Abstract
Pseudomonas aeruginosa exoenzyme S (ExoS) is a type III secretion (TTS) effector, which includes both a GTPase-activating protein (GAP) activity toward the Rho family of low-molecular-weight G (LMWG) proteins and an ADP-ribosyltransferase (ADPRT) activity that targets LMWG proteins in the Ras, Rab, and Rho families. The coordinate function of both activities of ExoS in J774A.1 macrophages was assessed by using P. aeruginosa strains expressing and translocating wild-type ExoS or ExoS defective in GAP and/or ADPRT activity. Distinct and coordinated functions were identified for both domains. The GAP activity was required for the antiphagocytic effect of ExoS and was linked to interference of lamellopodium and membrane ruffle formation. Alternatively, the ADPRT activity of ExoS altered cellular adherence and morphology and was linked to effects on filopodium formation. The cellular mechanism of ExoS GAP activity included an inactivation of Rac1 function, as determined in p21-activated kinase 1-glutathione S-transferase (GST) pull-down assays. The ADPRT activity of ExoS targeted Ras and RalA but not Rab or Rho proteins, and Ral binding protein 1-GST pull-down assays identified an effect of ExoS ADPRT activity on RalA activation. The results from these studies confirm the bifunctional nature of ExoS activity within macrophages when translocated by TTS.
Pseudomonas aeruginosa is an opportunistic pathogen that causes life-threatening infections in burn patients and exacerbates lung problems in individuals with cystic fibrosis. One of its virulence factors, exoenzyme S (ExoS), has been proposed to act as an antiphagocytic factor (9), thus enabling the bacteria to evade the host immune system. ExoS was characterized as an ADP-ribosylating enzyme by Iglewski et al. (18), and subsequent studies localized this activity to its carboxy terminus (20). After its translocation into human epithelial cells by the contact-dependent type III secretion (TTS) apparatus (40), the ADP-ribosyltransferase (ADPRT) activity of ExoS targets multiple substrates, including the low-molecular-weight G (LMWG) proteins Ras, RalA, certain Rab proteins, Rac1, and Cdc42 (6, 8, 17, 25). More recently, ExoS was found to also include a GTPase-activating protein (GAP) activity that targets the LMWG proteins Rho, Rac1, and Cdc42, which affect eukaryotic cell cytoskeletal structure (13). TTS-translocated ExoS can exert complex effects on eukaryotic cell function, including inhibition of DNA synthesis, alterations in cell morphology, microvillus effacement, and loss of cellular adherence, in addition to its antiphagocytic or anti-invasive effects (7, 9, 27, 28).
The GAP and ADPRT domains of ExoS both target LMWG proteins, which serve as molecular switches for a number of eukaryotic cell processes, such as growth, differentiation, phagocytosis, and apoptosis. LMWG proteins cycle between a GTP-bound, active state and a GDP-bound, inactive state. The low intrinsic GTP hydrolysis and GDP exchange reactions of G proteins are modulated by GAPs and guanine nucleotide exchange factors (GEFs), respectively. Many bacterial toxins have been found to modify the function of GTP binding proteins and, in turn, affect signal transduction in eukaryotic cells. Toxins can modulate LMWG protein function through modifications, such as ADP-ribosylation or glucosylation, or by acting as GEFs or GAPs, thus affecting the protein's GTP-bound state. Examples of such toxins include Clostridium botulinum C3-like exoenzymes, which ADP-ribosylate Rho (1); Clostridium difficile toxins, which monoglucosylate Rho, Rac1, and Cdc42 (19); and, as discussed here, P. aeruginosa ExoS, which ADP-ribosylates Ras, RalA, Rabs, and Rac1 (5, 8, 25). Other bacterial toxins, such as Salmonella enterica serovar Typhimurium SopE, act as GEFs for Rho proteins (15), while SptP, Yersinia YopE, and P. aeruginosa ExoS act as GAPs for Rho family LMWG proteins (10, 13, 39).
The dynamics of the eukaryotic cell actin cytoskeleton are controlled by the Rho family of LMWG proteins, with Rho regulating the formation of focal adhesions, Rac1 regulating the formation of membrane ruffles and lamellipodia, and Cdc42 regulating the formation of filopodia (14). While the association of these structures with the activation of specific LMWG proteins was initially described for fibroblasts, similar associations of Rac1 activation with lamellipodium formation and of Cdc42 activation with filopodium formation have been recognized in macrophages, while Rho regulates cell contractility and assembly of actin cables (2). Rho GTPases are also involved in phagocytosis. Two different types of phagocytosis have been described. Type I requires Rac1 and Cdc42 and is mediated by the Fc gamma receptor, and type II requires Rho and is mediated by the complement receptor type 3 (3). With specific regard to Pseudomonas, Rac1 and Cdc42 have been found to be required for phagocytosis of unopsonized P. aeruginosa by murine macrophages (23).
The bifunctionality of ExoS, coupled with its targeting of multiple LMWG proteins, provides an explanation for the complex and diverse effects of ExoS on cell function. In understanding the relative function of each domain of ExoS with regard to its cellular effects, it was recognized that the host cell can influence the toxic effects of ExoS when it is internalized by the TTS process (7, 34). For example, when GAP or ADPRT mutant forms of ExoS were internalized into HT-29 epithelial cells by TTS, minimal cytotoxic affects were linked to the GAP domain. Rather, it was the ADPRT domain that was required for the severe effects of ExoS on cell growth and morphology (7). The lack of a pronounced cytotoxic effect of ExoS GAP activity in epithelial cells prompted investigation of the bifunctional nature of ExoS activity in other cell types. Since previous studies linked the antiphagocytic effect of ExoS to its GAP activity (9), we chose to examine the bifunctionality of the ExoS GAP and ADPRT domains in J774A.1 macrophages internalized by the P. aeruginosa TTS process. By using P. aeruginosa strains expressing ExoS defective in the GAP and/or ADPRT activity, a dual effect of ExoS on J774A.1 macrophage function was identified, with the ADPRT activity of ExoS affecting morphology and adherence, while the GAP activity interfered with phagocytosis.
MATERIALS AND METHODS
Bacterial strains and culture media.
P. aeruginosa strain PA103 ΔexoU ΔexoT::Tc (TTS+ ExoS− ExoY− ExoU− ExoT− ETA+) (strain PA103ΔUT), a derivative of strain PA103 from which the exoU and exoT genes were deleted (37), was used as a host strain to translocate ExoS and ExoS mutants into eukaryotic cells by the TTS process. The pUCP vector (35), containing the exoS gene from P. aeruginosa strain 388 (18, 22) (pUCP-ExoS [ExoS]), was used for the construction of the ExoS mutants described in Table 1. Both bacterial strains were kindly provided by Dara Frank (Medical College of Wisconsin). All bacterial strains were grown in ExoS induction medium (TSBD-N) (18) for 16 h, as previously described (28). For bacterial-eukaryotic cell coculture experiments, bacteria were diluted, based on the culture optical density at 590 nm, to approximately 107 CFU/ml in Dulbecco's modified Eagle medium (DMEM) (Gibco-BRL, Gaithersburg, Md.) containing 0.6% bovine serum albumin (BSA) (Sigma, St. Louis, Mo.) (DMEM-BSA).
