. albicans. The C. albicans genome sequence contains a single gene, CMP1, encoding the calcineurin catalytic subunit (http://www-sequence.stanford.edu/group/candida/). To investigate the role of calcineurin signaling in this pathogenic yeast, we constructed cmp1 null mutants and analyzed the effect of CMP1 deletion on the growth, morphogenesis, and virulence of C. albicans.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are listed in Table 1. The strains were maintained on SD agar plates (6.7 g of yeast nitrogen base without amino acids [YNB; Bio 101, Vista, Calif.], 20 g of glucose, 0.77 g of complete supplement medium without uracil [CSM-URA; Bio 101], and 15 g of agar per liter). The strains were routinely grown in YPD liquid medium (20 g of peptone, 10 g of yeast extract, 20 g of glucose per liter) at 30°C. To support the growth of uridine-auxotrophic strains, 100 μg of uridine ml−1 was added to the media. ura3-negative derivatives of strains containing the URA3 flipper were isolated after induction of FLP expression by overnight growth in YCB-BSA (23.4 g of yeast carbon base, 2 g of yeast extract, 4 g of bovine serum albumin per liter, pH 4.0) containing 100 μg of uridine ml−1 and screening for smaller colonies after growth of the cells on SD agar plates containing 10 μg of uridine ml−1.
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Genotypea | Reference |
|---|---|---|---|
| SC5314 | Wild-type strain | 18 | |
| CAI4 | SC5314 | Δura3::imm434/Δura3::imm434 | 15 |
| CMP1M1A | CAI4 | CMP1/Δcmp1::URA3-FLIPb | This study |
| CMP1M1B | CAI4 | CMP1/Δcmp1::URA3-FLIP | This study |
| CMP1M2A | CMP1M1A | CMP1/Δcmp1::FRT | This study |
| CMP1M2B | CMP1M1B | CMP1/Δcmp1::FRT | This study |
| CMP1M3A | CMP1M2A | Δcmp1::URA3-FLIP/Δcmp1::FRT | This study |
| CMP1M3B | CMP1M2B | Δcmp1::URA3-FLIP/Δcmp1::FRT | This study |
| CMP1M4A | CMP1M3A | Δcmp1::FRT/Δcmp1::FRT | This study |
| CMP1M4B | CMP1M3B | Δcmp1::FRT/Δcmp1::FRT | This study |
| CMP1M5A | CMP1M4A | Δcmp1::FRT/Δcmp1::URA3 | This study |
| CMP1M5B | CMP1M4B | Δcmp1::FRT/Δcmp1::URA3 | This study |
| CMP1MK1A | CMP1M4A | Δcmp1::FRT/CMP1-URA3 | This study |
| CMP1MK1B | CMP1M4B | Δcmp1::FRT/CMP1-URA3 | This study |
Apart from the indicated features, all strains have the genotype of their parent.
URA3-FLIP, URA3 flipper cassette.
Plasmid construction.
A CMP1 deletion construct was generated in the following way. A KpnI-XhoI fragment containing CMP1 upstream sequences from positions −1058 to +77 with respect to the start codon was amplified from genomic DNA of C. albicans strain CAI4 with the primer pair CMP1 (5′-GCTAGACGTGAggTACCAACGGGTGG-3′) and CMP2 (5′-CAGTTCATTACCTcgaGTGGTTCGTCTATG-3′) (the lowercase letters represent nucleotide exchanges introduced to create the underlined KpnI and XhoI restriction sites). A SacII-SacI CMP1 downstream fragment from positions +1594 to +2316 was amplified with the primers CMP3 (5′-AATTAccgCGGGGATCATTACCCAAAGGTTC-3′) and CMP4 (5′-TTCATgaGCTCATTAATATATTGAATCAATAAG-3′). The CMP1 upstream and downstream fragments were cloned on both sides of the URA3 flipper cassette of plasmid pSFU1 (39), resulting in pCMP1M2, in which the CMP1 coding region from positions +78 to +1593 (245 bp before the stop codon) is replaced by the URA3 flipper (see Fig. 2A).
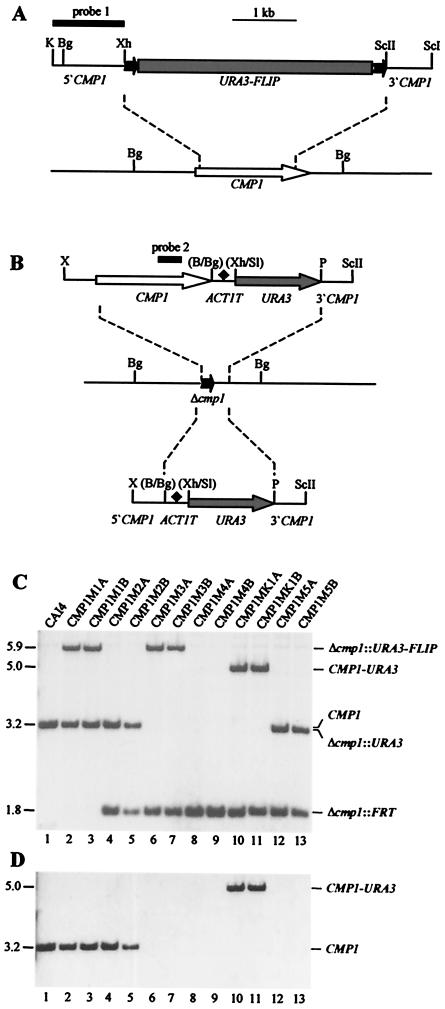
FIG. 2.
