Abstract
Ecteinascidin 743 (ET-743), a highly promising marine-based antitumor agent presently in phase II clinical trials, has been shown to interfere with the binding of minor-groove-interacting transcription factors, particularly NF-Y, with their cognate promoter elements in vitro. We have shown that NF-Y is a central mediator of activation of transcription of the human P glycoprotein gene (MDR1) by a variety of inducers and that NF-Y functions by recruiting the histone acetyltransferase PCAF to the MDR1 promoter. In the present study, we tested whether ET-743 could block activation of the MDR1 promoter by agents that mediate their effect through the NF-Y/PCAF complex. We report that physiologically relevant concentrations of ET-743 abrogate transcriptional activation of both the endogenous MDR1 gene and MDR1 reporter constructs by the histone deacetylase inhibitors as well as by UV light, with minimal effect on constitutive MDR1 transcription. Notably, this inhibition does not alter the promoter-associated histone hyperacetylation induced by histone deacetylase inhibitors, suggesting an in vivo molecular target downstream of NF-Y/PCAF binding. ET-743 is therefore the prototype for a distinct class of transcription-targeted chemotherapeutic agents and may be an efficacious adjuvant to the treatment of multidrug-resistant tumors.
Aberrant transcription plays a central role in the etiology and treatment of a number of diseases, including cancer. In particular, altered transcription of genes that mediate drug action and apoptosis can have a seriously negative impact on successful therapeutic intervention. Traditional clinical efforts have been directed at overcoming drug resistance once it has emerged. Although it stands to reason that preventing the onset of drug resistance may prove a more effective approach, this goal has been hampered by our limited knowledge of the mechanisms underlying transcriptional activation of drug resistance genes, as well as by the lack of agents that can block this activation specifically. Identifying the precise mechanisms underlying expression of drug resistance genes therefore offers opportunities for drug design.
Although the basis for anticancer drug resistance is multifaceted, the overexpression of P glycoprotein (Pgp), a membrane protein encoded in human cells by the multidrug resistance 1 (MDR1) gene, has been causally linked to the multidrug resistant phenotype in a variety of experimental and patient tumors (1). Long thought to confer resistance by mediating the efflux of drugs from the cell, more recent studies suggest that overexpression of Pgp also plays a general antiapoptotic role that extends beyond resistance to chemotherapeutics, because cells that overexpress Pgp are resistant to a wide range of caspase-dependent apoptotic inducers, including serum starvation, Fas ligand ligation, UV irradiation, and tumor necrosis factor (2, 3).
Two mechanisms have been described for the activation of MDR1 gene expression in resistant tumors. First, tumor cells can accumulate mutations that result in a high constitutive level of Pgp, conferring a growth advantage in the presence of MDR-associated drugs. This increased expression has most often resulted from gene amplification in cultured cells, although amplification of the MDR1 gene has not been documented in clinical samples. Recently, constitutively increased MDR1 expression was shown to be associated with gene rearrangements in some patients with drug-refractory acute lymphocytic leukemia (4). Given the instability of the tumor cell genome, preventing the occurrence of activating random mutations such as amplification or rearrangements would prove a daunting task, explaining why clinical efforts to date have been directed at deactivating the overexpressed Pgp rather than preventing its induction. However, a prophylactic approach has been reconsidered recently in light of our observation that, in addition to constitutive overexpression, some solid tumors can also mount a response to the onslaught of toxins by rapidly (within minutes) activating expression of the MDR1 gene (5). This latter mechanism provides the best opportunity for transcription-targeted therapeutic intervention.
We have previously shown that rapid induction of MDR1 transcription by multiple inducers, including histone deacetylase (HDAC) inhibitors (6) and UV irradiation (7), is mediated through an inverted CCAAT box within the proximal MDR1 promoter. The minor-groove-interacting transcription factor NF-Y binds to the MDR1 CCAAT box and orchestrates this activation through the recruitment of the coactivator, PCAF (6). PCAF, a factor involved in chromatin remodeling, in turn mediates transcriptional response through its ability to acetylate histones and possibly NF-Y itself (8). Therefore, NF-Y is a central mediator of MDR1 activation and likely functions, at least in part, by facilitating changes in chromatin structure in response to a variety of inducers. The identification of NF-Y as an integral component in MDR1 activation has prompted us to search for a transcriptional inhibitor that could suppress activation of MDR1 by these toxins.