TABLE 1.
Domain functions of ExoS plasmid constructs
| Plasmid | GAP activity | ADPRT activity | Reference |
|---|---|---|---|
| pUCP | − | − | 35 |
| ExoS | + | + | 22 |
| R146A | − | + | 7 |
| E379A/E381A | + | − | 7 |
| R146A/E379A/E381A | − | − | 7 |
Eukaryotic cell culture conditions and coculture with bacteria.
The J774A.1 murine macrophage-like cell line (ATCC TIB-67; American Type Culture Collection, Manassas, Va.) was maintained in DMEM supplemented with 10% fetal bovine serum (Gibco-BRL) at 37°C in 5% CO2-95% air. For bacterial infection studies, J774A.1 cells were seeded at a density of 1 × 105 to 2 × 105 cells/ml and grown for 72 h. At this time, the culture medium was removed, cells were washed once with phosphate-buffered saline (PBS), and DMEM-BSA containing no bacteria or containing 107 CFU of strain PA103ΔUT expressing either (i) wild-type ExoS, (ii) an R146A ExoS GAP mutant, (iii) an E379A/E381A ExoS ADPRT mutant, (iv) an R146A/E379A/E381A ExoS GAP/ADPRT mutant, or (v) a pUCP vector control (Table 1) per ml was added. Bacteria were added to cell monolayers at a multiplicity of infection of 100 and incubated for the indicated time periods at 37°C in 5% CO2-95% air.
Cell viability and adherence assays.
Following coculture with bacteria for 2, 4, or 6 h, nonadherent J774A.1 cells were recovered and washed once with PBS. Adherent cells were washed twice with PBS and then detached by scraping into PBS. Both nonadherent and adherent populations were quantified with a hemacytometer and assayed for viability based on trypan blue exclusion.
Cell morphology analysis.
To determine changes in cell morphology of J774A.1 cells due to ExoS activity, cells were analyzed by phase-contrast microscopy (Axiovert 200 microscope; Carl Zeiss Inc, Jena, Germany) after 5 h of coculture with the different strains. Briefly, bacteria were removed, cells were washed once with PBS, and then DMEM-10% fetal bovine serum containing 200 μg of gentamicin per ml was added to limit bacterial growth during microscopic examination.
SEM.
To examine differential effects of ExoS mutants on cell morphology by scanning electron microscopy (SEM), J774A.1 cells were seeded and cultured, as described above, on Thermanox coverslips (Nalge Nunc, Naperville, Ill.) and then cocultured with the different PA103ΔUT strains for 2.5 h. Following removal of medium containing bacteria, cells were fixed with 2% cacodylate glutaraldehyde for 30 min as the primary fixative and with 2% aqueous osmium tetroxide for 30 min as the secondary fixative. Samples were rinsed with distilled water, dehydrated with 100% ethanol, and incubated with hexamethyldisilazane (Sigma) until dry. The samples were mounted, sputter coated with gold palladium (20-nm coating), and examined with a JEOL 5410 scanning electron microscope.
Double-immunofluorescence assay of bacterial phagocytosis.
J774A.1 cells were seeded and cultured, as described above, in chamber slides (Nalge Nunc) prior to culture for 2.5 h with no bacteria or with PA103ΔUT expressing the plasmids described in Table 1. Cells were washed twice with PBS and fixed with 3% paraformaldehyde in PBS for 30 min at room temperature. Once fixed, the cells were washed twice with PBS and blocked for 1 h at 4°C in blocking buffer (PBS, 1% BSA, 5% normal goat serum [Vector Laboratories, Burlingame, Calif.]). Bacteria were first stained for 1 h at room temperature with guinea pig anti-P. aeruginosa immunoglobulin G (IgG) polyclonal antibody (Biogenesis, Kensington, N.H.) diluted 1:250 in blocking buffer. Cells were washed with PBS and incubated for 1 h at room temperature with goat anti-guinea pig IgG-SP-biotin conjugate (Jackson ImmunoResearch Laboratories, West Grove, Pa.) diluted 1:250 in antibody diluent (Dako, Carpinteria, Calif.). The cells were then washed with PBS and incubated for 1 h at room temperature with Extravidin-fluorescein isothiocyanate conjugate (Sigma) diluted 1:500 in antibody diluent. For the second staining, cells were first washed with PBS, refixed with 3% paraformaldehyde, washed twice with PBS, and permeabilized with chilled 100% methanol for 10 min at −20°C. Once permeabilized, the cells were blocked overnight at 4°C in blocking buffer. Bacteria were stained as described above, except that antibody and antibody conjugate were diluted 1:1,000 and the second conjugate used was Extravidin-tetramethyl rhodamine isothiocyanate (Sigma). Slides were mounted in mounting medium (Dako) and examined by fluorescence microscopy with an Axioplan research microscope (Carl Zeiss Inc.). Controls with nonpermeabilized cells were included to confirm the specificity of the second staining procedure.
Analysis of ADP-ribosylation of eukaryotic LMWG proteins by TTS-translocated ExoS.
ADP-ribosylation of endogenous LMWG proteins by ExoS was assessed based on alterations in protein mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as previously described (25). Following a 2.5- to 5-h coculture period with bacteria, J774A.1 cells were washed with PBS and lysed for 30 min on ice with lysis buffer (30 mM HEPES [pH 7.5], 1% Triton X-100, 10 mM NaCl, 10% glycerol, 1 mM EGTA, 25 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate, 10 mM benzamidine, 10 μg [each] of leupeptin and aprotinin per ml) (Sigma). For the analyses of Ras modification by ExoS, Ras was immunoprecipitated from cell lysates by using monoclonal Y13-259 Ras antibody (American Type Culture Collection) and protein G-agarose (Sigma) as previously described (25). All samples were resolved by SDS-12% PAGE, transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.), and immunoblotted for (i) Ras, using anti-pan-Ras LA045 antibody (Quality Biotech, Camden, N.J.); (ii) RalA, Rab8, Rab11, Rac1, Cdc42, or Rho (A, B, and C), using specific monoclonal antibodies against the respective proteins (Transduction Laboratories, Lexington, Ky.); or (iii) Rab5 and Rab7, using the respective polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.). Proteins were visualized by enhanced chemiluminescence (Amersham, Arlington Heights, Ill.). ExoS ADP-ribosylation of LMWG proteins in human HT-29 epithelial cells was determined after a 4- to 5-h coculture period, in a manner similar to that for J774A.1 cells, as previously described (8).
Analysis of Rac1 and RalA activation.