Construction of C. albicans cmp1 deletion mutants and complemented strains. (A) Structure of the deletion cassette from plasmid pCMP1M2 (top) and genomic structure of the CMP1 locus in the parent strain, CAI4 (bottom). The CMP1 coding region is represented by the open arrow, and the upstream and downstream sequences are represented by solid lines. Details of the URA3 flipper (shaded rectangle bordered by FRT sites [solid arrows]) have been presented elsewhere (39). The 34-bp FRT sites are not drawn to scale. The DNA fragment used for Southern hybridization analysis of the mutants is represented by the thick bar (probe 1). (B) Structures of the DNA fragments from pCMP1K1 (top) and pCMP1M4 (bottom), which were used for reintegration of an intact CMP1 copy (open arrow) or only the URA3 marker (shaded arrows), respectively, into one of the inactivated cmp1 alleles (middle). The ACT1 transcription termination sequence (ACT1T) is indicated by the solid diamond. Only relevant restriction sites are given. B, BamHI; Bg, BglII; K, KpnI; P, PstI; ScI, SacI; ScII, SacII; Sl, SalI; X, XbaI; Xh, XhoI. The restriction sites shown in parentheses were destroyed by the cloning procedure. The CMP1 internal fragment used for Southern hybridization analysis of the strains is indicated by the thick bar (probe 2). (C) Southern hybridization of BglII-digested genomic DNAs of the parent strain, CAI4, and mutant derivatives with the CMP1 probe 1. The sizes of the hybridizing fragments (in kilobases) are given on the left side of the blot, and their identities are indicated on the right. (D) Rehybridization of the same blot with a CMP1 internal fragment (probe 2).
For reintegration of an intact CMP1 copy into one of the inactivated CMP1 alleles of homozygous cmp1 deletion mutants, a PstI-SacII fragment containing CMP1 downstream sequences from positions +1825 to +2316 was first amplified with the primers CMP5 (5′-GAAGATTATCTgcAGTTAAACTTTC-3′) and CMP6 (5′-TGGTTCcgcgGCTCATTAATATATTGAATCAATAAG-3′) and cloned behind the URA3 selection marker of plasmid pGFP26 (38), from which the PstI site in the polylinker had been removed, resulting in pCMP1M3. A fragment containing the complete CMP1 open reading frame and upstream sequences was then amplified with the primers CMP1 and CMP8 (5′-CAGGGGCAAAAGgatccTTAACTTTGAGAT-3′) and digested at an XbaI site at position −528 and at the BamHI site introduced behind the stop codon. The CMP1 fragment was fused to a BglII-XhoI fragment containing the transcription termination sequence of the ACT1 gene, which was obtained by PCR with the primers ACT16 (5′-TTCTAAGAtctAAATTCTGGAAATCTGG-3′) and ACT21 (5′-atatactcgagGACATTTTATGATGGAATGAATGGG-3′), and cloned into the XbaI/SalI-digested pCMP1M3, resulting in pCMP1K1 (see Fig. 2B, top). The control construct pCMP1M4, which served for integration of only the URA3 selection marker, contains CMP1 upstream sequences amplified with the primers CMP1 and CMP7 (5′-CGTTGAACAGTggaTCCTGACATGATG-3′) and digested at the XbaI site at position −528 and at the BamHI site introduced at position +7 but not the CMP1 open reading frame in front of the ACT1 transcription termination sequence (see Fig. 2B, bottom).
C. albicans transformation.
C. albicans strains were transformed by electroporation (22) with the following gel-purified linear DNA fragments (see Fig. 2): the KpnI-SacI fragment from pCMP1M2 to delete the CMP1 wild-type alleles, the XbaI-SacII fragment from pCMP1K1 to reintroduce an intact CMP1 copy into one of the destroyed CMP1 alleles, and the XbaI-SacII fragment from pCMP1M4 to integrate only the URA3 marker but not the CMP1 open reading frame in the same way. Uridine prototrophic transformants were selected on SD agar plates without uridine.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. albicans was isolated as described previously (37). Ten micrograms of DNA was digested with BglII, separated on a 1% agarose gel, and, after ethidium bromide staining, transferred by vacuum blotting onto a nylon membrane and fixed by UV cross-linking. The gel-purified KpnI-XhoI CMP1 upstream fragment from pCMP1M2 and an internal SacI-HindIII fragment from the CMP1 coding region (positions +983 to +1365) were used as probes. Probe labeling, hybridization, washing, and signal detection were performed with the ECL labeling and detection kit provided by Amersham (Braunschweig, Germany) according to the instructions of the manufacturer.
Phenotypic assays.
For susceptibility tests, serial 10-fold dilutions of yeast suspensions were spotted on YPD plates and YPD plates containing 1.8 or 1.0 M NaCl, 10 mM LiCl, 0.3 M MnSO4, or 0.3 M CaCl2 and incubated at 30°C. In addition, YPD plates buffered with 155 mM Tris-HCl at pH 8.0 were used. For temperature sensitivity tests, growth on YPD plates was monitored at 30, 37, and 43°C.
