Abstract
Toll-like receptors (TLRs) play an important role in the innate immune response, particularly in the initial interaction between the infecting microorganism and phagocytic cells, such as macrophages. We investigated the role of TLR4 during infection of primary murine peritoneal macrophages with Salmonella enterica serovar Typhimurium. We found that macrophages from the C3H/HeJ mouse strain, which carries a functionally inactive Tlr4 gene, exhibit marked impairment of tumor necrosis factor alpha (TNF-α) secretion in response to S. enterica serovar Typhimurium infection. However, activation of extracellular growth factor-regulated kinase and NF-κB signaling pathways was relatively unaffected, as was increased expression of TNF-α mRNA. Furthermore, macrophage tolerance, which is associated with increased expression of the NF-κB p50 and p52 subunits, was induced by S. enterica serovar Typhimurium even in the absence of functional TLR4. These results indicate that during infection of macrophages by S. enterica serovar Typhimurium, TLR4 signals are required at a posttranscriptional step to maximize secretion of TNF-α. Signals delivered by pattern recognition receptors other than TLR4 are sufficient for the increased expression of the TNF-α transcript and at least some genes associated with macrophage tolerance.
Serovars of Salmonella enterica subspecies I cause two types of clinical problems in humans, both of which are important public health concerns. S. enterica serovar Typhimurium is a leading agent of food poisoning, an acute, generally self-limiting disease characterized by intestinal inflammation and diarrhea (42). S. enterica serovar Typhi causes typhoid, a systemic febrile illness that is responsible for significant morbidity and mortality worldwide, particularly in developing countries (21). S. enterica serovar Typhimurium also causes a typhoid-like disease in wild-type mice, and much of what we know about the host-pathogen interactions involved in Salmonella infections has been derived from studies with the mouse model. Like S. enterica serovar Typhi in human typhoid, S. enterica serovar Typhimurium invades the intestinal epithelium in the mouse and is then taken up by phagocytic cells in the gut-associated lymphoid tissue. From that site, the organism disseminates systemically and survives intracellularly for extended periods within splenic macrophages and hepatic Kupffer cells (45, 55). The interaction between S. enterica serovar Typhimurium and these macrophage populations results in activation of protective mechanisms, such as the production of reactive oxygen and nitrogen species, and increased expression of a number of proinflammatory genes, such as those encoding tumor necrosis factor alpha (TNF-α), interleukin-1, and interleukin-18 (4, 30, 50). Together with the development of adaptive immunity, these innate immune responses result in the generation of granulomatous foci that help to confine and control the infection.
A key aspect of the interaction between Salmonella and host macrophages is the activation of mammalian pattern recognition receptors (PRRs) that recognize conserved microbial molecules, often referred to as pathogen-associated molecular patterns (16). The best-studied of the mammalian PRRs are the surface proteins of the Toll-like receptor (TLR) family, and currently there are 10 known members of this family (33). TLR2, in a heterodimeric complex with either TLR1 or TLR6, responds to bacterial peptidoglycan (PG) and lipopeptides, while TLR4, TLR5, and TLR9 recognize lipopolysaccharide (LPS), flagellin, and hypomethylated CpG-rich DNA, respectively. In general, TLR4 has been considered to be the major receptor for gram-negative bacteria, while the recognition of gram-positive bacteria has been ascribed to TLR2 (48). TLRs are connected via the adaptor protein MyD88 to a signal transduction machinery that is utilized by all members of the family (33). Not surprisingly, they induce similar patterns of gene expression, including those involved in microbicidal and inflammatory responses. However, it is becoming increasingly clear that certain signaling mechanisms are specific to individual TLRs (20, 54), with the result that different patterns of gene expression are induced by different TLR agonists (8, 18, 23, 43, 49).
Prolonged interactions between microbial components and macrophages can lead to a state of subsequent hyporesponsiveness to the same stimulus or a related stimulus, a phenomenon known as tolerance. The mechanisms underlying the induction of macrophage tolerance have been the subject of intense study, particularly with respect to TLR ligands. Some of the mechanisms that have been implicated in this process include down-regulation of TLR4 (37); decreased expression of postreceptor signaling molecules, such as the interleukin-1 receptor-associated kinase (IRAK) (32); alterations in the association between TLR5 and downstream signaling molecules (34); increased expression of proteins that block TLR signal transduction, such as IRAK-M, the macrophage-specific, kinase-deficient member of the IRAK family (27), and SOCS-1 (suppressor of cytokine signaling 1) (26, 36); and, finally, upregulation of the p50 and p52 subunits of the NF-κB transcription factor (24, 51). Although the exact physiologic significance of macrophage tolerance is not clear, it may be important in limiting the extent and duration of inflammatory responses. In keeping with this idea, disruption of tolerance by genetically engineered mutations results in exaggerated in vitro and in vivo inflammatory responses to bacteria or bacterial products (3, 26, 27, 36). Furthermore, in mice with a disruption of the NF-κB p50 gene, oral infection with Helicobacter hepaticus results in a chronic inflammatory bowel disease, illustrating the importance of macrophage tolerance to normal immune homeostasis in the intestine (12).
Most studies of TLR-dependent macrophage activation and tolerance have been carried out with purified microbial components, and less is known about the role of the TLRs in these processes during the course of infection by whole bacteria, which express multiple PRR ligands. In particular, the role of PRRs other than TLR4 has been given relatively little attention in the context of the response to gram-negative bacteria, such as salmonellae. In order to understand and ultimately manipulate the Salmonella-macrophage interaction, it is necessary to know which responses to the organisms are in fact caused by activation of TLR4 and which are attributable to activation of receptors other than TLR4. To elucidate the role of TLR4 and other PRRs in macrophage responses to salmonellae, we examined the interaction between S. enterica serovar Typhimurium and primary peritoneal macrophages from either the C3H/HeOuJ mouse strain (wild type) or the C3H/HeJ strain, which carries a naturally occurring missense mutation in the TLR4 cytoplasmic domain that prevents signaling through the receptor (40). We report here that the production of TNF-α, a key cytokine involved in inflammatory and protective responses to salmonellae (30), is significantly compromised at a posttranscriptional step in macrophages that lack functional TLR4. However, activation of proximal signaling pathways and induction of genes associated with tolerance still occur in these cells, indicating that TLR4-independent mechanisms are operating.
MATERIALS AND METHODS
Reagents.
Standard laboratory chemicals were purchased from Sigma (St. Louis, Mo.) or from Fisher Scientific (Suwanee, Ga.). Highly purified LPS from Salmonella strain Minnesota R595 was obtained from List Biological Laboratories (Campbell, Calif.), and stock solutions were made in sterile water. Staphylococcus aureus PG was purchased from Fluka (Milwaukee, Wis.) and was dissolved in sterile water according to the manufacturer's instructions. Antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.) unless specified otherwise.
Preparation of peritoneal macrophages.
The TLR4 mutant C3H/HeJ strain (40) and the corresponding wild-type C3H/HeOuJ strain of mice were purchased from Jackson Laboratories, Bar Harbor, Maine. Thioglycollate-elicited macrophages were prepared by using a standard protocol (13). In brief, the mice were injected intraperitoneally with 1.5 ml of sterile 3% Brewer's thioglycollate (Becton-Dickinson Microbiology Systems, Sparks, Md.) per mouse. On day 5 after injection, macrophages were harvested from the peritoneal cavity by lavaging with sterile phosphate-buffered saline. The macrophages were centrifuged at 400 × g for 10 min, resuspended in Dulbecco's modified Eagle medium (Invitrogen Corporation, Carlsbad, Calif.) with 10% heat-inactivated fetal calf serum and penicillin-streptomycin, and distributed into the wells of 24-well plates. After 2 h of incubation at 37°C, nonadherent cells were washed away, which left wells with 80 to 90% confluent adherent macrophages behind (approximately 0.5 × 106 macrophages per well). The macrophages were rested overnight before stimulation. Before treatments were started or before Salmonella infection, the cells were washed and recultured in medium without antibiotics.
Bacterial culture and infections.
S. enterica serovar Typhimurium strains (wild-type strains SL1344 [53] and SL3201; flagellin-deficient strain SL3201 fliC/fljB, isogenic with SL3201, kindly provided by Andrew Gewirtz, Emory University [15]; HilA-deficient strain VV341, isogenic with SL1344 [22]) were grown to the stationary phase in Luria-Bertani (LB) medium at 37°C with constant shaking. Wild-type Bacillus subtilis strain PY79 was kindly provided by David Rudner, Harvard University, and was grown similarly. The bacteria were pelleted by centrifuging at 13,000 × g for 5 min, washed with sterile phosphate-buffered saline, and then resuspended in antibiotic-free medium at a density of 4 × 108 cells/ml. Twenty-five-microliter aliquots of each suspension (107 bacteria) were used to infect each well of macrophages (multiplicity of infection [MOI], 20:1). Heat-killed bacteria were prepared by placing a bacterial suspension in boiling water for 10 min. Bacterial culture supernatants were prepared from stationary-phase cultures of wild-type or flagellin-deficient S. enterica serovar Typhimurium. After centrifugation of the bacteria at 13,000 × g for 5 min, the supernatants were filtered through a 0.2-μm-pore-size syringe filter (Nalge, Rochester, N.Y.) and added to cell cultures at a final concentration of 5% (vol/vol).
ELISA for TNF-α.
Duplicate wells containing peritoneal macrophages were treated or infected as described below for the individual experiments, and supernatants were collected after 2 h. The supernatants were centrifuged at 13,000 × g for 5 min at 4°C to remove the bacteria, and 50-μl aliquots were analyzed in triplicate by an enzyme-linked immunosorbent assay (ELISA) by using a standard protocol (6). In brief, TNF-α in the supernatant was captured with purified rat anti-mouse TNF-α (Pharmingen, San Diego, Calif.) fixed to an Immulon 2HB microtiter plate (ThermoLab Systems, Franklin, Mass.). The captured TNF-α was detected with biotinylated rat anti-mouse TNF-α (Pharmingen), followed by streptavidin-conjugated horseradish peroxidase and o-phenylene diamine substrate (the latter two reagents were obtained from Zymed Laboratories, San Francisco, Calif.). The intensity of the color reaction was measured by determining the absorbance at 405 nm with an Emax microplate reader (Molecular Devices, Sunnyvale, Calif.). TNF-α concentrations were calculated based on a standard curve that was generated by using purified mouse recombinant TNF-α (Pharmingen).
After the supernatants were removed, the macrophages were washed and lysed in 1% Triton X-100. The protein concentrations of the lysates were determined with a DC protein assay kit (Bio-Rad, Hercules, Calif.) by following the manufacturer's recommendations. The normalized amount of TNF-α was derived by dividing the amount of TNF-α produced in each well by the total amount of protein in the cell lysate from that well.
Western blotting.
Macrophages were lysed in 1% Triton X-100 in a solution containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, and 2 mM phenylmethylsulfonyl fluoride. Aliquots of clarified cell lysate, normalized for total protein content, were separated on sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gels and transferred to nitrocellulose by using a semidry blotting apparatus as previously described (5). The blots were blocked with 5% nonfat dry milk and then incubated overnight at 4°C with the relevant primary antibody (mouse anti-phosphorylated extracellular growth factor-regulated kinase [anti-phospho-ERK], mouse anti-IκBα, goat anti-NF-κB p50, or mouse anti-NF-κB p52, all obtained from Santa Cruz Biotechnology). The bound primary antibodies were detected by enhanced chemiluminescence by using the appropriate horseradish peroxidase-conjugated anti-immunoglobulin antibodies (Zymed Laboratories) and the West Pico Super Signal substrate reagent (Pierce, Rockford, Ill.), as described in detail previously (5). After the appropriate autoradiographic exposures were made, the blots were stripped and reprobed with either rabbit anti-total ERK (in the case of the phospho-ERK blot) or goat anti-actin (in the case of the IκBα, p50, and p52 blots), both obtained from Santa Cruz Biotechnology.
For quantitation of the Western blots, the autoradiographs were scanned and analyzed with NIH Image software to obtain the mean pixel density of each band. To normalize the phospho-ERK signal, the pixel density of the phospho-ERK band was divided by the pixel density of the corresponding total ERK band. Similarly, the IκBα and the NF-κB p50 and p52 bands were normalized by using the corresponding actin bands.
RT-PCR analysis.
Total RNA was prepared from control or Salmonella-infected cells with the Trizol reagent (Invitrogen Corporation) by following the manufacturer's directions. Reverse transcription-PCR (RT-PCR) was carried out by using a GeneAmp kit (Roche, Nutley, N.J.) and following the protocol provided with the kit. In brief, 100-ng aliquots of total RNA were reverse transcribed at 42°C for 15 min with Moloney murine leukemia virus reverse transcriptase after annealing to random hexamers. One-quarter of each RT reaction mixture was used for PCR amplification with Taq polymerase by using commercially available primers specific for murine TNF-α or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (R & D Systems, Minneapolis, Minn.). Thirty cycles of amplification were carried out, and each cycle consisted of denaturation at 95°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 2 min. One-quarter of each amplification reaction mixture was electrophoresed on a 1.2% agarose gel and photographed under UV transillumination.
Real-time quantitative PCR analysis of TNF-α and GAPDH expression was carried out as described above, except that the reverse-transcribed RNA and primers were mixed with a SYBR Green master mixture (Applied Biosystems, Foster City, Calif.) instead of the usual PCR mixture components. Duplicate reaction mixtures were set up, and 40 cycles of amplification were carried out with an iCycler (Bio-Rad) by using the same conditions that were used for the standard PCR. A quantitative measure of the relative amounts of transcript was obtained based on the literature provided with the iCycler. In brief, SYBR Green is a double-stranded DNA-specific fluorescent dye, the fluorescence intensity of which is proportional to the amount of double-stranded DNA. The iCycler monitors the fluorescence of the reaction mixtures just before the denaturation step of each amplification cycle, and it records the cycle number at which the fluorescence crosses a specific threshold value in the exponential phase of amplification (the threshold cycle [Ct]). Ct is thus a measure of the quantity of the transcript of interest, and lower Ct values indicate larger starting quantities of transcript. The amount of TNF-α transcript was normalized to the amount of GAPDH transcript by subtracting the mean Ct value of the latter from the mean Ct value of the former (for each experimental condition). The difference between the normalized Ct values of the Salmonella-infected cells and the control, uninfected cells was plotted and used as a measure of the increase in TNF-α mRNA expression resulting from the infection. Because of the exponential nature of the PCR, a difference of n in Ct values represented a 2n-fold difference in the transcript levels.
Statistical analysis.
Statistical significance was determined by the Student t test, and a P value of <0.05 was considered significant.
RESULTS
Lack of a functional TLR4 significantly impairs TNF-α production during infection of macrophages by Salmonella.
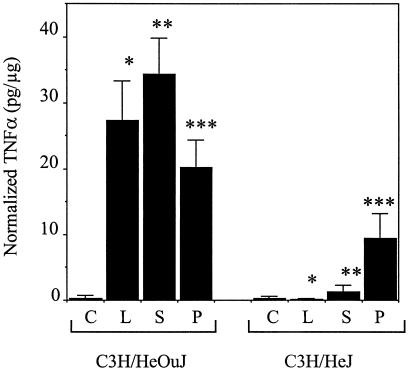
Infection of thioglycollate-elicited peritoneal macrophages from wild-type C3H/HeOuJ mice for 2 h with wild-type S. enterica serovar Typhimurium strain SL1344 at an MOI of 20 resulted in an approximately 100-fold increase in the amount of TNF-α secreted compared to the amount secreted by the control, uninfected cells, an effect that was similar to the effect observed after treatment with purified LPS (Fig. 1) (for control cells, 0.27 ± 0.44 pg/μg; for Salmonella-infected cells, 34.35 ± 5.51 pg/μg; for LPS-treated cells, 27.38 ± 6.00 pg/μg). In contrast, when macrophages from the C3H/HeJ strain, which has an inactivating point mutation in the TLR4 cytoplasmic domain (40), were used, the TNF-α responses to both LPS and S. enterica serovar Typhimurium were significantly lower than the corresponding C3H/HeOuJ responses (Fig. 1) (for control cells, 0.27 ± 0.42 pg/μg; for Salmonella-infected cells, 1.28 ± 1.01 pg/μg; for LPS-treated cells, 0.12 ± 0.12 pg/μg [P < 0.001 for the comparison of the responses to S. enterica serovar Typhimurium of C3H/HeOuJ and C3H/HeJ cells; P < 0.0005 for the comparison of the responses to LPS]). Treatment of the C3H/HeJ macrophages with the TLR2 ligand staphylococcal PG led to an approximately 35-fold increase in the amount of TNF-α secreted (Fig. 1) (for control cells, 0.27 ± 0.42 pg/μg; for PG-treated cells, 9.50 ± 3.68 pg/μg). This response indicated that the TLR2 signaling pathway was relatively preserved in the TLR4 mutant cells, even though the response was significantly reduced compared to the response of the C3H/HeOuJ macrophages to PG (20.19 ± 4.14 pg/μg; P < 0.0025). The response to the gram-positive bacterium B. subtilis was similar to the response obtained with PG (data not shown). There was no increase in TNF-α production from the C3H/HeJ macrophages even when the dose of Salmonella used for infection was increased 10-fold or when the analysis was extended to 6 h after infection. These results demonstrate the importance of the LPS-TLR4 interaction in inducing TNF-α during Salmonella infection of macrophages. In contrast, when C3H/HeOuJ macrophages were infected with specific mutants that were either deficient in a functional Salmonella pathogenicity island 1 type III secretion system or lacked both flagellin subunit genes, no difference in TNF-α production was noted when the data were compared to the data for infection with wild-type Salmonella. Similarly, when we used a previously documented approach to inhibit TLR9 function (i.e., treatment of cells with chloroquin [1, 31]), we did not observe any effect on the amount of TNF-α produced by C3H/HeOuJ macrophages in response to wild-type Salmonella infection (data not shown).
FIG. 1.
Production of TNF-α by peritoneal macrophages from C3H/HeOuJ and C3H/HeJ mice. Macrophages were not stimulated (C), were stimulated with 100 ng of LPS per ml (L) or 25 μg of PG per ml (P), or were infected with 107 wild-type S. enterica serovar Typhimurium strain SL1344 (S) for 2 h, and the supernatants were analyzed by ELISA for TNF-α. The amount of TNF-α produced was normalized to the protein content of the macrophage cell lysate. The data are the means of the results of at least four separate experiments, and the error bars indicate standard deviations. The P values for the differences between the responses of the two strains to LPS, Salmonella, and PG were <0.0005 (one asterisk), <0.001 (two asterisks), and <0.0025 (three asterisks), respectively.
Functional TLR4 is not required for activation of macrophage signaling pathways by Salmonella.
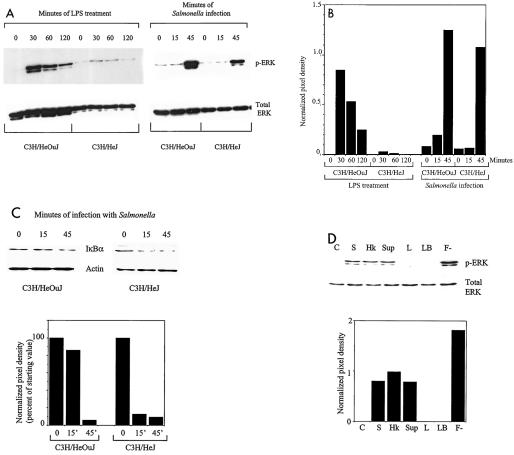
The impaired ability of C3H/HeJ macrophages to produce TNF-α in response to Salmonella infection could indicate either that there was exclusive activation of TLR4 during this process or that other PRRs were unable to induce high-level production of TNF-α. To evaluate these two possibilities, we examined signaling cascades that are known to be activated by all TLRs, viz., the ERK pathway and the NF-κB pathway (33). Macrophages from either C3H/HeOuJ or C3H/HeJ mice were infected with a wild-type Salmonella strain for different periods, and Western blotting of cell lysates was carried out to detect the phosphorylated (activated) form of the ERK protein or degradation of the IκBα protein (as a marker of activation of the NF-κB pathway). Figures 2A and B show that macrophages from the TLR4 mutant C3H/HeJ strain were defective in LPS-dependent activation of ERK when they were compared to the wild-type C3H/HeOuJ macrophages. As measured by the intensity of the phospho-ERK band (normalized to total ERK), the peak activation of ERK by LPS in the C3H/HeJ macrophages was only about 4% of that seen in the C3H/HeOuJ macrophages. In contrast, the activation of ERK in response to Salmonella infection was only slightly reduced by the TLR4 mutation (the peak activation in C3H/HeJ macrophages was about 86% of that in C3H/HeOuJ macrophages). Similarly, although activation of the NF-κB pathway in response to LPS was very markedly impaired in C3H/HeJ macrophages (data not shown), the degradation of IκBα in response to Salmonella infection was relatively unaffected in these cells (9.4% of the starting IκBα level was present at 45 min in the C3H/HeJ macrophages, compared with 5.7% in the C3H/HeOuJ macrophages [Fig. 2C]). These observations are consistent with the idea that Salmonella activates TLR4-independent signaling pathways during infection of macrophages.
FIG. 2.
(A) Activation of ERK in C3H/HeOuJ and C3H/HeJ peritoneal macrophages in response to LPS treatment or infection with wild-type S. enterica serovar Typhimurium. Macrophages were treated with 100 ng of LPS per ml or were infected with 107 SL1344 bacteria for the times indicated. Cell lysates were prepared and analyzed by Western blotting for the presence of phospho-ERK (p-ERK). The blots were then stripped and reprobed to detect total ERK. (B) Quantitation of the Western blot in panel A. The autoradiograph was scanned and analyzed by using NIH Image software. The mean pixel densities of the phospho-ERK bands (normalized to the pixel densities of the corresponding total ERK bands) are shown. (C) Degradation of IκBα in C3H/HeOuJ and C3H/HeJ peritoneal macrophages in response to infection with wild-type S. enterica serovar Typhimurium. Macrophages were infected with 107 SL1344 bacteria for the times indicated. Cell lysates were prepared and analyzed by Western blotting for the presence of IκBα. The blot was then stripped and reprobed for the presence of actin to confirm equivalent loading of lanes. Quantitation of the Western blot is shown at the bottom, and the data indicate the mean pixel densities of the IκBα bands (normalized to the pixel densities of the corresponding actin bands) expressed as percentages of the value at time zero for each strain. (D) Activation of ERK in C3H/HeJ peritoneal macrophages in response to S. enterica serovar Typhimurium or its products. Macrophages were left untreated (C) or were exposed to 107 wild-type S. enterica serovar Typhimurium strain SL1344 bacteria (S), 107 heat-killed SL1344 bacteria (Hk), a 5% (vol/vol) supernatant of a stationary culture of wild-type S. enterica serovar Typhimurium (Sup), a 5% (vol/vol) supernatant of a stationary culture of flagellin-deficient S. enterica serovar Typhimurium (F-), 5% (vol/vol) LB broth (LB), or 100 ng of LPS per ml (L) for 45 min. Cell lysates were prepared and analyzed by Western blotting for the presence of phospho-ERK. The blots were then stripped and reprobed to detect total ERK. Quantitation of the Western blot is shown at the bottom, and the data indicate the mean pixel densities of the phospho-ERK bands normalized to the pixel densities of the corresponding total ERK bands.
To further characterize TLR4-independent activation of signaling by Salmonella, we repeated the experiments described above using C3H/HeJ macrophages and either heat-killed bacteria or bacterial supernatants. As shown in Fig. 2D, treatment of the macrophages with heat-killed wild-type S. enterica serovar Typhimurium, as well as with culture supernatants of wild-type S. enterica serovar Typhimurium, resulted in robust activation of ERK. An equivalent volume of culture broth (LB broth) did not have this effect, nor did LPS at a concentration of 100 ng/ml. These results indicate that molecules that are present on viable and heat-killed bacteria or are released into the culture supernatant are able to activate signaling in macrophages independent of TLR4. Flagellin was not responsible for the activity, since culture supernatant from the flagellin-deficient strain S. enterica serovar Typhimurium SL3201 flicC/fljB activated ERK as well as, if not better than, supernatants from wild-type strain SL1344 (Fig. 2D) and SL3201 (data not shown).
Functional TLR4 is not required for induction of TNF-α mRNA in response to Salmonella.
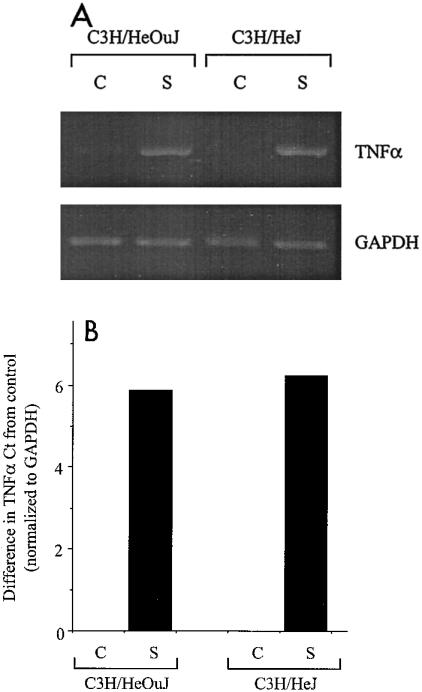
The data presented above indicate that PRRs other than TLR4 are activated during the course of Salmonella infection of macrophages. However, the signals resulting from such activation are insufficient for high-level production of secreted TNF-α. To determine at which level TNF-α production was blocked, we carried out a semiquantitative RT-PCR analysis to detect TNF-α mRNA in C3H/HeOuJ and C3H/HeJ macrophages after infection with wild-type S. enterica serovar Typhimurium. Figure 3A shows the result of this analysis and indicates that in both the wild-type and TLR4 mutant cells, infection with Salmonella led to a clear increase in TNF-α mRNA. This observation was confirmed by a quantitative, real-time PCR analysis, which indicated that the levels of induction of TNF-α mRNA in response to Salmonella infection were similar in macrophages from the C3H/HeOuJ and C3H/HeJ strains and were about 60-fold higher than the control value for C3H/HeOuJ mice and about 75-fold higher than the control value for C3H/HeJ mice (Fig. 3B). Thus, increased expression of TNF-α mRNA in response to Salmonella infection is independent of a functional TLR4, suggesting that the requirement for TLR4 signals is at a posttranscriptional step in the biosynthesis of the cytokine.
FIG. 3.
(A) Induction of TNF-α mRNA in C3H/HeOuJ and C3H/HeJ macrophages by Salmonella infection: semiquantitative RT-PCR analysis. Macrophages were not infected (lanes C) or were infected for 2 h with 107 wild-type S. enterica serovar Typhimurium SL1344 bacteria (lanes S). Total RNA was prepared and used for the RT-PCR with primers specific for either TNF-α or the housekeeping transcript GAPDH. Aliquots of the PCR products were analyzed by agarose gel electrophoresis and were visualized by UV transillumination. (B) Induction of TNF-α mRNA in C3H/HeOuJ and C3H/HeJ macrophages by Salmonella infection: quantitative real-time PCR analysis. Macrophages were not infected (bars C) or were infected for 2 h with 107 wild-type S. enterica serovar Typhimurium SL1344 bacteria (bars S). Total RNA was prepared, reverse transcribed, and amplified with an iCycler by using primers specific for either TNF-α or GAPDH in the presence of SYBR Green. Normalized Ct values were derived as described in Materials and Methods and are expressed as the differences from control, uninfected cells.
Functional TLR4 is not required for induction of macrophage tolerance by Salmonella.
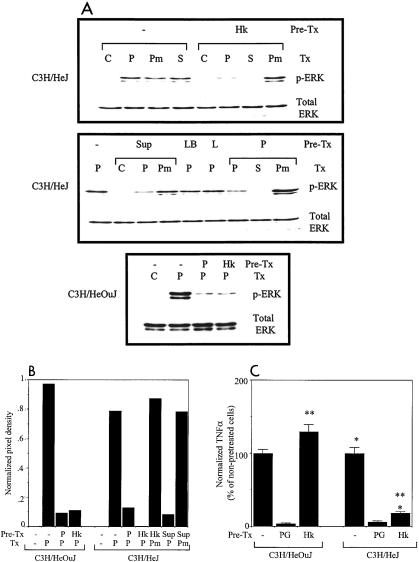
The results described above indicate that although PRRs other than TLR4 are engaged during the course of Salmonella infection of macrophages, the signals activated are poor inducers of secreted TNF-α. To determine whether this TLR4-independent engagement of PRRs has functional consequences other than the production of TNF-α, we examined the induction of macrophage tolerance. C3H/HeJ macrophages were pretreated overnight with either heat-killed wild-type Salmonella strain SL1344 or culture supernatants of this strain. After the initial stimulus was washed away, the cells were stimulated with PG, and activation of ERK was analyzed. As shown in Fig. 4A, pretreatment of C3H/HeJ macrophages with either the heat-killed bacteria (upper panel) or with the culture supernatant (middle panel) resulted in significant inhibition of ERK activation by PG, similar to that achieved by pretreatment with PG itself (middle panel), whereas activation by the unrelated stimulus phorbol myristate acetate was not affected. Quantitation of the Western blots confirmed these findings (Fig. 4B). The inhibition of PG-dependent ERK activation by pretreatment with heat-killed Salmonella was similar in C3H/HeJ and C3H/HeOuJ macrophages (Fig. 4A, lower panel, and Fig. 4B), indicating that the phenomenon is TLR4 independent. As expected, pretreatment of the C3H/HeJ macrophages with LPS did not affect the response to subsequent treatment with PG (Fig. 4A, middle panel). These results indicate that there is specific, TLR4-independent tolerization of PG-induced signaling by prior exposure to Salmonella products.
FIG. 4.
(A) Pretreatment of C3H/HeJ peritoneal macrophages with S. enterica serovar Typhimurium products results in tolerization of ERK activation by PG. Macrophages were not pretreated (−) or were pretreated overnight with 107 heat-killed wild-type S. enterica serovar Typhimurium SL1344 bacteria (Hk), 5% (vol/vol) culture supernatant of SL1344 (Sup), 5% (vol/vol) LB broth (LB), 100 ng of LPS per ml (L), or 25 μg of PG per ml (P). They were then left unstimulated (C) or were stimulated for 45 min with 107 live wild-type SL1344 bacteria (S), 25 μg of PG per ml (P), or 1 μg of phorbol myristate acetate per ml (Pm). Cell lysates were prepared and analyzed by Western blotting for the presence of phospho-ERK (p-ERK). Pre-Tx, pretreatment; Tx, treatment. (B) Quantitation of the Western blots shown in panel A. The autoradiographs were scanned and analyzed by using NIH Image software. The mean pixel densities of the phospho-ERK bands (normalized to the pixel densities of the corresponding total ERK bands) are shown. (C) Effect of pretreatment of peritoneal macrophages with heat-killed S. enterica serovar Typhimurium on subsequent TNF-α production in response to PG. Macrophages were not pretreated or were pretreated overnight with 25 μg of PG per ml or with 107 heat-killed wild-type S. enterica serovar Typhimurium SL1344 bacteria. After the pretreatment stimulus was washed away, all groups of macrophages were stimulated for 2 h with 25 μg of PG per ml, and the supernatants were analyzed by ELISA for TNF-α. The normalized amount of TNF-α was expressed as a percentage of the amount produced by the nonpretreated, PG-stimulated cells. The data are the means of the results of three separate experiments, and the error bars indicate standard deviations. The P values for the comparisons are <0.0005 (one asterisk) and <0.0005 (two asterisks).
To determine whether the TNF-α response to PG could also be tolerized by Salmonella in a TLR4-independent fashion, C3H/HeOuJ and C3H/HeJ macrophages were pretreated overnight with heat-killed bacteria and then stimulated the next morning with PG. As shown in Fig. 4C, pretreatment of the C3H/HeJ macrophages with heat-killed Salmonella resulted in an approximately 85% reduction in the amount of TNF-α produced compared with the amount obtained for cells that had not been pretreated (P < 0.0005), which was comparable to the approximately 95% reduction observed after pretreatment with PG itself. Pretreatment of C3H/HeJ macrophages with Salmonella culture supernatant had a similar inhibitory effect on the subsequent TNF-α response to PG (data not shown). Thus, in keeping with the data on ERK activation, tolerization of the PG-induced TNF-α response by Salmonella also does not require a functional TLR4. Interestingly, pretreatment of C3H/HeOuJ macrophages with heat-killed Salmonella did not lead to tolerization of the TNF-α response to subsequent treatment with PG and even increased the response to some extent (Fig. 4C). This significant difference (P < 0.0005) between the responses of the C3H/HeOuJ and C3H/HeJ macrophages is probably attributable to the fact that TLR4 signals can accentuate the subsequent response to TLR2 ligands rather than induce cross-tolerance (7).
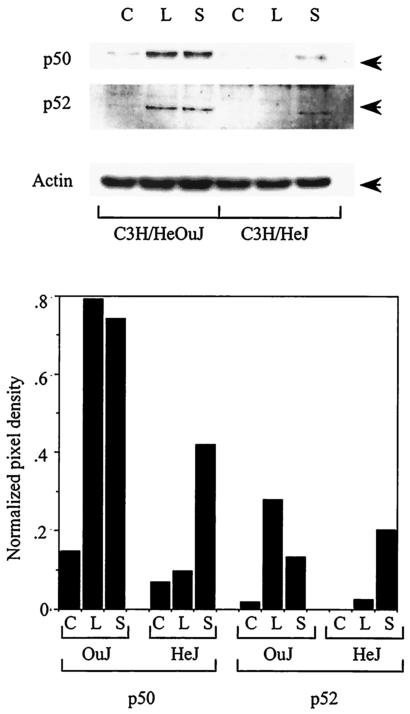
Various mechanisms have been proposed to explain the induction of macrophage tolerance, including increased expression of specific proteins, such as IRAK-M (27) and the NF-κB p50 and p52 subunits (24, 51). To determine the role of the NF-κB p50 and p52 proteins in the induction of tolerance by Salmonella, we examined the expression of these proteins after overnight incubation of wild-type and TLR4 mutant macrophages with either LPS or heat-killed Salmonella. As shown in Fig. 5, expression of both the NF-κB p50 and p52 proteins was increased by LPS and heat-killed Salmonella in the wild-type C3H/HeOuJ macrophages. As expected, induction of these proteins by LPS was not observed in the TLR4 mutant C3H/HeJ macrophages. However, there was an appreciable (albeit attenuated) increase in the expression of both NF-κB p50 and NF-κB p52 in the C3H/HeJ cells in response to Salmonella, indicating that the induction was at least partially independent of TLR4. Thus, our results indicate that macrophage tolerance can be induced by Salmonella even in the absence of a functional TLR4 and that this TLR4-independent tolerization is associated with increased expression of NF-κB p50 and p52.
FIG. 5.
Induction of genes associated with macrophage tolerance by Salmonella. Peritoneal macrophages from either C3H/HeOuJ or C3H/HeJ mice were left untreated (C) or were treated overnight with 100 ng of LPS per ml (L) or 107 heat-killed wild-type S. enterica serovar Typhimurium SL1344 bacteria (S). Protein lysates were prepared the next morning and analyzed by Western blotting with antibodies to NF-κB p50, NF-κB p52, or actin. The arrows indicate the relevant bands. Quantitation of the Western blots is shown at the bottom, and the data indicate the mean pixel densities of the p50 or p52 bands normalized to the pixel densities of the corresponding actin bands.
DISCUSSION
The data presented in this paper indicate that both TLR4 and PRRs other than TLR4 are activated during infection of macrophages by Salmonella. Activation of TLR4 is crucial to high-level production of TNF-α; in macrophages from the C3H/HeJ strain, which carries a mutated, functionally inactive Tlr4 gene (40), the amount of TNF-α protein detected in cell supernatants is markedly reduced (Fig. 1). Previously published work showed that expression of TNF-α is regulated at the level of transcription, mRNA stability, nucleocytoplasmic mRNA transport, translation, and secretion and that LPS-induced signals act at several of these steps (2, 9, 28, 29, 41, 47). Our results indicate that in the context of Salmonella infection, the major requirement for TLR4 signals is at a posttranscriptional step in TNF-α biosynthesis, since increased expression of the mRNA can be induced even in the absence of functional TLR4 (Fig. 3). Presumably, PRRs other than TLR4 are able to activate the signals necessary to increase the levels of the TNF-α transcript but are not sufficient for the subsequent steps involved in efficient translation and/or secretion of the protein. Consistent with this idea, we demonstrated that Salmonella activates at least two signaling pathways in macrophages independent of TLR4, the ERK pathway and the NF-κB pathway (Fig. 2). Furthermore, this TLR4-independent signaling is sufficient for the induction of macrophage tolerance (Fig. 4) associated with increased NF-κB p50 and p52 expression (Fig. 5). Based on our observations, we postulate a model of Salmonella-TLR interactions during macrophage infection (Fig. 6) in which both TLR4-dependent and TLR4-independent signals are activated. Both sets of signals are able to induce tolerance and to increase levels of the TNF-α mRNA, but the TLR4-dependent signal is essential for conversion of the TNF-α transcript into secreted protein. The precise nature of the TLR4-dependent signal necessary for high-level TNF-α expression remains to be identified. Recent studies have demonstrated that among the TLRs, TLR4 activates signaling mechanisms that are not activated by other members of the family, leading to patterns of gene expression that are unique to this receptor (8, 18, 23, 43, 49, 54). It is possible that these or similar TLR4-specific mechanisms function to increase TNF-α production.
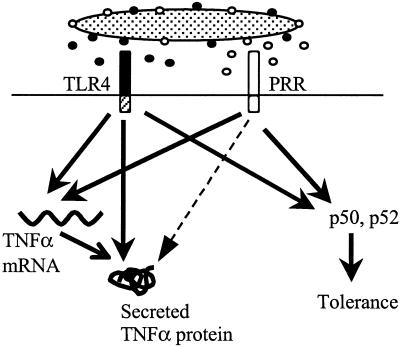
FIG. 6.
Model for the role of TLR4 and other PRRs in the Salmonella-macrophage interaction. Salmonella activates proximal signaling pathways via TLR4 and other PRRs. TLR4 signals are essential at a posttranscriptional step for high-level expression of secreted TNF-α protein. Signals provided by other PRRs are sufficient for induction of the TNF-α transcript and at least some genes associated with tolerance but not for production of high levels of secreted TNF-α. The dashed arrow indicates the signal required for high-level secreted TNF-α production that PRRs other than TLR4 are ineffective at providing.
The TLR4-independent induction of NF-κB p50, NF-κB p52, and tolerance clearly indicates that even in the case of gram-negative infection with relatively low numbers of organisms (the MOI was 20:1 in our experiments), ligands for PRRs other than TLR4 are available in amounts sufficient to have functional effects on macrophages. To generalize from these observations, it is likely that two types of genetic programs are activated as a result of the Salmonella-macrophage interaction; one of these, like the production of TNF-α, is TLR4 dependent, and the other, like the induction of tolerance, is TLR4 independent. This idea is in keeping with what is known about the in vivo response of the C3H/HeJ strain to Salmonella infection. This strain has increased susceptibility to infection with Salmonella, a trait that is associated with deficiencies in macrophage microbicidal activity and inflammatory cell recruitment (38, 39, 52). At the same time, Salmonella-dependent activation of macrophages for tumoricidal function and interleukin-6 production, Salmonella-induced dendritic cell maturation, and the development of anti-Salmonella adaptive immunity appear to be relatively unaffected in C3H/HeJ mice (10, 25, 44, 46, 56), indicating that there are TLR4-independent mechanisms. Our observations add macrophage tolerance to the list of functions that can be induced by Salmonella in a TLR4-independent manner and raise the possibility that tolerization in the context of compromised TLR4-dependent microbicidal and inflammatory mechanisms may contribute to the unusual susceptibility of the C3H/HeJ strain to Salmonella infection (38, 39).
It should be noted, however, that a deficiency of TLR4 function is not the only factor involved in the susceptibility of C3H/HeJ mice to Salmonella. LPS responsiveness has been clearly dissociated from Salmonella susceptibility in the C3H lineage (11), indicating that genes other than Tlr4 play a role in the latter characteristic. Interestingly, we found that peritoneal macrophages from C3HeB/FeJ mice, which are LPS responsive but Salmonella susceptible (11), produced significantly less TNF-α than C3H/HeOuJ macrophages produced in response to Salmonella (for C3HeB/FeJ, 14.77 ± 0.82 pg/μg; for C3H/HeOuJ, 34.35 ± 5.51 pg/μg [P < 0.001]), whereas there was not a significant difference between the two strains in terms of the amount of TNF-α produced in response to LPS (for C3HeB/FeJ, 23.90 ± 3.19 pg/μg; for C3H/HeOuJ, 27.38 ± 6.00 pg/μg [P < 0.25]). Further work is required to determine whether this observation is relevant to the susceptibility of the C3HeB/FeJ strain to Salmonella, but the observation suggests that variations in genes other than Tlr4 within the C3H lineage may contribute to differences in the TNF-α response to salmonellae. As an additional note of caution, it should be mentioned that all the studies described here were carried out with thioglycollate-elicited macrophages because of the difficulty in conveniently obtaining the numbers of resident peritoneal macrophages required for biochemical analyses. Differences between the responses of thioglycollate-elicited peritoneal macrophages and the responses of resident peritoneal macrophages have been documented (14, 19, 35), and this fact needs to be kept in mind when our findings are interpreted.
The identity of the Salmonella molecule or molecules that activate macrophages independent of TLR4 is the subject of on-going studies. The fact that salmonellae can induce tolerance to PG in C3H/HeJ macrophages (Fig. 4) is consistent with the involvement of a TLR ligand. The relevant bacterial molecule is unlikely to be flagellin based on the fact that flagellin-deficient Salmonella mutants activated signaling in C3H/HeJ macrophages as well as the wild-type strain (Fig. 2D). Bacterial DNA is also not likely to be involved since pretreatment of macrophages with chloroquin to inhibit activation of TLR9 did not affect signaling or TNF-α production (data not shown). A TLR2 ligand remains a reasonable candidate, and indeed, in preliminary experiments we detected TLR2 agonist activity in Salmonella culture supernatants (unpublished data). This activity may correspond to a previously described outer membrane lipoprotein released by several types of enterobacteria, including Salmonella, that was shown to activate C3H/HeJ macrophages to produce interleukin-6 (56). It may also be related to a recently identified PG-associated lipoprotein that is released by several gram-negative bacteria and that is able to activate C3H/HeJ macrophages (17). A membrane lipoprotein that is released in a soluble form would be consistent with our detection of TLR4-independent activity associated with both bacteria and bacterial supernatants. If the relevant Salmonella molecule does in fact engage TLR2, it is surprising that it is such a poor inducer of TNF-α production compared to PG, another TLR2 ligand (Fig. 1). This difference may be attributable to the amount of the molecule available for activation of TLR2. Alternatively, since it is known that different TLR2 ligands engage different TLR2 heterodimers (33), it is possible that PG activates one type of heterodimer, while the Salmonella molecule activates another. Future experiments will be directed at addressing some of these issues by identifying the Salmonella molecules involved in TLR4-independent activation of macrophages and clarifying their roles in pathogenesis.
Acknowledgments
We are grateful to Andrew Gewirtz and David Rudner for providing bacterial strains and to Beth McCormick, Cathryn Nagler-Anderson, Tor Savidge, and Shiv Pillai for critical reviews of the manuscript.
This work was supported by the National Institutes of Health (grant R01 AI48815).
Editor: A. D. O'Brien
REFERENCES
- 1.Ahmad-Nejad, P., H. Hacker, M. Rutz, S. Bauer, R. M. Vabulas, and H. Wagner. 2002. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur. J. Immunol. 32:1958-1968. [DOI] [PubMed] [Google Scholar]
- 2.Beutler, B., N. Krochin, I. W. Milsark, C. Luedke, and A. Cerami. 1986. Control of cachectin (tumor necrosis factor) synthesis: mechanism of endotoxin resistance. Science 232:977-980. [DOI] [PubMed] [Google Scholar]
- 3.Bohuslav, J., V. V. Kravchenko, G. C. N. Parry, J. H. Erlich, S. Gerondakis, N. Mackman, and R. J. Ulevitch. 1998. Regulation of an essential innate immune response by the p50 subunit of NF-κB. J. Clin. Investig. 102:1645-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherayil, B. J., and D. Antos. 2001. Inducible nitric oxide synthase and Salmonella infection. Microbes Infect. 3:771-776. [DOI] [PubMed] [Google Scholar]
- 5.Cherayil, B. J., B. A. McCormick, and J. Bosley. 2000. Salmonella enterica serovar Typhimurium-dependent regulation of iNOS expression in macrophages by invasins SipB, SipC, and SipD and effector SopE2. Infect. Immun. 68:5567-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, S. H., and H. E. Broxmeyer. 2000. Measurement of IL-3 and other hematopoietic cytokines such as GM-CSF, G-CSF, M-CSF, erythropoietin, Steele factor and Flt-3 ligand, p. 6.4.1-6.4.20. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology, vol. 1. John Wiley and Sons, New York, N.Y. [DOI] [PubMed]
- 7.Dobrovolskaia, M. A., A. E. Medvedev, K. E. Thomas, N. Cuesta, V. Toshchakov, T. Ren, M. J. Cody, S. M. Michalek, N. R. Rice, and S. N. Vogel. 2003. Induction of in vitro reprogramming by Toll-like receptor (TLR)2 and TLR4 agonists in murine macrophages: effects of TLR “homotolerance” versus “heterotolerance” on NF-κB signaling pathway components. J. Immunol. 170:508-519. [DOI] [PubMed] [Google Scholar]
- 8.Doyle, S. E., S. A. Vaidya, R. O. O'Connell, H. Dadgostar, P. W. Dempsey, T.-T. Wu, G. Rao, R. Sun, M. E. Haberland, R. L. Modlin, and G. Cheng. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17:251-263. [DOI] [PubMed] [Google Scholar]
- 9.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J.-H. Lin, C. Patriotis, N. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 10.Eisenstein, T. K., L. M. Killar, B. A. Stocker, and B. M. Sultzer. 1984. Cellular immunity induced by avirulent Salmonella in LPS-defective C3H/HeJ mice. J. Immunol. 133:958-961. [PubMed] [Google Scholar]
- 11.Eisenstein, T. K., L. W. Deakins, L. Killar, P. H. Saluk, and B. M. Sultzer. 1982. Dissociation of innate susceptibility to Salmonella infection and endotoxin responsiveness in C3HeB/FeJ mice and other strains in the C3H lineage. Infect. Immun. 36:696-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdman, S. E., J. G. Fox, C. A. Dangler, D. Feldman, and B. H. Horwitz. 2001. Cutting edge: typhlocolitis in NF-κB-deficient mice. J. Immunol. 166:1443-1447. [DOI] [PubMed] [Google Scholar]
- 13.Fortier, A. H., and L. A. Falk. 1994. Isolation of murine macrophages, p. 14.1.1-14.1.9. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology, vol. 3. John Wiley and Sons, New York, N.Y. [DOI] [PubMed]
- 14.Fortier, A. H., D. L. Hoover, and C. A. Nacy. 1982. Intracellular replication of Leishmania tropica in mouse peritoneal macrophages: amastigote infection of resident cells and inflammatory exudate macrophages. Infect. Immun. 38:1304-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gewirtz, A. T., P. O. Simon, Jr., C. K. Schmitt, L. J. Taylor, C. H. Hagedorn, A. D. O'Brien, A. S. Neish, and J. L. Madara. 2001. Salmonella Typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J. Clin. Investig. 107:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, S. 2002. Pattern recognition receptors: doubling up for the innate immune response. Cell 111:927-930. [DOI] [PubMed] [Google Scholar]
- 17.Hellman, J., J. D. Roberts, M. M. Tehan, J. E. Allaire, and H. S. Warren. 2002. Bacterial peptidoglycan-associated lipoprotein is released into the bloodstream in Gram-negative sepsis and causes inflammation and death in mice. J. Biol. Chem. 277:14274-14280. [DOI] [PubMed] [Google Scholar]
- 18.Hirschfeld, M., J. J. Weis, V. Toshchakov, C. A. Salkowski, M. J. Cody, D. C. Ward, N. Qureshi, S. M. Michalek, and S. N. Vogel. 2001. Signaling by Toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoover, D. L., and C. A. Nacy. 1984. Macrophage activation to kill Leishmania tropica: defective killing of amastigotes by macrophages elicited by sterile inflammatory agents. J. Immunol. 132:1487-1491. [PubMed] [Google Scholar]
- 20.Horng, T., G. M. Barton, R. A. Flavell, and R. Medzhitov. 2002. The adaptor molecule TIRAP provides signaling specificity for Toll-like receptors. Nature 420:329-333. [DOI] [PubMed] [Google Scholar]
- 21.House, D., A. Bishop, C. Parry, G. Dougan, and J. Wain. 2001. Typhoid fever: pathogenesis and disease. Curr. Opin. Infect. Dis. 14:573-578. [DOI] [PubMed] [Google Scholar]
- 22.Hueck, C. J., M. J. Hantman, V. Bajaj, C. Johnston, C. A. Lee, and S. I. Miller. 1995. Salmonella Typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol. Microbiol. 18:479-490. [DOI] [PubMed] [Google Scholar]
- 23.Jones, B. W., T. K. Means, K. A. Heldwein, M. A. Keen, P. J. Hill, J. T. Belisle, and M. J. Fenton. 2001. Different Toll-like receptor agonists induce distinct macrophage responses. J. Leukoc. Biol. 69:1036-1044. [PubMed] [Google Scholar]
- 24.Kastenbauer, S., and H. W. L. Ziegler-Heitbrock. 1999. NF-κB1 (p50) is upregulated in lipopolysaccharide tolerance and can block tumor necrosis factor gene expression. Infect. Immun. 67:1553-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killar, L. M., and T. K. Eisenstein. 1985. Immunity to Salmonella Typhimurium infection in C3H/HeJ and C3H/HeNCrlBR mice: studies with an aromatic-dependent live S. Typhimurium strain as a vaccine. Infect. Immun. 47:605-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinjyo, I., T. Hanada, K. Inagaki-Ohara, H. Mori, D. Aki, M. Ohishi, H. Yoshida, M. Kubo, and A. Yoshimura. 2002. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 17:583-591. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, K., L. D. Hernandez, J. E. Galan, C. A. Janeway, Jr., R. Medzhitov, and R. Flavell. 2002. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 110:191-202. [DOI] [PubMed] [Google Scholar]
- 28.Kontoyiannis, D., A. Kotlyarov, E. Carballo, L. Alexopoulou, P. J. Blackshear, M. Gaestel, R. Davis, R. Flavell, and G. Kollias. 2001. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 20:3760-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lalmanach, A.-C., and F. Lantier. 1999. Host cytokine response and resistance to Salmonella infection. Microbes Infect. 1:719-726. [DOI] [PubMed] [Google Scholar]
- 31.Leadbetter, E. A., I. R. Rifkin, A. M. Hohlbaum, B. C. Beaudette, M. J. Shlomchik, and A. Marshak-Rothstein. 2002. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 416:603-607. [DOI] [PubMed] [Google Scholar]
- 32.Li, L., S. Cousart, J. Hu, and C. E. McCall. 2000. Characterization of interleukin-1 receptor-associated kinase in normal and endotoxin-tolerant cells. J. Biol. Chem. 275:23340-23345. [DOI] [PubMed] [Google Scholar]
- 33.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135-145. [DOI] [PubMed] [Google Scholar]
- 34.Mizel, S. B., and J. A. Snipes. 2002. Gram-negative flagellin-induced self-tolerance is associated with a block in interleukin-1 receptor-associated kinase release from Toll-like receptor 5. J. Biol. Chem. 277:22414-22420. [DOI] [PubMed] [Google Scholar]
- 35.Nacy, C. A., C. N. Oster, S. L. James, and M. S. Meltzer. 1984. Activation of macrophages to kill Rickettsia and Leishmania: dissociation of intracellular microbicidal activities and extracellular destruction of neoplastic and helminth targets. Contemp. Top. Immunobiol. 13:147-153. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa, R., T. Naka, H. Tsutsui, M. Fujimoto, A. Kimura, T. Abe, E. Seki, S. Sato, O. Takeuchi, K. Takeda, S. Akira, K. Yamanishi, I. Kawase, K. Nakanishi, and T. Kishimoto. 2002. SOCS1 participates in negative regulation of LPS responses. Immunity 17:677-687. [DOI] [PubMed] [Google Scholar]
- 37.Nomura, F., S. Akashi, Y. Sakao, S. Sato, T. Kawai, M. Matsumoto, K. Nakanishi, M. Kimoto, K. Miyake, K. Takeda, and S. Akira. 2000. Cutting edge: endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface Toll-like receptor 4 expression. J. Immunol. 164:3476-3479. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien, A. D., E. S. Metcalf, and D. L. Rosenstreich. 1982. Defect in macrophage effector function confers Salmonella Typhimurium susceptibility on C3H/HeJ mice. Cell. Immunol. 67:325-333. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien, A. D., D. L. Rosenstreich, I. Scher, G. H. Campbell, R. P. MacDermott, and S. B. Formal. 1980. Genetic control of susceptibility to Salmonella Typhimurium in mice: role of the LPS gene. J. Immunol. 124:20-24. [PubMed] [Google Scholar]
- 40.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in the Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 41.Raabe, T., M. Bukrinsky, and R. A. Currie. 1998. Relative contribution of transcription and translation to the induction of tumor necrosis factor-alpha by lipopolysaccharide. J. Biol. Chem. 273:974-980. [DOI] [PubMed] [Google Scholar]
- 42.Rabsch, W., H. Tschäpe, and A. J. Bäumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237-247. [DOI] [PubMed] [Google Scholar]
- 43.Re, F., and J. L. Strominger. 2001. Toll-like receptor 2 (TLR2) and TLR4 differentially activate human dendritic cells. J. Biol. Chem. 276:37692-37699. [DOI] [PubMed] [Google Scholar]
- 44.Rescigno, M., M. Urbano, M. Rimoldi, B. Valzasina, G. Rotta, F. Granucci, and P. Ricciardi-Castagnoli. 2002. Toll-like receptor 4 is not required for the full maturation of dendritic cells or for the degradation of Gram-negative bacteria. Eur. J. Immunol. 32:2800-2806. [DOI] [PubMed] [Google Scholar]
- 45.Salcedo, S. P., M. Noursadeghi, J. Cohen, and D. W. Holden. 2001. Intracellular replication of Salmonella Typhimurium strains in specific subsets of splenic macrophages in vivo. Cell. Microbiol. 3:587-597. [DOI] [PubMed] [Google Scholar]
- 46.Schafer, R., C. A. Nacy, and T. K. Eisenstein. 1988. Induction of activated macrophages in C3H/HeJ mice by avirulent Salmonella. J. Immunol. 140:1638-1644. [PubMed] [Google Scholar]
- 47.Shurety, W., A. Merino-Trigo, D. Brown, D. A. Hume, and J. L. Stow. 2000. Localization and post-Golgi trafficking of tumor necrosis factor-alpha in macrophages. J. Interferon Cytokine Res. 20:427-438. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 49.Toshchakov, V., B. W. Jones, P.-Y. Perera, K. Thomas, J. M. Cody, S. Zhang, B. R. G. Williams, J. Major, T. A. Hamilton, M. J. Fenton, and S. N. Vogel. 2002. TLR4, but not TLR2, mediates IFN-β-induced STAT1α/β-dependent gene expression in macrophages. Nat. Immunol. 3:392-398. [DOI] [PubMed] [Google Scholar]
- 50.Vazquez-Torres, A., and F. C. Fang. 2001. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 9:29-33. [DOI] [PubMed] [Google Scholar]
- 51.Wedel, A., M. Frankenberger, G. Sulski, I. Petersmann, D. Kuprash, S. Nedospasov, and H. W. Ziegler-Heitbrock. 1999. Role of p52 (NF-κB2) in LPS tolerance in a human B cell line. Biol. Chem. 380:1193-1199. [DOI] [PubMed] [Google Scholar]
- 52.Weinstein, D. L., C. R. Lissner, R. N. Swanson, and A. D. O'Brien. 1986. Macrophage defect and inflammatory cell recruitment dysfunction in Salmonella susceptible C3H/HeJ mice. Cell. Immunol. 102:68-77. [DOI] [PubMed] [Google Scholar]
- 53.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella Typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 54.Yamamoto, M., S. Sato, H. Hemmi, H. Sanjo, S. Uematsu, T. Kaisho, K. Hoshino, O. Takeuchi, M. Kobayashi, T. Fujita, K. Takeda, and S. Akira. 2002. Essential role for TIRAP in activation of the signaling cascade shared by TLR2 and TLR4. Nature 420:324-328. [DOI] [PubMed] [Google Scholar]
- 55.Young, D., T. Hussell, and G. Dougan. 2002. Chronic bacterial infections: living with unwanted guests. Nat. Immunol. 3:1026-1032. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, H., D. W. Niesel, J. W. Peterson, and G. R. Klimpel. 1998. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect. Immun. 66:5196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]