To analyze effects of ExoS on Rac1 activation, J774A.1 cells were seeded in 100-mm-diameter dishes and cocultured with bacteria for 5 h, after which cells were washed and lysed as described above. Lysates were cleared by centrifugation at 16,000 × g for 5 min, and supernatants were incubated with 10 μl of p21 activated kinase 1 (PAK)-glutathione S-transferase (GST) bound to glutathione-Sepharose (GSH) beads for 1 h at 4°C with rocking. PAK-GST was constructed in our laboratory, by D. Greene, by amplifying a region encoding amino acids 50 to 134 of the PAK binding domain (12) and ligating it into the pGEX-4T-1 cloning vector (Amersham Pharmacia) for expression as a GST fusion protein. PAK-GST beads incubated with cell lysates were washed three times with wash buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 0.1% Triton X-100, 10% glycerol, 20 mM NaF), resuspended in Laemmli electrophoresis sample buffer, and heated at 95°C for 2 min. Samples were resolved by SDS-12% PAGE and immunoblotted for Rac1 as described above. To analyze effects of ExoS on RalA activation, J774A.1 cells were lysed, following a 5-h coculture period, on ice for 20 min in Ral binding domain (RBD) pull-down lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 20 mM MgCl2, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg [each] of leupeptin and aprotinin per ml [Sigma]). Lysates were cleared, and supernatants were incubated for 2 h with 8 μl of the RBD of Ral binding protein 1 (RalBP1) bound to GST and precipitated with glutathione Sepharose (RBD-GST-GSH) beads, prepared as previously described (6). The beads were washed twice with lysis buffer, resuspended in Laemmli electrophoresis sample buffer, resolved, and immunoblotted for RalA as described above.
Statistical analyses.
The statistical analyses were performed with the Student t test.
RESULTS
Cytotoxic effect of ExoS on macrophages.
Previous studies found that P. aeruginosa strain 388 was cytotoxic for primary murine bone marrow-derived macrophages, based on increased membrane permeability (4). This cytotoxic effect was dependent on the P. aeruginosa TTS system but independent of ExoS. To assess whether ExoS might exert other effects on macrophage function, J774A.1 cells were compared for alterations in morphology and adherence following coculture with no bacteria (control) or with PA103ΔUT strains expressing pUCP plasmid-encoded wild-type ExoS or GAP and/or ADPRT mutant forms of ExoS (Table 1). These mutants were constructed based on previous in vitro and in vivo ExoS structure-function studies that identified residue Arg146 to be integral to ExoS GAP activity (13, 30) and residues Glu379 and Glu381 to contribute to ExoS ADPRT activity (7, 24, 32). Strain PA103ΔUT was used in these studies, rather than strain 388, since it was able to be genetically manipulated to allow selective TTS translocation of ExoS or ExoS mutants in the absence of other known type III effectors (37). Comparable production and secretion of ExoS and ExoS mutants by strain PA103ΔUT have been previously confirmed (7). While the exoS gene remained under the control of its endogenous promoter in the pUCP vector, levels of plasmid-encoded product are predicted to be higher than those for the chromosomally encoded gene.
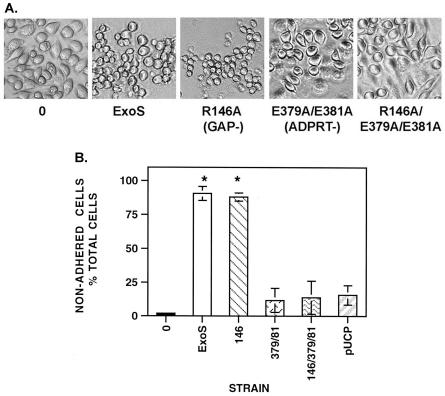
No obvious alterations in J774A.1 cell properties were evident during the first 4 h of coculture, with cells treated with the different bacterial strains closely resembling untreated control cells in regard to morphology, adherence, and viability (data not shown). When the coculture period was extended to 5 h, cells exposed to ADPRT-active ExoS and the R146A-GAP mutant showed a predominantly rounded appearance, which was not apparent in cells exposed to other ExoS mutants (Fig. 1A). Further extension of the culture period to 6 h resulted in a significant loss of adherence to the matrix of cells treated with ExoS and the R146A-GAP mutant compared to control cells, as indicated in Fig. 1B. In contrast, no alteration in morphology or significant loss of adherence was detected compared to untreated control cells following a 6-h exposure to the E379A/E381A-ADPRT mutant (P = 0.35), the R146A/E379A/E381A-GAP/ADPRT mutant (P = 0.49), or the pUCP vector control (P = 0.24). Since the pUCP vector control was found to behave similarly to the R146A/E379A/E381A-GAP/ADPRT ExoS mutant, the GAP/ADPRT ExoS mutant was included as a negative control in subsequent assays to specifically recognize the effects of ExoS GAP and ADPRT functions. When cell viability was compared to loss of adherence, a decrease in viability was evident in cells treated with all PA103ΔUT strains (data not shown), indicating that a non-ExoS-related bacterial effect was responsible for loss of viability, as previously reported (4, 16). These studies identify toxic effects of ExoS ADPRT activity on J774A.1 cell adherence and morphology, which occur in addition to TTS-related effects on cell viability.
FIG. 1.
Cytotoxic effects of ExoS GAP and ADPRT activities on the J774A.1 cell line. J774A.1 cells were seeded at 105 cells/ml and cultured for 72 h, to 70% confluency, prior to coculture for 2, 4, and 6 h, with no bacteria (0) or with 107 CFU of strain PA103ΔUT expressing the indicated ExoS construct (described in Table 1) per ml. (A) Effect on morphology. After 5 h of coculture, bacteria were removed, cells were washed once with PBS, and medium containing antibiotic was added to limit bacterial growth. Cells were visualized by phase-contrast microscopy. Cell rounding was more evident in cells cocultured with bacteria expressing ADPRT-active ExoS (ExoS and R146A). Magnification, ×32. (B) Effect on adherence. At 6 h, when loss of cell adherence occurred, nonadherent and attached cells were recovered, washed with PBS and quantified by using a trypan blue stain. Results are expressed as the percentage of total cells that are nonadherent. The means and standard errors from five independent experiments are shown, and an asterisk indicates a statistically significant difference in adherence (P = 0.0002 for ExoS and P = 0.0001 for R146A relative to control cells not treated with bacteria, P = 0.01 for ExoS and P = 0.003 for R146A relative to the pUCP vector control, and P = 0.0001 for ExoS and R146A relative to E379A/E381A and P = 0.02 for ExoS and R146A relative to R146A/E379A/E381A).
Analysis of macrophage morphology by SEM.
The Rho family of LMWG proteins regulates eukaryotic actin cytoskeletal structure, with Rho controlling cell contractility and assembly of actin cables, Rac1 promoting the organization of actin filaments into lamellipodium and ruffle structures, and Cdc42 regulating filopodium structures (2, 14). While these structures are all known to participate in macrophage locomotion, it is also recognized that lamellipodia, membrane ruffles, and filopodia participate in the phagocytic process (3).
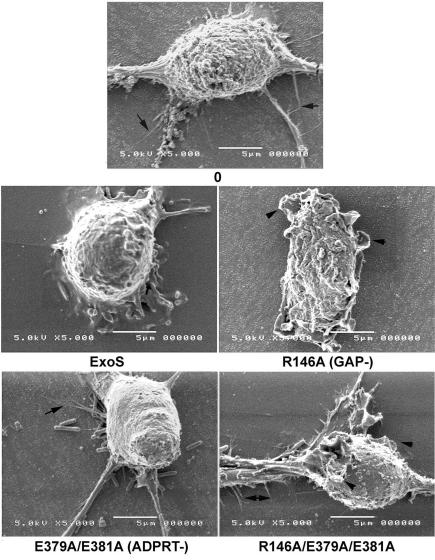
To assess the influence of the ExoS GAP and ADPRT domains on the formation of these functional cytoskeletal structures, bacteria expressing mutant forms of ExoS were examined for their effect on J774A.1 cell surface structure by SEM. A 2.5-h coculture period was chosen for these experiments, since at this time minimum J774A.1 cell detachment was observed, as determined in assays described above. As shown in Fig. 2, untreated control J774A.1 cells exhibited normal macrophage morphology, characterized by cell surface ruffles and lamellipodia, as well as the presence of filopodia and pseudopods. When the cells were cocultured with the ExoS-producing strain, a reduction in the formation of all surface structures was evident. In comparison, cells exposed to the R146A-GAP ExoS mutant exhibited enhanced ruffling and lamellipodium formation but minimal to no fliopodium formation. Conversely, cells exposed to the E379A/E381A-ADPRT mutant showed minimal or no ruffle or lamellipodium formation, but exhibited filopodia. The combined ExoS GAP/ADPRT mutant maintained cell surface characteristics similar to those of control cells. These results provided an indication of the differential effects of ExoS GAP and ADPRT functions on J774A.1 cytoskeletal structure, with the GAP domain of ExoS linked to suppression of lamellipodium and membrane ruffle formation and the ADPRT domain linked to suppression of fliopodium formation.
FIG. 2.
Analysis of J774A.1 cell morphology by SEM. J774A.1 cells were grown on Thermanox coverslips and cocultured for 2.5 h with the indicated ExoS-expressing PA103ΔUT strain. Cells were fixed with 2% cacodylate glutaraldehyde, followed by 2% osmium tetroxide. Samples were rinsed, dehydrated with ethanol, and then incubated with hexamethyldisilazane until dry, mounted, sputter coated with gold palladium, and examined with a JEOL 5410 scanning electron microscope. Representative pictures of the predominant phenotype associated with each mutant are shown. Control cells (0) showed normal macrophage morphology. Cells cocultured with PA103ΔUT expressing ExoS showed decreased lamellipodia and membrane ruffling (70.5%; 12 of 17 cells) and decreased filopodia (58.8%; 10 of 17 cells). Cells cocultured with the R146A-GAP mutant showed enhanced lamellipodia and ruffles (100%; 10 of 10 cells) and no filopodia (70%; 7 of 10 cells). Cells cocultured with the E379A/E381A-ADPRT mutant showed no lamellipodia and ruffles (60%; 6 of 10 cells) and restored filopodia (70%; 7 of 10 cells). Cells cocultured with the R146A/E379A/E381A-GAP/ADPRT mutant showed restored lamellipodia, membrane ruffles, and filopodia (100%; nine of nine cells). Arrowheads identify lamellipodia, and arrows identify filopodia. Bar, 5 μm.
Roles of the GAP and ADPRT activities in the antiphagocytic effect of ExoS.
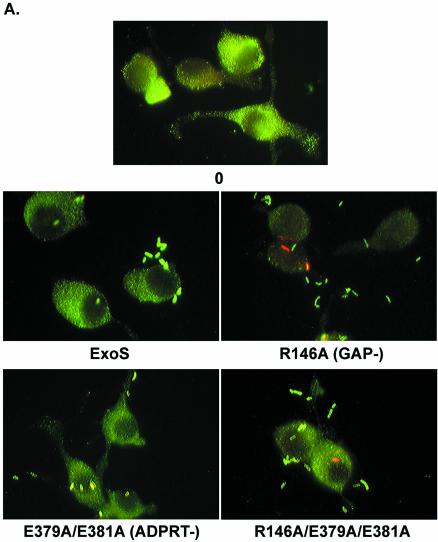
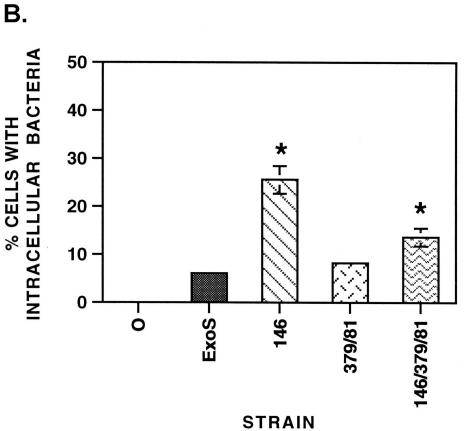
Previous studies performed with an E381A-ADPRT ExoS mutant, expressed and secreted by the TTS system of Yersinia pseudotuberculosis, identified an antiphagocytic activity of ExoS (9). To further assess the contributing roles of the GAP and ADPRT domains in the antiphagocytic function of ExoS, PA103ΔUT strains expressing ExoS mutants were assayed for their ability to resist phagocytosis by J774A.1 macrophages in antibiotic protection studies using both gentamicin and amikacin aminoglycosides (400 μg/ml). The results from these studies proved to be inconclusive, since unlysed J774A.1 cells included as controls (cells exposed to bacteria and treated with antibiotics but not lysed) revealed numbers of associated bacteria comparable to those of similarly treated lysed cells (data not shown). The studies indicated that the antibiotic treatment was not effective in killing externally associated PA103ΔUT strains, and as a consequence this methodology did not provide an accurate means of assaying J774A.1 phagocytic activity. Based on these results, a double-immunofluorescence assay was developed to assess the antiphagocytic properties of PA103ΔUT strains expressing ExoS. After 2.5 h of infection (a time point selected again to minimize influences of bacterial toxicity), an increased number of intracellular bacteria (red bacteria) were evident, in relation to extracellular bacteria (green bacteria), in cells exposed to the R146A-GAP or R146A/E379A/E381A-GAP/ADPRT mutant (Fig. 3A). In comparison, cells infected with strains expressing ExoS or the E379A/E381A-ADPRT mutant had very few or no intracellular bacteria. When cells with internalized bacteria were quantified, a statistically significant increase in bacterial internalization (P ≤ 0.05) was detected in cells exposed to the R146A-GAP and the R146A/E379A/E381A-GAP/ADPRT mutants relative to cells exposed to strains expressing ExoS or the E379A/E381A-ADPRT mutant (Fig. 3B). Also apparent in these analyses was a statistically significant difference in the percentage of cells with internalized bacteria expressing the GAP mutant (26.56% ± 2.91%) compared to the double GAP/ADPRT mutant (13.63% ± 1.86%) (P = 0.009). These studies support the role of the GAP domain in the antiphagocytic function of ExoS and provide an indication, through studies of the GAP/ADPRT mutant, of the potential of the ADPRT domain to modulate this antiphagocytic activity.
FIG. 3.