Filamentous growth was tested at 37°C on agar plates containing Lee's medium (26), synthetic low-ammonium dextrose (SLAD) medium (19), or 10% fetal calf serum (FCS). To analyze hyphal growth in liquid media, cells from a YPD overnight culture were inoculated into Lee's medium buffered at pH 7.0 with 50 mM sodium citrate, Spider medium (27), SLAD medium, or RPMI 1640 medium with 10% FCS and incubated at 37°C.
Virulence tests.
Eight- to 10-week-old female BALB/c mice (Charles River Breeding Laboratories, Sulzfeld, Germany) (n = 5 to 8 for each group) were infected with 5 × 105 C. albicans cells by intravenous injection as described previously (25, 46). Survival curves were calculated according to the Kaplan-Meier method using the PRISM program (GraphPad Software, San Diego, Calif.) and compared using the log-rank test. A P value of <0.05 was considered significant. For microscopical examination, C. albicans cells were extracted from potassium hydroxide-solubilized kidneys and stained with calcofluor white (Sigma) as described previously (25, 46). For enumeration of CFU, kidneys were homogenized in phosphate-buffered saline, and replicates of serial dilutions were plated on rich solid media. The kidney fungal burden was calculated as CFU per milligram (wet weight) of kidney.
Sequence analysis.
The overall similarity of the CaCmp1 protein with the ScCna1 and ScCna2 proteins was determined using the program GAP from the Genetics Computer Group, Madison, Wis. Multiple alignment of the proteins was performed with the CLUSTALW program (http://www.ebi.ac.uk/clustalw/).
RESULTS
Identification of the CMP1 gene encoding the calcineurin catalytic subunit of C. albicans.
A BLAST search identified an open reading frame (6.3161) in the C. albicans genome sequence encoding a protein with high similarity to ScCna1p (69% similarity; 61% identity) and ScCna2p (69% similarity; 59% identity), the two calcineurin catalytic subunits of S. cerevisiae. The gene is located on contig 6-2213 and has been annotated as CMP1 by the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida/). The CMP1 open reading frame is 1,830 bp in length and encodes a protein of 609 amino acids with a theoretical molecular mass of 69.9 kDa. An alignment of CaCmp1p with ScCna1p and ScCna2p is shown in Fig. 1. The presence of the highly conserved calcineurin B and calmodulin binding domains in CaCmp1p indicates that calcineurin may also be regulated by these proteins in C. albicans. No other gene with comparable homology is present in the C. albicans genome sequence, suggesting that C. albicans possesses only one catalytic subunit for calcineurin, as has been found for C. neoformans (43).
FIG. 1.
Alignment of the calcineurin catalytic subunits Cna1p and Cna2p from S. cerevisiae (Sc) and Cmp1p from C. albicans (Ca). The amino acid sequences of the proteins are displayed in standard single-letter code using the BOXSHADE program (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html). Identical residues are on solid backgrounds, and similar residues are on shaded backgrounds. Amino acid positions are shown on the right. The highly conserved calcineurin B binding, calmodulin binding, and autoinhibitory domains are indicated by labeled bars, and their delineation is according to the work of Odom et al. (43).
Generation of C. albicans cmp1 deletion mutants.
To analyze the role of calcineurin in C. albicans, we inactivated the CMP1 gene using the URA3-flipping strategy, which is based on the repeated use of the URA3 selection marker for integrative transformation and its subsequent excision from the genome by the site-specific recombinase FLP (39). The ura3-negative strain CAI4 was transformed with a deletion cassette in which almost all of the CMP1 coding sequence had been replaced by the URA3 flipper (see Materials and Methods) (Fig. 2A). From two transformants in which the deletion cassette had been correctly inserted in one of the CMP1 alleles (strains CMP1M1A and CMP1M1B [Fig. 2C, lanes 2 and 3]), the URA3 flipper was excised by FLP-mediated recombination, resulting in the uridine-auxotrophic strains CMP1M2A and CMP1M2B (Fig. 2C, lanes 4 and 5). When these strains were transformed again with the same deletion cassette, integration was successfully targeted to the remaining wild-type CMP1 allele in several transformants of both parent strains, demonstrating that CMP1 is not an essential gene in C. albicans. The URA3 flipper was excised from the two independent homozygous cmp1 mutants CMP1M3A and CMP1M3B (Fig. 2C, lanes 6 and 7), generating the uridine-auxotrophic derivatives CMP1M4A and CMP1M4B (Fig. 2C, lanes 8 and 9). These strains served as hosts for reintroduction of a complete copy of the CMP1 gene or the URA3 selection marker alone into one of the inactivated Δcmp1 alleles (see Materials and Methods) (Fig. 2B). This resulted in the generation of the two independent, prototrophic, homozygous cmp1 mutants CMP1M5A and CMP1M5B (Fig. 2C, lanes 12 and 13) and the corresponding complemented strains, CMP1MK1A and CMP1MK1B (Fig. 2C, lanes 10 and 11).
When the blot shown in Fig. 2C was rehybridized with a probe from the CMP1 coding region, there also was only one hybridizing band in the parental strain, CAI4, which disappeared after two rounds of allelic replacement in the homozygous Δcmp1 mutants (Fig. 2D), providing additional evidence that C. albicans possesses only one gene encoding the calcineurin catalytic subunit.
Deletion of CMP1 causes hypersensitivity to ionic and alkaline pH stresses.
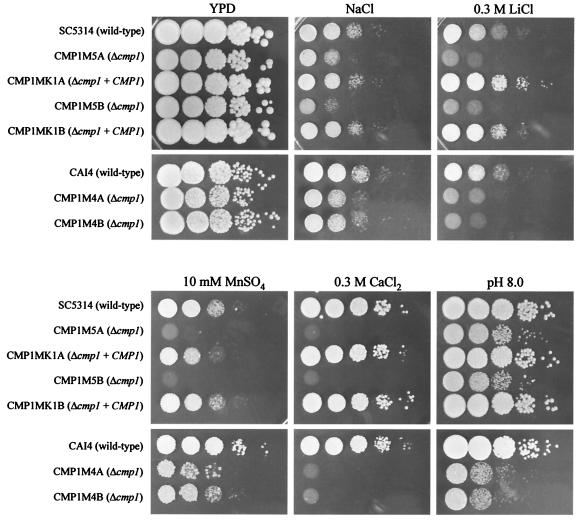
We first tested whether deletion of CMP1 in C. albicans resulted in phenotypes that have been described for calcineurin mutants of S. cerevisiae. As shown in Fig. 3, the Δcmp1 mutants exhibited increased sensitivity to cationic stress, as they grew more slowly than the wild-type strain SC5314 in the presence of increased concentrations of sodium, lithium, or manganese. In addition, deletion of CMP1 resulted in enhanced susceptibility to alkaline conditions, since growth of the mutants was reduced at pH 8.0. These results provided further evidence that CMP1 indeed encodes the C. albicans orthologue of the calcineurin catalytic subunit.
FIG. 3.
Deletion of CMP1 causes hypersensitivity to cationic and alkaline stresses. Serial 10-fold dilutions of the indicated strains were spotted on a YPD plate or YPD plates containing NaCl (1.8 M for URA3+ strains and 1.0 M for ura3 mutant strains), 0.3 M LiCl, 10 mM MnSO4, or 0.3 M CaCl2 and incubated at 30°C for 2 days. Alkaline pH sensitivity was tested on YPD plates buffered at pH 8.0.
In S. cerevisiae, inactivation of calcineurin results in reduced susceptibility to high Ca2+ concentrations. Interestingly, deletion of the CMP1 gene in C. albicans caused the opposite phenotype. Whereas the presence of 300 mM CaCl2 was well tolerated by the wild-type strain, growth of the Δcmp1 mutants was inhibited under these conditions, demonstrating that there are also differences between the calcineurin-mediated responses to certain stress conditions in the two species. All these phenotypes were observed in both URA3+ and ura3 Δcmp1 mutants, and they were complemented by reintroduction of an intact CMP1 copy.
CMP1 is not required for growth at elevated temperatures and hyphal formation in C. albicans.
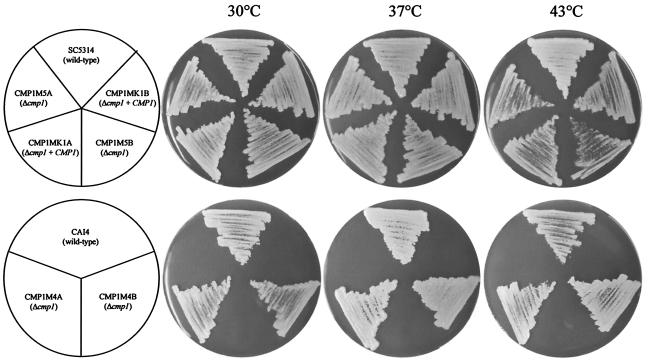
In the pathogenic yeast C. neoformans, inactivation of calcineurin results in the inability of the mutants to grow at 37°C. However, in C. albicans, deletion of CMP1 did not cause such a temperature-sensitive phenotype, since the Δcmp1 mutants grew as well as the wild-type and complemented strains at 30 or 37°C (Fig. 4). We noted slightly reduced growth of both the mutants and the complemented strains at 43°C in comparison with the wild-type strain SC5314 (Fig. 4, upper row). This phenotype apparently was caused by the genetic background of the CAI4 strain, from which the mutants were constructed, and not by the transformation procedure, since no growth difference was observed in a direct comparison of the ura3 Δcmp1 mutants CMP1M4A and -B and their parental strain, CAI4 (Fig. 4, lower row).
FIG. 4.
Deletion of CMP1 does not cause temperature sensitivity in C. albicans. The indicated strains were streaked on YPD plates and grown for 2 days at 30 or 37°C or for 7 days at 43°C. The upper row shows the growth of URA3 strains, and the lower row shows the growth of ura3 strains, on uridine-supplemented plates.
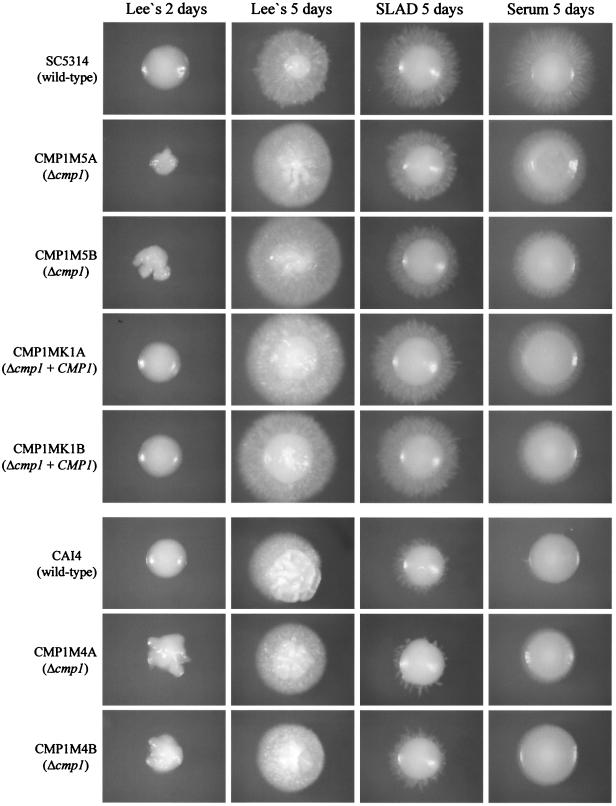
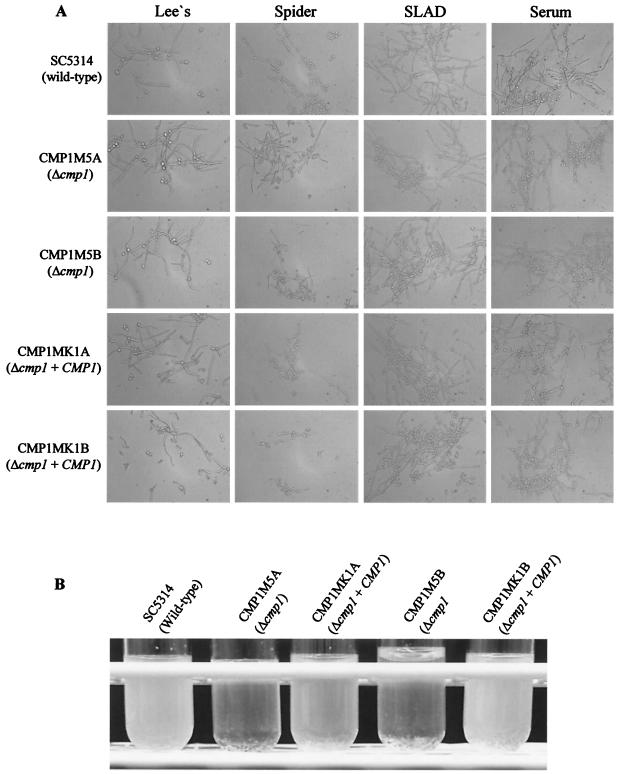
Since C. neoformans calcineurin mutants are defective in filamentous growth during mating and haploid fruiting, we tested whether deletion of CMP1 also affected hyphal formation in C. albicans. However, in several solid and liquid media known to induce hyphal growth in C. albicans, the Δcmp1 mutants formed hyphae as efficiently as the control strains (Fig. 5 and 6). Interestingly, under some conditions, hyphal growth was even increased in the Δcmp1 mutants. After 2 days of growth on Lee's agar plates, the mutants formed wrinkled colonies, whereas those of the wild-type and complemented strains were smooth at this time point (Fig. 5). However, after prolonged growth on these plates, the filamentations of mutants and control strains were similar. The enhanced hyphal formation of the Δcmp1 mutants was also observed in liquid Lee's medium, in which the mutants aggregated more strongly and settled to the bottom of the culture tubes more rapidly than the control strains (Fig. 6B). These results demonstrated that calcineurin signaling is not required for hyphal formation in C. albicans under the conditions tested.
FIG. 5.
Filamentous growth of C. albicans Δcmp1 mutants and control strains on hypha-inducing solid media. The colonies were photographed after 2 or 5 days as indicated.
FIG. 6.
Hyphal growth of C. albicans Δcmp1 mutants and control strains in liquid media at 37°C. (A) Microscopic appearance of the indicated strains after 7 h in Lee's medium (pH 7.0), 7 h in Spider medium, 8 h in SLAD medium, or 4 h in RPMI medium with 10% FCS (serum). (B) Aggregation phenotypes of Δcmp1 mutants after overnight growth in Lee's medium (pH 7.0). The cultures were briefly shaken and allowed to settle for 1 min before the photograph was taken.
Calcineurin is essential for virulence of C. albicans.
Calcineurin mutants of C. neoformans are avirulent (8, 16, 43). Although calcineurin signaling is involved in many different cellular reactions, the avirulent phenotype of C. neoformans calcineurin mutants is not surprising because of their inability to grow at body temperature. Since the C. albicans Δcmp1 mutants grew well at 37°C, it was important to test whether calcineurin signaling is necessary for the capacity of C. albicans to infect a mammalian host.