Ecteinascidin (ET)-743 (Fig. 1A) is a highly promising, exceedingly potent antitumor agent isolated from the marine tunicate Ecteinascidia turbinata and is currently in phase II clinical trials in Europe and the United States (9, 10). Preclinical studies have shown that ET-743 is toxic to most tumor cell lines in the nanomolar to subnanomolar range; indeed, antitumor effects were observed in phase I trials with concentrations of less than 2 mg/m2 body weight. Although ET-743 has been shown to bend and alkylate DNA in the minor groove (11, 12), cause microtubule disruption (13), and target topoisomerase I (14, 15), its mechanism of action in vivo is unknown. A recent study showed that ET-743 interfered with the interaction of minor-groove-binding proteins, particularly NF-Y, with their cognate DNA elements in vitro (16). In light of this observation, we have investigated the possibility that physiologically relevant concentrations of ET-743 could target NF-Y-mediated transcription in vivo by using MDR1 as a model NF-Y-regulated promoter.
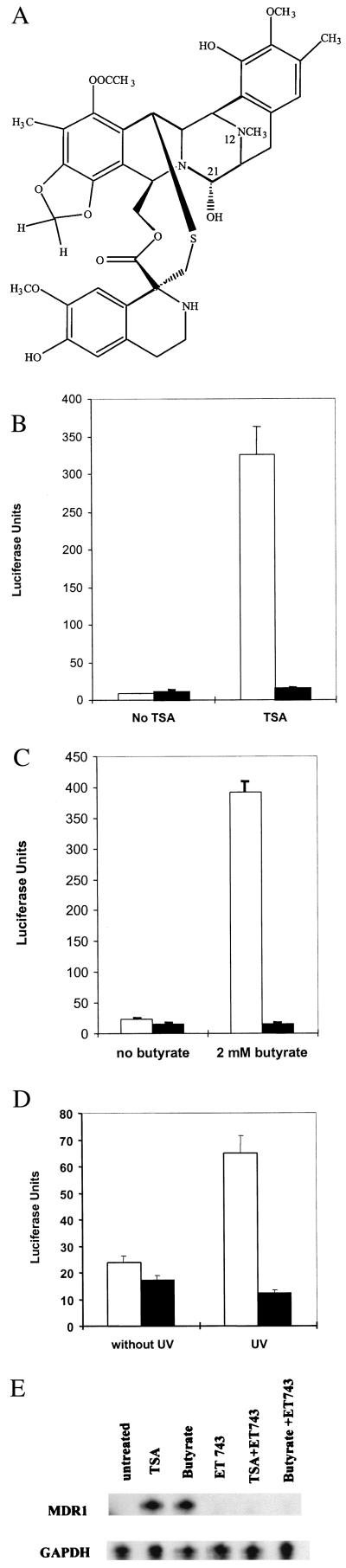
Figure 1.
ET-743 inhibits activation of the transfected and endogenous MDR1 promoter by multiple inducers. (A) Structure of ET-743. (B–D) SW620 cells stably transfected with an MDR1 promoter/luciferase construct were treated with 100 ng/ml trichostatin A (TSA; B), 2 mM sodium butyrate (C), or 10 J/m2 UV irradiation (D) without (white bars) or with (black bars) 50 nM ET-743. The stably transfected cells were treated as indicated for 24 h, and luciferase activity was determined. The data represent the results of three independent experiments performed in triplicate. (E) Nuclease protection analysis of MDR1 RNA from untreated SW620 cells (lane 1) or cells treated with 100 ng/ml TSA (lane 2), 2 mM sodium butyrate (lane 3), 50 ng/ml ET-743 (lane 4), 50 nM TSA and 50 ng/ml ET-743 (lane 5), or 2 mM sodium butyrate and 50 ng/ml ET-743 (lane 6). Total RNA was extracted from cells, and nuclease protection assays were performed with MDR1 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene-specific ribonucleotide probes (6) by using 20 μg and 0.66 μg RNA, respectively.
Materials and Methods
Cell Lines and Reagents.
The human colon carcinoma cell line SW620 (American Type Culture Collection; CCL 227) was stably transfected as described (6) with the pMDR1(−1202) reporter construct, in which the MDR1 promoter sequence (−1202 to +118) was inserted upstream of the luciferase gene in the pGL2B vector (Promega). Cells were maintained in RPMI medium 1640 supplemented with 10% (vol/vol) FCS, 2.0 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. TSA was purchased from Wako Biochemicals (Osaka, Japan) and dissolved in 100% (vol/vol) ethanol; sodium butyrate was purchased from Sigma; and ET-743 was obtained from PharmaMar (Madrid) and dissolved in 100% (vol/vol) ethanol.
MDR1 Promoter Assays.
Cells stably transfected with the MDR1 reporter construct were seeded into 6-well plates at a density of 2 × 105 to 5 × 105 cells per well. After 24 h, the cells were treated as indicated (TSA, 100 ng/ml; sodium butyrate, 2 mM; ET-743, 50 nM; UV irradiation, 10 J/m2; wavelength 254 nm) and incubated for an additional 24 h before harvesting for luciferase and protein assays (6). Luciferase activity was normalized to protein concentration. For nuclease protection assays, SW620 cells were grown in 150-mm-diameter plates to 50% confluence and then treated with different combination of drugs as indicated for 24 h. Total RNA was extracted from the cells, and nuclease protection assays were performed as described (6). Total RNA was used to detect MDR1 and glyceraldehyde-3-phosphate dehydrogenase transcripts.
Cell-Cycle Analysis.
SW620 cells were grown in 150-mm-diameter plates to 50% confluence, treated as indicated for 24 h, and then washed with PBS. Cell nuclei were then prepared from adherent cells as described and subjected to flow cytometric analysis (17).
Histone Acetylation and Chromatin Immunoprecipitation Assays.
For analysis of histone acetylation, cells were grown in 150-mm-diameter plates and treated for 24 h as indicated. Total histones were isolated from cells as described (18) and subjected to Western blot analysis with an anti-acetylated H4 antibody (Upstate Biotechnology, Lake Placid, NY). For chromatin immunoprecipitation analyses, stably transfected cells were seeded in 150-mm plates (1 × 107 cells per 10 plates) and subjected to various treatments as indicated (untreated, 100 ng/ml TSA, 50 nM ET-743, or 100 ng/ml TSA with 50 nM ET-743) for 24 h. After treatment, DNA–protein crosslinking was performed by incubating cells in 1% formaldehyde for 10 min at 37°C, after which cells were sonicated on ice (15 10-s pulses) to shear chromosomal DNA. DNA (300 μg) from each treatment was used in the assays. Input DNA was precleared for 1 h at 4°C in the presence of a salmon sperm DNA/protein A agarose slurry (Upstate Biotechnology, Lake Placid, NY). For each treatment, chromatin was immunoprecipitated overnight with anti-acetyl H4 antibody (Upstate Biotechnology), mouse IgG (negative control; Santa Cruz Biotechnology), or no antibody (negative control), after which additional salmon sperm DNA/protein A agarose slurry was added to collect the immune complexes. Beads were washed, and proteins were eluted according to the method provided by the vendor (Upstate Biotechnology). DNA was recovered by phenol/chloroform extraction and ethanol precipitation. Immunoprecipitated DNA was subject to PCR with MDR1 promoter-specific primers and visualized on a 1% agarose gel. Under these conditions, the majority, if not all, of the signal detected was from the transfected MDR1 promoter. Quantitation of PCR products was performed with Real-Time PCR Sequence Detection (TaqMan, Perkin–Elmer).
Results
In light of the observation that ET-743 inhibited NF-Y/DNA interactions in vitro, we tested whether ET-743 could inhibit transcriptional activation of the MDR1 promoter by inducers that we have previously shown to require NF-Y for their transcriptional effect (6, 7). Colon carcinoma SW620 cells stably transfected with an MDR1 promoter/luciferase construct were exposed to the HDAC inhibitors TSA or sodium butyrate or to UV irradiation in the presence or absence of 50 nM ET-743 (Fig. 1 B–D). As previously observed, exposure to each of the inducers led to an increase in MDR1 promoter activity (Fig. 1 B–D, right side, white bars), which depended on an intact inverted CCAAT box at position −72 (as described; ref. 6). However, cotreatment with 50 nM ET-743 abrogated promoter activation by each of the inducers (Fig. 1 B–D, right side, black bars); concentrations as low as 10 nM were able to overcome activation partially (data not shown). Interestingly, ET-743 had little effect on basal MDR1 promoter activity by this assay (Fig. 1 B–D, left side, black bars), consistent with the previous observation that mutation of the inverted CCAAT box within the MDR1 promoter had little effect on basal activity in SW620 cells (6).
We next examined the effect of ET-743 on endogenous MDR1 gene expression. Previously, we had shown that a 24-h exposure of SW620 cells to either TSA or butyrate resulted in a 20- to 40-fold increase in steady-state MDR1 mRNA levels (6). To examine the effect of ET-743 on this activation, SW620 cells were incubated with TSA alone, butyrate alone, ET-743 alone, or a combination of each of the HDAC inhibitors and ET-743. After 24 h, RNA was isolated and assayed by nuclease protection (Fig. 1E). As expected, both HDAC inhibitors dramatically induced MDR1 gene expression. ET-743 blocked this induction without significantly affecting basal MDR1 RNA levels or control glyceraldehyde-3-phosphate dehydrogenase RNA levels, consistent with the results obtained from analysis of the MDR1 reporter constructs.
Because our investigation into the effects of ET-743 on MDR1 expression was prompted by an earlier study indicating that, at micromolar concentrations, the drug interfered with NF-Y/DNA interactions, we performed gel-shift analyses of the interaction of the MDR1 CCAAT box element with the NF-Y present in nuclear extracts from either untreated or ET-743-treated cells. No difference in complex formation was observed between these two extracts, indicating that the concentrations of ET-743 that were achieved intracellularly (nanomolar) were not sufficient to impede the interaction of NF-Y with its cognate binding site irreversibly (data not shown).
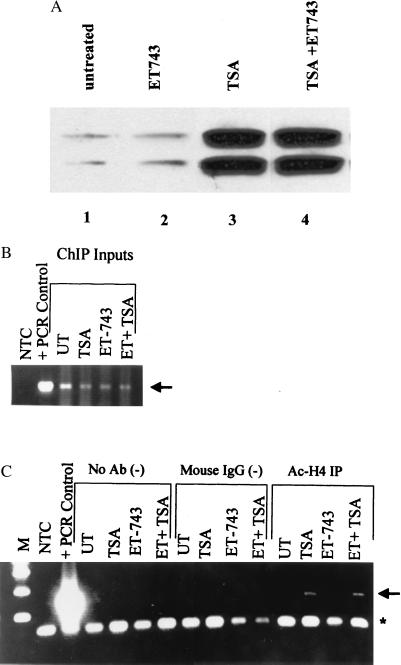
To address this issue more directly, we assessed the effect of ET-743 on the known function of NF-Y on the MDR1 promoter, the recruitment of the histone acetyltransferase PCAF and subsequent acetylation of promoter-associated histones. First, we determined the effect of ET-743 on the general histone hyperacetylation observed when HDACs are inhibited by TSA. Total histones were isolated from untreated cells and cells that had been exposed to TSA and were analyzed by Western blotting with an antibody that specifically recognizes the acetylated tail of histone H4. As shown in Fig. 2A, a 24-h exposure to 100 ng/ml TSA resulted in marked hyperacetylation of histone H4; ET-743 had no effect on either basal acetylation or TSA-induced hyperacetylation of this histone. We next evaluated the effect of ET-743 on acetylation of H4 histones localized at the MDR1 promoter by using a chromatin immunoprecipitation assay (Fig. 2 B and C). The SW620 MDR1 reporter transfectants were either untreated or exposed to 100 ng/ml TSA for 24 h in the presence or absence of 50 nM ET-743. After treatment, cells were exposed to 1% formaldehyde to crosslink chromosomal DNA to interacting proteins. Chromatin was then isolated, sonicated, and quantitated. Similar amounts of chromatin from each treatment (see Fig. 2B for input) were immunoprecipitated with the anti-acetylated H4 antibody, a control (IgG) antibody, or no antibody. Immunoprecipitated DNA was recovered, amplified by PCR, and visualized by agarose gel electrophoresis. As shown in Fig. 2C, association of the MDR1 promoter with hyperacetylated histone H4 can be observed after exposure to TSA (arrow); quantitative PCR analysis with the Perkin–Elmer TaqMan assay showed this increase in MDR1-associated acetylated H4 to be 10- to 15-fold (data not shown). Importantly, ET-743 had no effect on TSA-mediated hyperacetylation of this promoter region. Because we had shown that NF-Y binding was required for modulation of the MDR1 promoter by the chromatin modifying enzymes, these results suggest that ET-743 mediates its effect subsequent to or independent of NF-Y binding and MDR1 promoter acetylation.
Figure 2.
ET-743 does not affect TSA-induced global histone hyperacetylation or accumulation of MDR1 promoter-associated acetylated histone H4. Stably transfected SW620 cells were incubated for 24 h in the presence of TSA alone (100 ng), ET-743 alone (50 nM), or a combination of both. (A) After incubation, total histones were isolated and subjected to Western analysis with an anti-acetylated histone H4 antibody. The two bands observed represent different acetylated forms of histone H4. (B and C) For chromatin immunoprecipitation assays, cells were treated as above, and then chromatin was immunoprecipitated with mouse IgG, anti-acetylated H4 antibody, or no antibody (negative control). MDR1 promoter DNA associated with hyperacetylated H4 was visualized after PCR amplification. The arrow indicates the specific MDR1 promoter PCR product, and the asterisk denotes the input primers. (B) Input DNA. (C) Immunoprecipitated DNA. ChIP, chromatin immunoprecipitation; NTC, negative control; UT, untreated; M, marker.
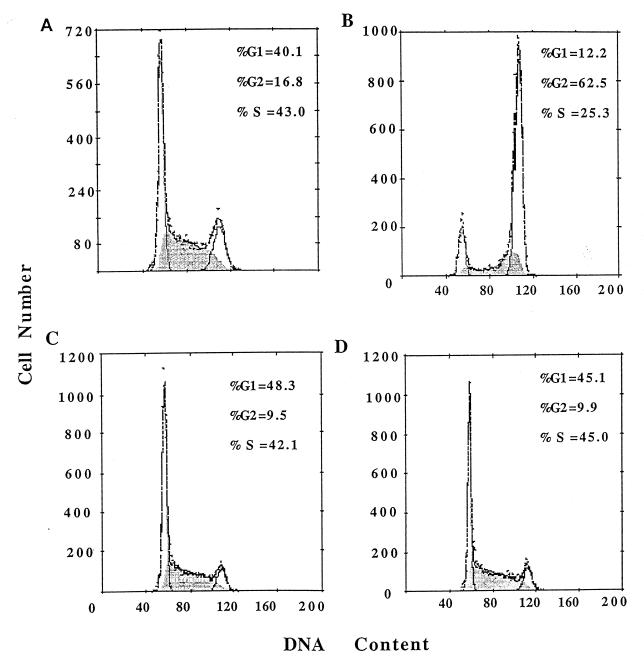
Taken together, our results indicate that ET-743 blocks activation of the MDR1 promoter by multiple inducers with only a minimal effect on constitutive expression. Although this observation could impact the treatment of drug-resistant tumors significantly, it seems unlikely that MDR1 is the cytotoxic target of ET-743. As a first step in identifying other genes that are regulated by ET-743, we evaluated its effect on cell-cycle progression of SW620 cells in the presence and absence of TSA. A 24-h exposure to 50 nM ET-743 alone had little significant effect on the cell-cycle progression of SW620 cells (Fig. 3, compare A and C). However, 100 ng/ml TSA induced a strong G2/M block (Fig. 3B), which was abolished in the presence of ET-743 (Fig. 3D). This result suggests that both drugs may share at least a subset of target genes whose function is required for cell-cycle progression.
Figure 3.
ET-743 abrogates TSA-induced cell-cycle arrest. Cell nuclei from untreated SW620 cells (A) or cells treated with 100 ng/ml TSA (B), 50 nM ET-743 (C), or TSA and ET-743 (D) were prepared and subjected to flow cytometric cell-cycle analysis (17).
Discussion
We have shown that ET-743, a highly promising marine-based anticancer agent, blocks activation of the MDR1 promoter by multiple inducers. Our data suggest that this inhibition is selective for activated transcription, because constitutive MDR1 promoter activity is not repressed significantly at these physiologically relevant concentrations. Moreover, it seems that the mechanism by which this inhibition is accomplished is more complex than simply the inhibition of NF-Y binding suggested by previous in vitro studies (16), because (i) the concentrations required for in vivo repression (nanomolar) are logs below what was required for in vitro inhibition of NF-Y binding (micromolar); (ii) nuclear extracts prepared from ET-743-treated cells supported NF-Y complex formation that was indistinguishable from that observed in extracts from untreated cells; and (iii) promoter-localized hyperacetylation, which depends on the recruitment of the histone acetylase PCAF by promoter-bound NF-Y, was not affected by ET-743.
The implications of this study are far-reaching. First, ET-743 is the first pharmacologically relevant agent that prevents the activation of MDR1 transcription by multiple stress inducers. This finding could be particularly important in light of two recent studies indicating that MDR1 expression, and consequently Pgp levels, can be activated rapidly in human tumors during the course of cytotoxic therapy. Our laboratory has shown that, in five of five patients with metastatic sarcoma, tumor MDR1 RNA levels increased up to 10-fold within 50 min of exposure to the DNA-damaging agent doxorubicin (5). In a separate study, an increase in Pgp expression was observed in a patient with acute myeloid leukemia after 4 and 12 h of administration of daunorubicin/AraC (19). Thus, inducible expression of MDR1 by toxic agents such as chemotherapy and radiation may play a major role in the development of clinical drug resistance, and ET-743 is the first agent identified with the potential for blocking this activation.
The observation that ET-743 blocks NF-Y-mediated transcription has ramifications that extend beyond regulation of MDR1 expression. Mantovani and colleagues (20) demonstrate, in an accompanying paper, that ET-743 also blocks heat activation of the NF-Y-dependent HSP70 promoter; similar to what we observed with the MDR1 promoter, constitutive expression of HSP70 is not affected appreciably. Taken together, these studies suggest that ET-743 targets inducible NF-Y-regulated genes, including those that are required for a rapid “survive or die” decision in the face of genotoxic stress. Certainly, MDR1 and HSP70 both fit this profile. Moreover, both our study and theirs suggest a role for ET-743 in the regulation of genes that are induced during cell-cycle progression. In light of our observation that ET-743 abrogates the TSA-induced G2/M block in SW620 cells, it is interesting to note that NF-Y is a critical regulator of many cell-cycle genes, including cyclin B1 and cyclin B2, which are required for transit through G2/M.
Although the present study and the accompanying report by Mantovani and colleagues (20) strongly support a role for NF-Y as a direct target of ET-743, this role has not yet been proven, and the precise mechanism by which ET-743 exerts its transcriptional effects remains to be determined. For the reasons delineated above, it is unlikely that ET-743 at the concentrations used is inhibiting NF-Y binding to the MDR1 CCAAT box. An alternative possibility is that ET-743 affects the NF-Y/PCAF complex through a mechanism independent of DNA binding, possibly by altering interaction of other, as yet unidentified, coactivators. Indeed, PCAF has been shown to be part of a large multiprotein complex that includes factors involved in stress response and recognition of DNA damage (21), which is interesting in light of our observation that genotoxic agents such as UV light induce MDR1 transcription through this complex, and that this induction is also blocked by ET-743. Exactly how ET-743 could affect interaction with other factors is an open question. However, Hurley and colleagues (11) have shown that ET-743 bends DNA toward the major groove, which is a unique feature among DNA-interactive agents that occupy the minor groove. One could envision how the interaction of ET-743 with DNA could alter local promoter architecture, preventing functional complex formation or inhibiting interactions with other components of the transcriptional machinery. A second possibility that cannot be ruled out is that the target of ET-743 is not NF-Y or NF-Y alone but includes another factor involved in MDR1 induction. Indeed, we have shown recently that an Sp1 site downstream of the MDR1 CCAAT box is also required for transcriptional activation by HDAC inhibitors and UV irradiation (S.J., Z. Hu, and K.W.S., unpublished data and ref. 7), suggesting Sp1 as a putative ET-743 target for abrogation of MDR1 induction. Further studies are needed to distinguish among these possibilities.
In conclusion, we have shown that the antitumor agent ET-743 can inhibit activation of the MDR1 promoter by multiple inducers at clinically achievable concentrations. Moreover, constitutive MDR1 transcription is not affected appreciably under these conditions, suggesting that, in the clinic, ET-743 may selectively inhibit activation of MDR1 expression in tumor cells without affecting constitutive expression in normal cells. From a broader perspective, ET-743 seems to target a subset of inducible genes, some of which are involved in stress response and cell-cycle progression. In addition to its promising role as an antitumor agent, ET-743 will prove a useful tool in dissecting the transcriptional regulation of this gene class.
Acknowledgments
The authors thank R. Mantovani for sharing unpublished information. We thank P. G. Magro for establishing the stable cell lines used for these studies. We also acknowledge R. Mantovani, J. R. Bertino, and members of the Scotto laboratory for fruitful discussions. This work was supported by National Cancer Institute Grants P30-CA-08748 (to the Memorial Sloan–Kettering Cancer Center) and RO1-CA-57307 (to K.W.S.) and by the Horsfall Award for Graduate Studies (to S.J.).
Abbreviations
- Pgp
P glycoprotein
- ET-743
ecteinascidin 743
- MDR
multidrug resistance
- TSA
trichostatin A
- HDAC
histone deacetylase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Germann U A, Pastan I, Gottesman M M. Semin Cell Biol. 1993;4:63–76. doi: 10.1006/scel.1993.1008. [DOI] [PubMed] [Google Scholar]
- 2.Smyth M J, Krasovskis E, Sutton V R, Johnstone R W. Proc Natl Acad Sci USA. 1998;95:7024–7029. doi: 10.1073/pnas.95.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnstone R W, Cretney E, Smyth M J. Blood. 1999;93:1075–1085. [PubMed] [Google Scholar]
- 4.Knutsen T, Mickley L A, Ried T, Green E D, du Manoir S, Schrock E, Macville M, Ning Y, Robey R, Polymeropoulos M, Torres R, Fojo T. Genes Chromosomes Cancer. 1998;23:44–54. doi: 10.1002/(sici)1098-2264(199809)23:1<44::aid-gcc7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Abolhoda A, Wilson A, Ross H, Danenberg P, Burt M, Scotto K. Clin Cancer Res. 1999;5:3352–3356. [PubMed] [Google Scholar]
- 6.Jin S, Scotto K W. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Jin S, Scotto K W. J Biol Chem. 2000;275:2979–2985. doi: 10.1074/jbc.275.4.2979. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izbicka E, Lawrence R, Raymond E, Eckhardt G, Faircloth G, Jimeno J, Clark G, Von Hoff D D. Ann Oncol. 1998;9:981–987. doi: 10.1023/A:1008224322396. [DOI] [PubMed] [Google Scholar]
- 10.Valoti G, Nicoletti M I, Pellegrino A, Jimeno J, Hendriks H, D'Incalci M, Faircloth G, Giavazzi R. Clin Cancer Res. 1998;4:1977–1983. [PubMed] [Google Scholar]
- 11.Zewail-Foote M, Hurley L H. J Med Chem. 1999;42:2493–2497. doi: 10.1021/jm990241l. [DOI] [PubMed] [Google Scholar]
- 12.Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn K W. Biochemistry. 1996;35:13303–13309. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Rocha M, Garcia-Gravalos M D, Avila J. Br J Cancer. 1996;73:875–883. doi: 10.1038/bjc.1996.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez E J, Owa T, Schreiber S L, Corey E J. Proc Natl Acad Sci USA. 1999;96:3496–3501. doi: 10.1073/pnas.96.7.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takebayashi Y, Pourquier P, Yoshida A, Kohlhagen G, Pommier Y. Proc Natl Acad Sci USA. 1999;96:7196–7201. doi: 10.1073/pnas.96.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonfanti M, La Valle E, Fernandez Sousa Faro J M, Faircloth G, Caretti G, Mantovani R, D'Incalci M. Anticancer Drug Des. 1999;14:179–186. [PubMed] [Google Scholar]
- 17.Giaretti W, Nusse M. Methods Cell Biol. 1994;41:389–400. doi: 10.1016/s0091-679x(08)61730-6. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 19.Hu X F, Slater A, Kantharidis P, Rischin D, Juneja S, Rossi R, Lee G, Parkin J D, Zalcberg J R. Blood. 1999;93:4086–4095. [PubMed] [Google Scholar]
- 20.Minuzzo M, Marchini S, Broggini M, Faircloth G, D'Incalci M, Mantovani R. Proc Natl Acad Sci USA. 2000;97:6780–6784. doi: 10.1073/pnas.97.12.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati M L, Qin J, Nakatani Y. Mol Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]