Antiphagocytic activity of ExoS determined by a double-immunofluorescence assay. (A) Fluorescence microscopy. J774A.1 cells were seeded on slides, cultured as described for Fig. 1, and then cocultured for 2.5 h with no bacteria (0) or with 107 CFU of the indicated PA103ΔUT strain per ml. Cells were washed, fixed with 3% paraformaldehyde, and blocked. Extracellular bacteria were stained first with guinea pig anti-P. aeruginosa IgG polyclonal antibody and visualized with a goat anti-guinea pig IgG-SP-biotin conjugate, followed by Extravidin-FITC conjugate (green bacteria). Intracellular bacteria were stained second by permeabilizing cells with 100% methanol at −20°C and staining as described above, with the exception that Extravidin-tetramethyl rhodamine isocyanate was used (red bacteria). Slides were mounted and examined by fluorescence microscopy for cells with associated extracellular bacteria (green) and/or intracellular bacteria (red). Magnification, ×100. (B) Quantification of intracellular bacteria. The experimental results from panel A were quantified as the percentage of cells with internalized bacteria, with the means and standard errors of analyses performed in triplicate in three independent experiments represented. An asterisk indicates a statistically significant difference for the R146A-GAP and R146A/E379A/E381A-GAP/ADPRT mutants relative to control cells (0), as well as to all of the other strains analyzed (P ≤ 0.05).
Analysis of LMWG protein modification by bacterially translocated ExoS ADPRT activity.
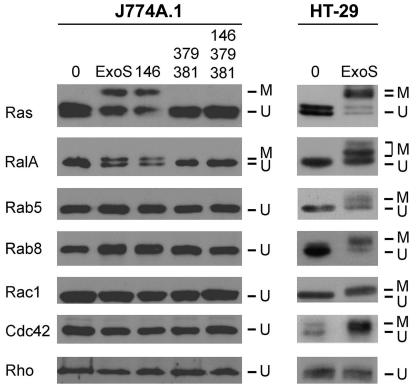
Previous studies performed with human epithelial cells found multiple, but selective, endogenous LMWG proteins to be ADP-ribosylated by bacterially translocated ExoS (6, 8, 25). Specifically, Ras, RalA, Rab5, Rab7, Rab8, Rab11, Rac1, and Cdc42 were identified as substrates of TTS-translocated ExoS, while Rab4 and Rho (A, B, and C) were not substrates. To understand the cellular mechanism associated with the effects of bacterially translocated ExoS on J774A.1 macrophage function, endogenous LMWG protein substrate modification was examined following coculture with strains expressing GAP and/or ADPRT mutant forms of ExoS. Of the ADPRT substrates examined, only Ras and RalA exhibited a shift in molecular mass by SDS-PAGE analysis following exposure to ADPRT-active forms of ExoS (ExoS and the R146A-GAP mutant). Modification of Ras and RalA was detected as early as 2.5 h of coculture (data not shown), and no modification of additional substrates, including Rab5 and Rab8 (shown) and Rab7 and Rab11 (not shown) or Rho proteins Rac1, Cdc42, and Rho (A, B, and C), was detected when the coculture period was extended to 5 h (Fig. 4). Differences observed in the apparent efficiencies of Ras and RalA modification in J774A.1 cells are related to the properties of the two proteins when analyzed by SDS-PAGE rather than to the multiple ADP-ribosylation of Ras. The shift in the masses of Ras and RalA in J774A.1 cells is consistent with the addition of one ADP-ribose moiety to each, as previously assessed by two-dimensional electrophoresis (6-8, 38). As expected, no LMWG protein modification was detected in J774A.1 macrophages cocultured with ADPRT mutant forms of ExoS (E379A/E381A or R146A/E379A/E381A).
FIG. 4.
Substrate modification by ExoS ADPRT activity in J774A.1 cells. J774A.1 cells were seeded and cocultured for 5 h with the indicated ExoS-expressing PA103ΔUT strain, as described for Fig. 1. For the analysis of Ras modification, cells were lysed in TBS-TDS buffer (10 mM Tris [pH 7.4], 140 mM NaCl, 1% Triton X 100, 0.5% sodium deoxycholate, 0.1% SDS) for 30 min on ice. Lysates were cleared by centrifugation, and Ras was immunoprecipitated with monoclonal Y13-259 Ras antibody. For analyses of all the other LMWG proteins, cells were scraped, washed with PBS and lysed as described in Materials and Methods. Samples were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with antibodies specific for the indicated LMWG protein. Blots were visualized by enhanced chemiluminescence. The mobilities of modified (M) and unmodified (U) proteins are indicated. The results are representative of those from analyses performed in multiple independent studies. TTS-translocated ExoS substrate modification of J774A.1 cells is compared with the more extensive substrate modification previously reported for HT-29 human epithelial cells (8).
A comparison of ExoS ADPRT substrate modification of J774A.1 macrophages to that of HT-29 human epithelial cells (Fig. 4) highlights the more limited modification exhibited by J774A.1 cells. Noteworthy, in addition to the different pattern of ExoS substrate specificity, was that the shift in molecular mass of Ras and RalA in J774A.1 cells due to ExoS ADPRT activity was consistently less efficient than that observed in HT-29 cells. A similar, more limited pattern of ExoS ADPRT substrate modification as that observed in J774A.1 macrophages has been previously observed in other rodent cell lines (34). The differences observed in substrate modification of Ras and RalA between HT-29 and J774A.1 cells have been found to relate to the addition of multiple ADP-ribose moieties, with Ras being ADP-ribosylated twice and RalA being ADP-ribosylated two or three times in HT-29 cells, as previously shown by two-dimensional electrophoresis (7, 8, 25). These studies support that ADPRT substrate modification by TTS-translocated ExoS is more limited in J774A.1 macrophages than in human epithelial cells, with only a limited modification of Ras and RalA detected.
Effect of ExoS on Rac1 and RalA function.
The apparent distinct effects of ExoS GAP and ADPRT activities on J774A.1 surface structures directed our analysis of the effects of ExoS mutants on the activation of LMWG proteins that regulate these structures. Rac1 and Cdc42, which are targeted by ExoS GAP activity (13, 21), are known to affect lamellipodium and fliopodium formation, respectively (2, 14). However, RalA, which is targeted by ExoS ADPRT activity, has also been implicated in fliopodium formation in conjunction with the exocyst complex, a multisubunit complex, conserved in mammalian and yeast cells, that regulates vesicle trafficking (26, 36). Within this complex, activated RalA has been found to induce fliopodium formation upon binding to activated Cdc42 and Sec5. To assess the coordinated effects of TTS-translocated ExoS on Rac1 and RalA function, the activation of both proteins was determined in pull-down assays, based on their ability to interact with binding domains in their respective downstream effectors. In these analyses, the Cdc42-Rac-interacting binding (CRIB) domain of PAK fused to GST was used to detect activated Rac1, and the RBD of RalBP1 fused to GST was used to detect activated RalA. Analyses of Rho and Cdc42, which are also targets of ExoS GAP, are not included in this study.
When the activation state of Rac1 was examined following coculture with PA103ΔUT expressing wild-type ExoS, minimal Rac1-GTP was detected in cell lysates, indicating that ExoS interfered with Rac1 activation and/or binding to its downstream effector PAK (Fig. 5A). In support of the interference in Rac1 activation relating to the GAP activity of ExoS, increased levels of activated Rac1 (Rac1-GTP) were detected in lysates of cells exposed to strains expressing mutant GAP (R146A-GAP and R146A/E379A/E381A-GAP/ADPRT mutants). Also apparent in Fig. 5A is the consistent finding of residual levels of activated Rac1 in the lysates of cells infected with the E379A/E381A-ADPRT mutant, despite the presence of an intact GAP domain. This finding suggests that the GAP activity of ExoS may be more efficient in inactivating Rac1 when coordinated with a functional ADPRT domain. The results from these studies support that the GAP activity of ExoS interferes with Rac1 activation in J774A.1 macrophages, which correlates with SEM results that link lamellipodium and membrane ruffle formation to interference with GAP function.
FIG. 5.
Analysis of Rac1 and RalA activation. J774A.1 cells were grown and cocultured with the indicated PA103ΔUT strains for 5 h, as for Fig. 1. Bacteria were removed and the cells treated as follows. (A) To detect GTP-active Rac1, cells were lysed and incubated with PAK-GST bound to GSH beads. Proteins bound to beads were resolved by SDS-PAGE, and Rac1 was detected by immunoblot analyses with a Rac1-specific antibody. (B) To detect GTP-active RalA, cells were lysed in RBD-lysis buffer and incubated with the RBD-GST bound to GSH beads. RalA binding to beads was examined as described above but detected with a RalA-specific antibody. The results are representative of those from three independent studies. The ratio of active Rac1-GTP or RalA-GTP to total Rac1 or RalA, respectively, in each lysate was quantified based on densitometry analysis and normalized relative to untreated control cells. The mean (standard deviation) for each condition is represented. Ratios of greater than one, relative to untreated control cells, observed in response to some bacterial strains are predicted to reflect the activation of the LMWG proteins in J774A.1 cells upon exposure to bacteria.
When the activation state of RalA of J774A.1 macrophages was analyzed by using RalBP-1-GST, RalA activation was impaired in cells exposed to strains expressing ADPRT-active forms of ExoS (ExoS and R146A) but not in cells exposed to ADPRT mutant forms of ExoS (E379A/E381A and R146A/E379A/E381A) or in control cells (Fig. 5B). The results indicate that ADP-ribosylation of RalA by ExoS interferes with RalA activation and binding to its downstream effector in J774A.1 macrophages and suggest the potential of ExoS ADPRT activity to interfere with fliopodium formation through the inactivation of RalA and downstream effects mediated through the exocyst complex.
DISCUSSION
ExoS has been characterized as a bifunctional, Rho GAP and ADP-ribosylating toxin, but its bifunctional activity appears to be modulated when ExoS is translocated by the P. aeruginosa TTS process. When expressed by transient transfection, the GAP activity of ExoS caused cytoskeletal alterations in epithelial cells, due to effects on Rho, Rac, and Cdc42 (21, 31). Alternatively, the ADPRT domain of ExoS was cytotoxic to epithelial cells when expressed by transient transfection, as monitored by the uptake of trypan blue and a decrease in reporter gene expression (29). In comparison, when ExoS was internalized into HT-29 epithelial cells by the P. aeruginosa TTS process, its GAP activity was found to contribute minimally to alterations in cell morphology, while the ADPRT activity was required for effects of ExoS on cell growth and morphology (7, 27). Also in HT-29 cells, TTS-translocated ExoS ADPRT activity was associated with only limited effects on cell viability. Alterations observed in the effects of the ExoS GAP and ADPRT domains on epithelial cell function when internalized by TTS, including the detection of only limited effects of ExoS GAP activity on cell morphology, led to our investigation of the bifunctional nature of ExoS activity in J774A.1 macrophages, where an antiphagocytic function of ExoS GAP activity had previously been reported (9).
Structure-function studies of ExoS have identified an arginine residue at position 146 to be integral to ExoS GAP activity in vitro and to contribute to GAP function within the cell (13, 21, 31). Within the ADPRT domain, Glu381 was identified as a major catalytic residue in in vitro ADPRT reactions (24). ExoS cytotoxicity was also found to be diminished when an E381A mutant form of ExoS was expressed by transient transfection (29). Potential differences in the functionality of ExoS ADPRT activity within the cell and in vitro became apparent when the ExoS E381A mutant was internalized into HT-29 epithelial cells by the TTS process (7). In these studies, E381A ExoS exhibited limited ADP-ribosylation of cellular Ras, an activity not recognized in vitro. This residual activity was eliminated when a secondary catalytic residue, E379, was also mutated (7). Based on these findings of ExoS functional domains, our studies used an R146A-GAP mutant and an E379A/E381A-ADPRT mutant to examine the independent and coordinate functions of the GAP and ADPRT domains of ExoS upon its TTS-mediated translocation into J774A.1 macrophages.
In analyses of J774A.1 macrophages, we were able to identify distinct, but not necessarily independent, functions of both the GAP and ADPRT domains of ExoS. The introduction of an R146A mutation in the GAP domain of ExoS resulted in increased bacterial uptake by J774A.1 cells, as assessed in a double-immunofluorescence assay. In contrast, no alteration in the efficiency of bacterial internalization was detected with the E379A/E381A-ADPRT mutant. These results are consistent with previous reports which support an antiphagocytic function for ExoS, independent of its ADPRT domain (9). In examining the function of the ExoS ADPRT domain, toxic effects of ExoS on J774A.1 cell rounding and adherence were decreased by an E379A/E381A-ADPRT mutant, but no decrease in these toxic effects was detected with the ExoS GAP mutant. These results support the ability of the ADPRT domain of ExoS to exert a toxic effect on J774A.1 macrophages, independent of its GAP function. A possible functional interrelationship between the GAP and ADPRT domains of ExoS was recognized in analyses of the effects of the combined R146A/E379A/E381A-GAP/ADPRT mutant on phagocytosis. While this mutant was associated with a significant increase in bacterial uptake compared to that of ExoS or the ExoS ADPRT mutant, the efficiency of phagocytosis was consistently less than that observed for the single R146A-GAP mutant. These data suggest an influence of ExoS ADPRT activity on the antiphagocytic function of the GAP domain. One possible explanation for this influence might be the recently described auto-ADPRT activity of ExoS, which has been found to down-regulate ExoS GAP function (33). Our studies, unlike that original report, must attribute the further down-regulation of ExoS GAP function to the auto-ADP-ribosylation of residues other than R146, since the modulatory effect that we observed was detected in conjunction with an R146A-GAP mutation. Together, these studies support a functional role of ExoS GAP in the antiphagocytic activity of ExoS, while the ADPRT domain of ExoS is able to exert independent toxic effects on J774A.1 macrophage morphology and adherence.
The distinct functions of the GAP and ADPRT domains of ExoS, and their link to specific LMWG protein function, became further apparent in SEM analyses of J774A.1 cytoskeleton-linked cell surface structures. Normal cell surface features of J774A.1 macrophages include membrane ruffles, lamellipodia, and filopodia, and loss of these structures was evident upon exposure to ExoS-producing bacteria. In examining the roles of the GAP and ADPRT activities of ExoS in alterations of cell surface structures, exposure to the R146A-GAP mutant resulted in enhanced lamellipodium and membrane ruffle formation and minimal to no fliopodium formation, while exposure to an E379A/E381A-ADPRT mutant caused loss of lamellipodia and membrane ruffles but restored fliopodium formation. Lamellipodium and membrane ruffle formation is controlled by Rac1 in both fibroblasts and macrophages (2, 14), and expression of these structures upon exposure to an ExoS GAP mutant links the GAP activity of ExoS to interference with Rac1 function. Alternatively, fliopodium formation is controlled by Cdc42 (2, 14), and the presence of these structures upon exposure to an ExoS ADPRT mutant links the ADPRT function of ExoS with interference of fliopodium formation. SEM analyses thus provided an indication that the GAP and ADPRT activities of ExoS may exert independent effects on the function of Rho family proteins.
In analyzing the molecular mechanisms underlying the effects of ExoS on macrophage morphology, pull-down assays, using the CRIB domain of PAK to detect activated Rac1, found bacterially translocated ExoS to interfere with Rac1 activation. In determining which domain of ExoS was affecting Rac1 function, the R146A-GAP mutant was found to be most effective in reversing the effects of ExoS on Rac1 activation, confirming the importance of the GAP function of ExoS in Rac1 inactivation. Notably, residual Rac1 activity was consistently detected following exposure to the E379A/E381A-ADPRT ExoS mutant, which maintains an active GAP. While the precise explanation for residual active Rac1 in J774A.1 cells treated with the E379A/E381A-ADPRT mutant remains unknown, the results again indicate a role of ExoS ADPRT activity (via possibly auto-ADP-ribosylation) in the down-regulation of ExoS GAP function. Taken together, our studies of J774A.1 cells find the inactivation of Rac1 by ExoS GAP to be consistent with SEM analyses, which link the GAP activity of ExoS to interference with lamellipodium and membrane ruffle formation, and also to support the notion that the antiphagocytic activity of ExoS GAP is mediated by Rac1 inactivation.
Functionally, the ADPRT domain of ExoS was toxic to J774A.1 cell morphology, adherence, and fliopodium formation, but no significant effect of ExoS on J774A.1 viability was detected, consistent with previous reports (4, 16). Analyses of ADPRT substrate modification in J774A.1 macrophages identified only Ras and RalA as substrates of ExoS upon its TTS translocation. ADP-ribosylation of Ras and RalA by TTS-translocated ExoS has been previously found to interfere with the signaling of both proteins to their respective downstream effectors in human epithelial cells (6, 11, 17, 38). While the cellular function of RalA is less well characterized than that of Ras, recent studies link activated RalA to the exocyst complex, which in turn affects fliopodium formation upon binding to activated Cdc42 (26, 36). The functional association between RalA and Cdc42 is notable, since SEM analyses revealed a negative influence of ExoS ADPRT activity on fliopodium formation in J774A.1 macrophages. Consistent with the ability of the ADPRT domain of ExoS to interfere with fliopodium formation by inactivating RalA, TTS-translocated ExoS ADPRT activity was found to inhibit RalA binding to the RBD of RalBP1, a downstream effector of RalA. However, it should be noted that we have been unable to confirm a direct interference with Cdc42 activation by TTS-translocated ExoS ADPRT activity, due to our inability to efficiently detect activated Cdc42 in J774A.1 cells by using commercially available CRIB domains, from either PAK or Wiscott-Aldrich syndrome protein, as Cdc42 activation probes. Our present data thus remain consistent with the potential of ExoS ADPRT activity to interfere with fliopodium formation through the ADP-ribosylation of and interference with RalA function and with the idea that this, coupled with effects of ExoS ADPRT activity on Ras function (11, 38), likely contributes to the toxic effects of ExoS on J774A.1 morphology and adherence.
The identification of Ras and RalA, but not Rac1, as substrates of ExoS ADPRT activity in J774A.1 macrophages differs from results previously reported for human epithelial cells, where Ras, RalA, Rac1, Cdc42, and certain Rab proteins were all identified as LMWG protein substrates of TTS-translocated ExoS (8). Recent studies of a variety of cell lines have identified two patterns of LMWG protein substrate modification (34). One is a more limited pattern of Ras and RalA ADP-ribosylation, as detected in J774A.1 macrophages and common to rodent cell lines. The second is a more extensive pattern of Ras, RalA, Rab, and Rac1 ADP-ribosylation, detected in human and simian cell lines, and in human epithelial cells this pattern coincided with limited detectable effects of ExoS GAP activity on cell function. The targeting of ExoS GAP, but not ADPRT, activity to Rac1 in J774A.1 cells, in conjunction with effects of ExoS GAP on Rac1 activation and function, highlights differences in the coordinated function of the ExoS GAP and ADPRT domains in macrophages and human epithelial cells. While the cellular mechanism for these differences has yet to be determined, recent studies have linked the targeting of ExoS GAP and ADPRT domains to Rac1 to opposite effects on Rac1 function. ExoS GAP activity down-regulates Rac1 function (21, 31), while the ADP-ribosylation of Rac1 by ExoS is associated with Rac1 activation (8, 17). It might therefore be predicted that in cell lines, such as in J774A.1 cells, where ExoS ADPRT does not target Rac1, the down-regulation of Rac1 function by ExoS GAP might be more evident.
The mechanisms used by pathogens to overcome eukaryotic defenses are an integral part of our understanding of the disease process and therefore of our ability to prevent disease. The studies presented in this paper further support the ability of an opportunistic pathogen, P. aeruginosa, to manipulate the host primary line of defense, by interfering with the normal function of phagocytes. Unlike previous reports of human epithelial cells (7), studies of J774A.1 macrophages have allowed the recognition of distinct, but coordinated, functions of the GAP and ADPRT domains of ExoS when translocated by the P. aeruginosa TTS process. ExoS GAP activity is antiphagocytic and affects lamellipodium and membrane ruffle formation, in association with the inactivation of Rac1. ExoS ADPRT activity affects cell adherence, morphology, and fliopodium formation, in association with the ADP-ribosylation of RalA and Ras. The results confirm the bifunctional activity of ExoS within J774A.1 macrophages and provide insight into the coordinate targeting and function of its GAP and ADPRT domains that lead to the down-regulation of macrophage function.
Acknowledgments
We thank and recognize the contribution of Anna D. Tischler in the standardization of the double-immunofluorescence assay. We also thank Dara Frank for providing bacterial cell lines; Timothy Vincent for helpful suggestions and advice; Jennifer Fraylick, Jeannine La Rocque, and Deanne Greene for their contributions to assays included in this paper; Jim Nicholson for helping with the immunofluorescence microscopy; Carol Moskos for invaluable guidance with the scanning electron microscopy; and Craig D. Easterbrook for encouragement.
This study was supported by Public Health Service grant NIH-NIAID 45569.
Editor: D. L. Burns
REFERENCES
- 1.Aktories, K., C. Mohr, and G. Koch. 1992. Clostridium botulinum C3 ADP-ribosyltransferase. Curr. Top. Microbiol. Immunol. 175:115-131. [DOI] [PubMed] [Google Scholar]
- 2.Allen, W. E., G. E. Jones, J. W. Pollard, and A. J. Ridley. 1997. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J. Cell Sci. 110:707-720. [DOI] [PubMed] [Google Scholar]
- 3.Caron, E., and A. Hall. 1998. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282:1717-1721. [DOI] [PubMed] [Google Scholar]
- 4.Coburn, J., and D. W. Frank. 1999. Macrophages and epithelial cells respond differently to the Pseudomonas aeruginosa type III secretion system. Infect. Immun. 67:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coburn, J., R. T. Wyatt, B. H. Iglewski, and D. M. Gill. 1989. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J. Biol. Chem. 264:9004-9008. [PubMed] [Google Scholar]
- 6.Fraylick, J. E., M. J. Riese, T. S. Vincent, J. T. Barbieri, and J. C. Olson. 2002. ADP-ribosylation and functional effects of Pseudomonas aeruginosa exoenzyme S on cellular Ral. Biochemistry 41:9680-9687. [DOI] [PubMed] [Google Scholar]
- 7.Fraylick, J. E., J. R. La Rocque, T. S. Vincent, and J. C. Olson. 2001. Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function. Infect. Immun. 69:5318-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraylick, J. E., E. A. Rucks, D. M. Greene, T. S. Vincent, and J. C. Olson. 2002. Eukaryotic cell determination of ExoS ADP-ribosyltransferase substrate specificity. Biochem. Biophys. Res. Commun. 291:91-100. [DOI] [PubMed] [Google Scholar]
- 9.Frithz-Lindsten, E., Y. Du, R. Rosqvist, and A. Forsberg. 1997. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 25:1125-1139. [DOI] [PubMed] [Google Scholar]
- 10.Fu, Y., and J. E. Galan. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401:293-297. [DOI] [PubMed] [Google Scholar]
- 11.Ganesan, A. K., T. S. Vincent, J. C. Olson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor catalyzed nucleotide exchange. J. Biol. Chem. 274:21823-21829. [DOI] [PubMed] [Google Scholar]
- 12.Glaven, J. A., I. Whitehead, S. Bagrodia, R. Kay, and R. A. Cerione. 1999. The Dbl-related protein, Lcf, localizes to microtubules and mediates the activation of Rac signaling pathways in cells. J. Biol. Chem. 274:2279-2285. [DOI] [PubMed] [Google Scholar]
- 13.Goehring, U.-M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase activating protein for Rho-GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 14.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-513. [DOI] [PubMed] [Google Scholar]
- 15.Hardt, W.-D., L.-M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galan. 1988. Salmonella typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815-826. [DOI] [PubMed] [Google Scholar]
- 16.Hauser, A. R., and J. N. Engel. 1999. Pseudomonas aeruginosa induces type III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect. Immun. 67:5530-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriksson, M. L., C. Sundin, A. L. Jansson, A. Forsberg, R. H. Palmer, and B. Hallberg. 2002. Exoenzyme S shows selective ADP-ribosylation and GAP activities towards small GTPases in vivo. Biochem. J. 367:617-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iglewski, B. H., J. Sadoff, M. J. Bjorn, and E. S. Maxell. 1978. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc. Natl. Acad. Sci. USA 75:3211-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Just, I., J. Selzer, M. Wilm, C. V. Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500-503. [DOI] [PubMed] [Google Scholar]
- 20.Knight, D. A., V. Finck-Barbancon, S. M. Kulich, and J. T. Barbieri. 1995. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 63:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krall, R., J. Sun, K. J. Pederson, and J. T. Barbieri. 2002. In vivo Rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulich, S. M., D. W. Frank, and J. T. Barbieri. 1995. Expression of recombinant exoenzyme S of Pseudomonas aeruginosa. Infect. Immun. 63:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, D. J., D. Cox, J. Li, and S. Greenberg. 2000. Rac1 and Cdc42 are required for phagocytosis, but not NF-KB-dependent gene expression, in macrophages challenged with Pseudomonas aeruginosa. J. Biol. Chem. 275:141-146. [DOI] [PubMed] [Google Scholar]
- 24.Liu, S., S. M. Kulich, and J. T. Barbieri. 1996. Identification of glutamic acid 381 as a candidate active site residue of Pseudomonas aeruginosa exoenzyme S. Biochemistry 35:2754-2758. [DOI] [PubMed] [Google Scholar]
- 25.McGuffie, E. M., D. W. Frank, T. S. Vincent, and J. C. Olson. 1998. Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 66:2607-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moskalenko, S., D. O. Henry, C. Rosse, G. Mirey, J. H. Camonis, and M. A. White. 2002. The exocyst is a Ral effector complex. Nat. Cell Biol. 4:66-72. [DOI] [PubMed] [Google Scholar]
- 27.Olson, J. C., J. E. Fraylick, E. M. McGuffie, K. M. Dolan, T. L. Yahr, D. W. Frank, and T. S. Vincent. 1999. Interruption of multiple cellular processes in HT-29 epithelial cells by Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 67:2847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson, J. C., E. M. McGuffie, and D. W. Frank. 1997. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect. Immun. 65:248-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pederson, K. J., and J. T. Barbieri. 1998. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas aeruginosa exoenzyme S is cytotoxic to eukaryotic cells. Mol. Microbiol. 30:751-759. [DOI] [PubMed] [Google Scholar]
- 30.Pederson, K. J., S. Pal, A. J. Vallis, D. W. Frank, and J. T. Barbieri. 2000. Intracellular localization and processing of Pseudomonas aeruginosa ExoS in eukaryotic cells. Mol. Microbiol. 37:287-299. [DOI] [PubMed] [Google Scholar]
- 31.Pederson, K. J., A. J. Vallis, K. Aktories, D. W. Frank, and J. T. Barbieri. 1999. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol. Microbiol. 32:393-401. [DOI] [PubMed] [Google Scholar]
- 32.Radke, J., K. J. Pederson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect. Immun. 67:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riese, M. J., U.-M. Goehring, M. E. Ehrmantraut, J. Moss, J. T. Barbieri, K. Aktories, and G. Schmidt. 2002. AutoADP ribosylation of Pseudomonas aeruginosa ExoS. J. Biol. Chem. 277:12082-12088. [DOI] [PubMed] [Google Scholar]
- 34.Rucks, E. A., J. E. Fraylick, L. M. Brandt, T. S. Vincent, and J. C. Olson. 2003. Cell line differences in bacterially translocated ExoS ADP-ribosyltransferase substrate specificity. Microbiology 149:319-331. [DOI] [PubMed] [Google Scholar]
- 35.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vector derived from pUC18/19. Gene 97:109-112. [DOI] [PubMed] [Google Scholar]
- 36.Sugihara, K., S. Asano, K. Tanaka, A. Iwamatsu, K. Okawa, and Y. Ohta. 2002. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat. Cell Biol. 4:73-78. [DOI] [PubMed] [Google Scholar]
- 37.Vallis, A. J., V. Finck-Barbancon, T. L. Yahr, and D. W. Frank. 1999. Biological effects of Pseudomonas aeruginosa type III-secreted proteins in CHO cells. Infect. Immun. 67:2040-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent, T. S., J. E. Fraylick, E. M. McGuffie, and J. C. Olson. 1999. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 39.von Pawel-Rammingen, U., M. V. Telpnev, G. Schmidt, K. Aktories, H. Wolf-Watz, and R. Rosqvist. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737-748. [DOI] [PubMed] [Google Scholar]
- 40.Yahr, T. L., J. Goranson, and D. W. Frank. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol. Microbiol. 22:991-1003. [DOI] [PubMed] [Google Scholar]