To address the role of calcineurin in C. albicans virulence, mice were intravenously injected either with one of the two independent Δcmp1 mutants (CMP1M5A or -B) or with the respective complemented strain (CMP1MK1A or -B). As illustrated in Fig. 7A, all of the mice succumbed to CMP1MK1A or -B infection within 17 days, while the clinical course of Δcmp1 mutant infection was much less severe: during follow-up, all animals infected with the Δcmp1 mutants survived beyond day 63 postinfection (p.i.) (mutant versus complemented strains, P ≤ 0.026). The kidneys of CMP1MK1B-infected animals presented with increasing signs of hyperemia (day 6 p.i.), followed by complete tissue destruction (day 8 p.i.) (Fig. 7B), and both hyphae and mycelia were readily detected after calcofluor white staining (data not shown). In contrast, the kidneys of Δcmp1 mutant-infected mice showed no sign of disease (days 6 and 14 p.i.) (Fig. 7B), and upon microscopic examination of KOH-solubilized organs, neither C. albicans hyphae nor blastoconidia were detectable. Accordingly, enumeration of C. albicans CFU in infected kidneys showed that the Δcmp1 mutant was cleared very rapidly after infection by the host immune defense. As illustrated in Fig. 7C, we could recover only a few mutant cells from infected kidneys on day 7 and none on day 11, whereas increasing numbers of CFU were obtained from the kidneys of mice infected with the complemented strain CMP1MK1B, which outnumbered those of the mutant by several orders of magnitude. These results demonstrate that calcineurin is essential for virulence in C. albicans.
FIG. 7.
C. albicans Δcmp1 mutants are avirulent. (A) Survival curves of mice intravenously infected with 5 × 105 cells of the Δcmp1 mutant strains CMP1M5A and -B or the respective complemented strains, CMP1MK1A and -B. (B) Macrographs of kidneys from animals infected with strains CMP1MK1B (Δcmp1 + CMP1) (left) or CMP1M5B (Δcmp1) (right). The duration of infection is indicated for each frame (d, day p.i.). (C) Fungal organ burden for kidneys. Shown are average numbers of CFU (+ standard deviations) after infection with 5 × 105 cells of either the Δcmp1 mutant strain CMP1M5B (open bars) or the complemented strain CMP1MK1B (solid bars) calculated for two or three pairs of kidneys. n.d., not detectable.
DISCUSSION
Our analysis of C. albicans Δcmp1 mutants demonstrates similar requirements for calcineurin signaling for adaptation to certain stress conditions, e.g., cationic stress and alkaline pH, in C. albicans and S. cerevisiae. However, there are also some differences between the calcineurin-mediated cellular responses in these two yeast species. High Ca2+ concentrations are not tolerated by wild-type S. cerevisiae cells. This phenotype seems to be due to inadequate activation of calcineurin, since calcineurin mutation in S. cerevisiae results in resistance to high Ca2+ concentrations (9). In contrast, C. albicans tolerated high Ca2+ concentrations well, but growth under these conditions was inhibited after deletion of CMP1. Therefore, calcineurin seems to be required for adaptation of C. albicans to high Ca2+ concentrations. Opposite effects of calcineurin inactivation have also been described for S. cerevisiae and C. neoformans. Whereas calcineurin mutants of S. cerevisiae show increased sensitivity to Mn2+, inactivation of calcineurin resulted in Mn2+ tolerance in C. neoformans (14, 43). Therefore, the roles of calcineurin in the adaptation to specific stress conditions are not identical in different yeast species.
Inhibitors of ergosterol biosynthesis stimulate Ca2+ influx and activate calcium signaling pathways that are essential for cell survival in S. cerevisiae (2). Disturbance of calcium signaling by calcium chelators or by pharmacological or genetic inhibition of calcineurin increases the sensitivity of yeast cells to azoles and terbinafine, antifungal agents that inhibit ergosterol biosynthesis (12). In line with this, fluconazole, a normally fungistatic drug, kills C. albicans cells in the presence of the calcineurin inhibitor cyclosporine (32), and the combination of fluconazole and cyclosporine is effective in eliminating C. albicans from infected animals (31). During the progress of our study, Cruz et al. reported that C. albicans calcineurin mutants, which were obtained by deleting the CNB1 gene encoding the regulatory subunit of calcineurin, were hypersensitive to membrane stress caused by the presence of sodium dodecyl sulfate or ergosterol biosynthesis inhibitors (7). We confirmed that the growth of our C. albicans Δcmp1 mutants was also inhibited by the presence of sodium dodecyl sulfate or fluconazole and that the mutants were rapidly killed in the presence of these agents (data not shown).
Of particular interest for us was the role of calcineurin in the pathogenicity of C. albicans, since calcineurin mutants of C. neoformans have been shown to be avirulent. In fact, as we demonstrate here, calcineurin is essential for virulence in C. albicans as well. However, in contrast to C. neoformans, calcineurin is not required for growth of C. albicans at 37°C. Therefore, the virulence defect of the Δcmp1 mutants cannot be explained simply by temperature sensitivity. Another important characteristic of C. albicans related to its pathogenic potential is the ability to switch between yeast and hyphal growth forms. Since filamentous growth is abolished after inactivation of calcineurin in C. neoformans, we tested if deletion of CMP1 would also affect hyphal formation in C. albicans. Hyphal growth in C. albicans is stimulated by different environmental signals, e.g., neutral pH, elevated temperature, nutrient starvation, or the presence of serum (13), and calcium has been implicated as a second messenger in the induction of hyphal formation by environmental signals (20). Interestingly, calcineurin was recently shown to be involved in the transcriptional response to alkaline pH in S. cerevisiae (47). Our results demonstrate that hyphal induction by various environmental signals was not affected by inactivation of calcineurin. Deletion of CMP1 even resulted in an increased stimulation of hyphal formation under certain conditions. However, it is possible that some physical or biochemical properties of hyphae are altered in the Δcmp1 mutants compared with the wild type, and this may have impacts on the fungus-host interaction. We note that Sanglard and colleagues recently also reported disruption of the calcineurin catalytic subunit in C. albicans (D. Sanglard, F. Ischer, O. Marchetti, and J. Bille, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. J-1845, 2001) and found reduced virulence of the mutants in a rat model of Candida endocarditis, which corroborates our findings of virulence attenuation in a mouse model of systemic candidiasis.
It has recently become evident that the use of the URA3 marker for the genetic engineering of C. albicans can cause problems in interpreting mutant phenotypes. The expression level of an ectopically inserted URA3 gene depends on the integration locus (24), and since uridine auxotrophy renders C. albicans avirulent (4, 21), this may affect virulence and virulence-related traits (1). Sundstrom and colleagues have recently shown that mutant phenotypes unrelated to inactivation of the target gene are obtained when the URA3 marker is inserted at a different genomic site or in a different way in the homozygous mutants than in the complemented strains (50). To avoid such problems, we paid particular attention to strain construction in our study. In both mutants and complemented strains, the URA3 marker was inserted at the same genomic site in exactly the same way. To shield the URA3 gene from the possible influence of upstream sequences, the ACT1 termination sequence was inserted in front of the URA3 marker. The only difference between mutants and complemented strains is the presence or the absence of the target gene open reading frame. In addition, in all in vitro phenotypic assays, we compared not only the URA3+ Δcmp1 mutants with the URA3+ control strains, but also the ura3 Δcmp1 mutants with the ura3 parent, CAI4. A phenotypic difference between mutants and control strains that is caused by differences in URA3 expression should not be observed in the latter comparison. All the phenotypic effects of CMP1 deletion were observed in both comparisons, demonstrating that they were specifically caused by the inactivation of calcineurin. For the virulence tests in mice, we included only those strains that allow optimal comparison, i.e., strains with and without a functional CMP1 copy, all of which contain one copy of the URA3 gene integrated in the same way. A comparison of the mutants with the wild-type strain SC5314 is less reliable, since the latter contains two copies of the URA3 gene and because the URA3 deletion in strain CAI4 also affects the flanking genes (17). Two independently constructed pairs of mutants and complemented strains, all derived at the same time from the same parent strain, pro-duced identical results, providing convincing evidence that loss of virulence in the mutants was due to deletion of the CMP1 gene.
The virulence defect of Δcmp1 mutants may be explained by an inability of the C. albicans cells to adapt to stressful environmental conditions within the host. The fact that calcineurin is essential for virulence not only in C. neoformans (8, 16, 43) but also in C. albicans suggests that calcineurin represents an attractive target for novel antimycotic agents. Cruz et al. (7) proposed that nonimmunosuppressive analogues of the calcineurin inhibitors FK506 and CsA may be useful in combination with azoles to treat Candida infections. Although such agents did not inhibit the growth of C. albicans in standard media in vitro, and the growth of calcineurin mutants is not significantly affected under these conditions, the virulence defect of Δcmp1 mutants indicates that inhibition of calcineurin by itself might be sufficient to block an infection.
Acknowledgments
This study was supported by a grant from the Bayerische Forschungsstiftung. Joachim Morschhäuser is the recipient of a Heisenberg fellowship from the Deutsche Forschungsgemeinschaft. Barbara Bodendorfer was supported by the Interdisciplinary Center for Clinical Research at the University of Erlangen (A15), and Klaus Schröppel was supported by the Deutsche Forschungsgemeinschaft (Schr450/4-2).
We thank Kai Michaelis for help with the preparation of Fig. 1. Sequence data for C. albicans was obtained from the Stanford Genome Technology Center website (http://www-sequence.stanford.edu/group/candida). Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
Editor: T. R. Kozel
REFERENCES
- 1.Bain, J. M., C. Stubberfield, and N. A. Gow. 2001. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol Lett. 204:323-328. [DOI] [PubMed] [Google Scholar]
- 2.Bonilla, M., K. K. Nastase, and K. W. Cunningham. 2002. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21:2343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breuder, T., C. S. Hemenway, N. R. Movva, M. E. Cardenas, and J. Heitman. 1994. Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc. Natl. Acad. Sci. USA 91:5372-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, M. F., W. H. Bowen, X. J. Zhao, and R. L. Cihlar. 1995. Avirulence of Candida albicans auxotrophic mutants in a rat model of oropharyngeal candidiasis. FEMS Microbiol Lett. 126:177-180. [DOI] [PubMed] [Google Scholar]
- 5.Cruz, M. C., M. Del Poeta, P. Wang, R. Wenger, G. Zenke, V. F. Quesniaux, N. R. Movva, J. R. Perfect, M. E. Cardenas, and J. Heitman. 2000. Immunosuppressive and nonimmunosuppressive cyclosporine analogs are toxic to the opportunistic fungal pathogen Cryptococcus neoformans via cyclophilin-dependent inhibition of calcineurin. Antimicrob. Agents Chemother. 44:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz, M. C., D. S. Fox, and J. Heitman. 2001. Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J. 20:1020-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz, M. C., A. L. Goldstein, J. R. Blankenship, M. Del Poeta, D. Davis, M. E. Cardenas, J. R. Perfect, J. H. McCusker, and J. Heitman. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz, M. C., R. A. Sia, M. Olson, G. M. Cox, and J. Heitman. 2000. Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect. Immun. 68:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham, K. W., and G. R. Fink. 1994. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124:351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cyert, M. S., R. Kunisawa, D. Kaim, and J. Thorner. 1991. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. USA 88:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cyert, M. S., and J. Thorner. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12:3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edlind, T., L. Smith, K. Henry, S. Katiyar, and J. Nickels. 2002. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol. Microbiol. 46:257-268. [DOI] [PubMed] [Google Scholar]
- 13.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 14.Farcasanu, I. C., D. Hirata, E. Tsuchiya, F. Nishiyama, and T. Miyakawa. 1995. Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese, in blocking the entry of ions into the cells. Eur. J. Biochem. 232:712-717. [PubMed] [Google Scholar]
- 15.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox, D. S., M. C. Cruz, R. A. Sia, H. Ke, G. M. Cox, M. E. Cardenas, and J. Heitman. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol. Microbiol. 39:835-849. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, M. G., J. E. O'Connor, L. L. Garcia, S. I. Martinez, E. Herrero, and L. del Castillo Agudo. 2001. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast 18:301-311. [DOI] [PubMed] [Google Scholar]
- 18.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 19.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 20.Gow, N. A. 1994. Growth and guidance of the fungal hypha. Microbiology 140:3193-3205. [DOI] [PubMed] [Google Scholar]
- 21.Kirsch, D. R., and R. R. Whitney. 1991. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect. Immun. 59:3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köhler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuno, T., H. Tanaka, H. Mukai, C. D. Chang, K. Hiraga, T. Miyakawa, and C. Tanaka. 1991. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 180:1159-1163. [DOI] [PubMed] [Google Scholar]
- 24.Lay, J., L. K. Henry, J. Clifford, Y. Koltin, C. E. Bulawa, and J. M. Becker. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schröppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H., J. Köhler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807-815. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., S. Ishii, M. Tokai, H. Tsutsumi, O. Ohki, R. Akada, K. Tanaka, E. Tsuchiya, S. Fukui, and T. Miyakawa. 1991. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol. Gen. Genet. 227:52-59. [DOI] [PubMed] [Google Scholar]
- 30.Lo, H. J., J. R. Köhler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 31.Marchetti, O., J. M. Entenza, D. Sanglard, J. Bille, M. P. Glauser, and P. Moreillon. 2000. Fluconazole plus cyclosporine: a fungicidal combination effective against experimental endocarditis due to Candida albicans. Antimicrob. Agents Chemother. 44:2932-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchetti, O., P. Moreillon, M. P. Glauser, J. Bille, and D. Sanglard. 2000. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 44:2373-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matheos, D. P., T. J. Kingsbury, U. S. Ahsan, and K. W. Cunningham. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11:3445-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazur, P., N. Morin, W. Baginsky, M. el-Sherbeini, J. A. Clemas, J. B. Nielsen, and F. Foor. 1995. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol. 15:5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendizabal, I., G. Rios, J. M. Mulet, R. Serrano, and I. F. de Larrinoa. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 425:323-328. [DOI] [PubMed] [Google Scholar]
- 36.Mendoza, I., F. Rubio, A. Rodriguez-Navarro, and J. M. Pardo. 1994. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 269:8792-8796. [PubMed] [Google Scholar]
- 37.Millon, L., A. Manteaux, G. Reboux, C. Drobacheff, M. Monod, T. Barale, and Y. Michel-Briand. 1994. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J. Clin. Microbiol. 32:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morschhäuser, J., S. Michel, and J. Hacker. 1998. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 257:412-420. [DOI] [PubMed] [Google Scholar]
- 39.Morschhäuser, J., S. Michel, and P. Staib. 1999. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol. Microbiol. 32:547-556. [DOI] [PubMed] [Google Scholar]
- 40.Moser, M. J., J. R. Geiser, and T. N. Davis. 1996. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 16:4824-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura, T., Y. Liu, D. Hirata, H. Namba, S. Harada, T. Hirokawa, and T. Miyakawa. 1993. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 12:4063-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro-Garcia, F., M. Sanchez, C. Nombela, and J. Pla. 2001. Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol. Rev. 25:245-268. [DOI] [PubMed] [Google Scholar]
- 43.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prokisch, H., O. Yarden, M. Dieminger, M. Tropschug, and I. B. Barthelmess. 1997. Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol. Gen. Genet. 256:104-114. [DOI] [PubMed] [Google Scholar]
- 45.Schreiber, S. L., and G. R. Crabtree. 1992. The mechanism of action of cyclosporin A and FK506. Immunol. Today 13:136-142. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer, A., S. Rupp, B. N. Taylor, M. Röllinghoff, and K. Schröppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 47.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Ariño. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46:1319-1333. [DOI] [PubMed] [Google Scholar]
- 48.Stathopoulos, A. M., and M. S. Cyert. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11:3432-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stathopoulos-Gerontides, A., J. J. Guo, and M. S. Cyert. 1999. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 13:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundstrom, P., J. E. Cutler, and J. F. Staab. 2002. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect. Immun. 70:3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]