Abstract
A gene encoding a major secreted antigen, SagA, was identified in Enterococcus faecium by screening an E. faecium genomic expression library with sera from patients with E. faecium-associated endocarditis. Recombinant SagA protein showed broad-spectrum binding to extracellular matrix (ECM) proteins, including fibrinogen, collagen type I, collagen type IV, fibronectin, and laminin. A fibrinogen-binding protein, purified from culture supernatants of an E. faecium clinical isolate, was found to match the N-terminal sequence of the predicted SagA protein and to react with the anti-SagA antibody, confirming that it was the SagA protein; this protein appeared as an 80- to 90-kDa smear on a Western blot that was sensitive to proteinase K and resistant to periodate treatment and glycoprotein staining. When overexpressed in E. faecium and Escherichia coli, the native and recombinant SagA proteins formed stable oligomers, apparently via their C-terminal domains. The SagA protein is composed of three domains: (i) a putative coiled-coil N-terminal domain that shows homology to the N-terminal domain of Streptococcus mutans SagA protein (42% similarity), previously shown to be involved in cell wall integrity and cell shape maintenance, and to the P45 protein of Listeria monocytogenes (41% similarity); (ii) a central domain containing direct repeats; and (iii) a C-terminal domain that is similar to that found in various proteins, including P45 (50% similarity) and P60 (52% similarity) of L. monocytogenes. The P45 and P60 proteins both have cell wall hydrolase activity, and the latter has also been shown to be involved in virulence, whereas cell wall hydrolase activity was not detected for SagA protein. The E. faecium sagA gene, like the S. mutans homologue, is located in a cluster of genes encoding proteins that appear to be involved in cell wall metabolism and could not be disrupted unless it was first transcomplemented, suggesting that the sagA gene is essential for E. faecium growth and may be involved in cell wall metabolism. In conclusion, the extracelluar E. faecium SagA protein is apparently essential for growth, shows broad-spectrum binding to ECM proteins, forms oligomers, and is antigenic during infection.
Enterococci are the second to third most commonly isolated pathogens in nosocomial infections and the third most common pathogens associated with nosocomial bloodstream infection (22, 34). The genus Enterococcus consists of at least 23 species, two of which account for >95% of the clinically important isolates: Enterococcus faecalis (80 to 90%) and Enterococcus faecium (5 to 15%). More attention has recently been focused on E. faecium because of an increasing number of infections caused by this organism and its resistance to multiple antibacterial agents, including ampicillin and vancomycin (23). In some hospitals, E. faecium now accounts for >30% of enterococcal infections. However, until very recently, relatively little has been known about this organism other than its antibiotic resistance (26, 32).
Bacterial surface and secreted proteins play important roles in interactions with the host, including adherence, internalization, toxin synthesis, adaptive responses to changes in the environment, and escaping from the host immune system, among others. Previously in our laboratory, an immunoscreening method had been used to identify E. faecalis antigens expressed during infection; several surface and/or secreted proteins were identified, some of which have been shown to be involved in E. faecalis virulence (42, 49-51). In the current study, we adopted a similar approach to the study of E. faecium, and one of the major secreted antigens identified, SagA, was characterized.
Earlier studies of enterococcal adhesion to human extracellular matrix (ECM) and serum proteins have shown that some isolates are able to bind to some proteins, but the nature of a binding agent and the interaction, as well as an influence on enterococcal virulence, have not been confirmed (17, 40, 41, 48, 52). Most of the studies have been focused on E. faecalis, which is generally thought to be the more virulent of two clinically important species. Previously, our laboratory identified a collagen-binding adhesin, Ace, from E. faecalis, and found that it attached to collagen types I and IV, as well as to laminin (24, 25, 31, 33) and, more recently, we identified Acm, a specific collagen-binding adhesin from E. faecium, which did not attach to other ECM proteins (26). We show in the present study that a major secreted antigen, SagA, in addition to apparently being essential for E. faecium growth, exhibits broad-spectrum binding to ECM proteins, including fibrinogen, collagens, fibronectin, and laminin.
MATERIALS AND METHODS
Bacterial strains and antisera.
Strains and plasmids used in the present study are listed in Table 1. Escherichia coli was grown in Luria-Bertani broth or agar with appropriate antibiotics at 37°C. Enterococci were grown in brain heart infusion (BHI) broth or agar (Difco) at 37°C. Sera from two patients with E. faecium endocarditis were collected and used individually. The transformable E. faecium strain TX1330 (38) was used for genetic manipulation because we have been unable to electroporate plasmids or generate disruptions in TX16.
TABLE 1.
Strains and plasmids
| Plasmid or Strain | Relevant propertiesa | Reference or source |
|---|---|---|
| Plasmids | ||
| pAT18 | Plasmid vector; Ermr | 45 |
| pQE31 | His6 expression vector; Ampr | