Abstract
FkpA is a peptidylprolyl isomerase whose expression is regulated by the alternative sigma factor, sigma factor E (σE). In contrast to the results of a previous report, inactivation of fkpA was found to have only a minor effect on the ability of Salmonella enterica serovar Typhimurium to invade and survive within epithelial and macrophage cell lines and cause infection in mice. However, an effect of the fkpA mutation on serovar Typhimurium virulence was seen if the mutation was combined with mutations in surA or htrA, two other σE-regulated genes, which encode proteins involved in protein folding and/or degradation in the periplasm.
Salmonella spp. possess a number of regulatory systems that allow them to sense and adapt to adverse conditions, primarily by altering the expressions of particular genes. One such system that is important for survival of Salmonella enterica serovar Typhimurium in vivo and in the environment is controlled by the alternative sigma factor RpoE (σE) (10). Two σE-regulated genes have been shown to be involved in Salmonella virulence, htrA (also known as degP) (10, 11) and surA (10, 14). Inactivation of rpoE has a far greater effect on serovar Typhimurium in vitro and in vivo than does inactivation of htrA or surA (10, 14). This indicates that other genes in the RpoE regulon may have a role in serovar Typhimurium virulence.
A promising candidate for another σE-regulated gene involved in Salmonella virulence is fkpA. FkpA is a periplasmic peptidylprolyl isomerase (PPIase). PPIases assist protein folding by catalyzing the normally slow cis-trans isomerization of prolyl residues in polypeptides (7). FkpA, like HtrA and SurA, has recently been shown to possess chaperone-like activity independent of its enzymatic activity (1, 2, 13). FkpA homologues are important for infection of host cells by other intracellular pathogens such as Legionella pneumophila and Chlamydia trachomatis (7, 9). In a previous study, an fkpA mutant of serovar Typhimurium biotype Copenhagen survived less well in macrophages and epithelial cells in vitro than did the parental wild-type (WT) strain (8). However, the role of fkpA in the virulence of serovar Typhimurium was not studied. Here we investigate the effect of inactivation of fkpA, alone or in combination with mutations in other σE-regulated genes, on the survival and invasion of serovar Typhimurium in different cell lines and in mice.
The complete serovar Typhimurium fkpA gene was isolated by PCR using primers designed from the Escherichia coli fkpA sequence. The predicted serovar Typhimurium FkpA protein exhibited 90% amino acid identity with its E. coli counterpart (data not shown). This finding was confirmed with the publication of the complete genome sequence of serovar Typhimurium LT2 (12). The −35, −10, and intervening sequences of the E. coli σE-dependent promoter of fkpA were 100% conserved in the upstream region of the serovar Typhimurium fkpA gene (4) (data not shown). The fkpA gene was insertionally inactivated by introduction of a Km resistance cassette via a unique PstI site within fkpA (nucleotide 306, amino acid 102). This construct was used to produce a serovar Typhimurium SL1344 fkpA mutant (GVB387) by allelic exchange using the suicide vector pRDH10 (10). The disruption of fkpA was confirmed by PCR and Southern hybridization (data not shown). We also constructed a serovar Typhimurium SL1344 htrA fkpA double mutant (GVB388) by P22 transduction of fkpA::Kmr into the serovar Typhimurium SL1344 ΔhtrA strain BRD915 (6).
To examine whether FkpA is involved in the virulence of serovar Typhimurium, we determined whether the 50% lethal dose of GVB387 (fkpA mutant) following oral and parenteral (intravenous) infection differed significantly from that of the WT strain. There was no significant difference in the 50% lethal doses, or the times to death, of the WT and fkpA mutant strains by either route of infection (data not shown). To analyze further whether FkpA is involved in serovar Typhimurium virulence, the abilities of the WT and fkpA mutant strains to compete for growth in murine tissues were compared by competition assay (3). Three days after intraperitoneal inoculation, there were approximately twice as many CFU of the WT strain as of the fkpA mutant in the livers and spleens of mice, although this difference was not statistically significant (Table 1). However, the htrA fkpA double mutant exhibited a significant (P < 0.05) increase in attenuation over the single htrA mutant (competitive index [CI], 0.289) (Table 1).
TABLE 1.
Competition assay with serovar Typhimurium strainsa
| Strains | CI |
|---|---|
| fkpA mutant versus SL1344 | 0.6c |
| fkpA htrA mutant versus htrA mutant | 0.289b |
| fkpA mutant versus C5 | 1.1c |
| surA fkpA mutant versus C5 | 0.0121b |
| surA fkpA mutant versus surA mutant | 0.464b |
Mice were infected intraperitoneally with an inoculum containing ∼103 CFU of both strains. Three days later, mice were killed and the number of CFU of each strain in the liver and spleen was determined. The CI was determined using the following formula: (input CFU of first strain/input CFU of second strain)/(output CFU of first strain/output CFU of second strain). A Student's t test was used to determine whether the difference in the mean number of CFU recovered compared with the number of CFU inoculated for the pair of strains in each CI test was statistically significant. This was calculated by comparing the means of the ratio of the number of CFU recovered/the number of CFU administered for each strain.
The difference in the recoveries of the two strains from murine tissue was significant (P < 0.05).
The difference in the recoveries of the two strains from murine tissue was not significant.
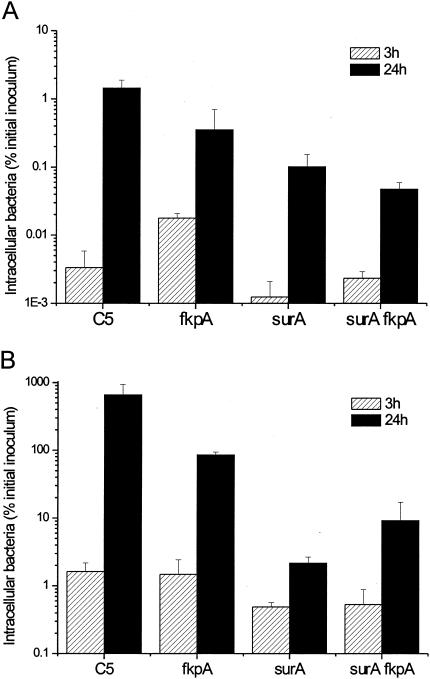
Previously, it has been reported that FkpA was required for the survival of a serovar Typhimurium Copenhagen strain in macrophage (J774.A1) and epithelial (Caco-2) cell lines (8). We investigated whether our serovar Typhimurium fkpA mutant exhibited a defect in invasion or intracellular survival in the murine macrophage cell line RAW264.7 and the human epithelial cell lines Hep-2 and Caco-2. Both the fkpA and htrA fkpA mutants were found to invade RAW264.7 cells at a similar rate as SL1344 and BRD915 (htrA mutant), and thereafter all of the aforementioned strains grew intracellularly and there was no major difference in the numbers of bacteria isolated from the cells after 24 h (data not shown). A similar result was obtained with Hep-2 and Caco-2 cells (data not shown). The effect of inactivation of fkpA on the interaction of serovar Typhimurium with eukaryotic cells is contrary to that reported by Horne et al. (8). This may be due to the different genetic backgrounds of the bacterial strains used in the two studies. To ascertain whether this was the case, the fkpA::Kmr mutation was transduced into a second WT serovar Typhimurium strain, C5, to generate strain GVB844. The survival of this mutant was analyzed in Caco-2 and RAW264.7 cells. In this case, there was a small, but significant (P < 0.05), effect of the fkpA mutation, as in both cell lines the number of GVB844 cells recovered after 24 h was lower than the number for C5 (Fig. 1).
FIG. 1.
Effect of mutations in fkpA and surA on the ability of serovar Typhimurium C5 to invade and replicate in epithelial and macrophage cell lines. The abilities of the different serovar Typhimurium strains to invade and grow in eukaryotic cells were examined using Caco-2 (A) and RAW264.7 (B) cells. Cells were infected with bacteria at a multiplicity of infection of ∼1:1, and the assay was performed as described previously (10). The graphs indicate the viable bacteria recovered from inside the cells (as a percentage of the initial inoculum) at 3 and 24 h after infection. Each bar represents the mean number of CFU from triplicate experiments, and the error bars indicate standard deviations.
E. coli possesses four genes encoding periplasmic PPIases, fkpA, surA, ppiA, and ppiD. In a search of the serovar Typhimurium genome database, a homologue for each of these genes was identified. It is possible that loss of FkpA may be compensated by the presence of other periplasmic PPIases. SurA has been shown to be involved in serovar Typhimurium pathogenesis, and recently its expression has been found to be regulated by σE in E. coli (5). We wished to examine whether a role for FkpA in serovar Typhimurium physiology in vivo and in vitro would be revealed in a strain that also lacked SurA. A surA fkpA double mutant was constructed by P22 transduction of the fkpA::Kmr mutation from GVB387 into the C5 ΔsurA strain BRD115 (14). The surA mutant invaded and survived less well than did C5 in both Caco-2 and RAW264.7 cells (24 h; P < 0.05) (Fig. 1), which is in agreement with the results of previous work (14). Inactivation of fkpA did not further reduce the ability of the surA mutant to invade or grow within eukaryotic cells (3 and 24 h; P < 0.05) (Fig. 1).
A competition assay was used to examine whether inactivation of fkpA decreased the ability of a surA mutant to grow and survive in murine tissues. The CI for the surA fkpA mutant versus the surA mutant was 0.464 (P < 0.05). This indicates that lack of fkpA in a strain that also lacks surA does have a small effect on the ability of serovar Typhimurium to grow in vivo. The CI for the surA fkpA mutant versus C5 was 0.0121 (Table 1), indicating that the double mutant is highly attenuated (P < 0.05) compared with its parental WT strain. It was not possible to perform a competition experiment with the WT and surA mutant strains because the mutant lacks a selectable marker.
Contrary to the results of a previous report, this study indicates that FkpA is not crucial for normal survival or growth of serovar Typhimurium within phagocytic or nonphagocytic cells in vitro. FkpA is also dispensable for oral or parenteral infection of mice. A small effect on the in vivo growth of a serovar Typhimurium fkpA mutant was evident if the strain also lacked either SurA or HtrA, both of which are also involved in protein folding in the periplasm of serovar Typhimurium. We are currently identifying other members of the σE regulon of serovar Typhimurium and investigating their importance for Salmonella pathogenesis.
Acknowledgments
This work was supported by grants from the BBSRC (no. 17/PRS12222) and the NSF (no. MCB-9985987).
Editor: B. B. Finlay
REFERENCES
- 1.Arie, J. P., N. Sassoon, and J. M. Betton. 2001. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol. Microbiol. 39:199-210. [DOI] [PubMed] [Google Scholar]
- 2.Behrens, S., R. Maier, H. de Cock, F. X. Schmid, and C. A. Gross. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 4.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 5.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 6.Everest, P., G. Frankel, J. Li, P. Lund, S. Chatfield, and G. Dougan. 1995. Expression of LacZ from the htrA, nirB and groE promoters in a Salmonella vaccine strain: influence of growth in mammalian cells. FEMS Microbiol. Lett. 126:97-101. [DOI] [PubMed] [Google Scholar]
- 7.Hacker, J., and G. Fischer. 1993. Immunophilins: structure-function relationship and possible role in microbial pathogenicity. Mol. Microbiol. 10:445-456. [DOI] [PubMed] [Google Scholar]
- 8.Horne, S. M., T. J. Kottom, L. K. Nolan, and K. D. Young. 1997. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium Copenhagen. Infect. Immun. 65:806-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horne, S. M., and K. D. Young. 1995. Escherichia coli and other species of the Enterobacteriaceae encode a protein similar to the family of Mip-like FK506-binding proteins. Arch. Microbiol. 163:357-365. [DOI] [PubMed] [Google Scholar]
- 10.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, E, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 12.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 13.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 14.Sydenham, M., G. Douce, F. Bowe, S. Ahmed, S. Chatfield, and G. Dougan. 2000. Salmonella enterica serovar Typhimurium surA mutants are attenuated and effective live oral vaccines. Infect. Immun. 68:1109-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]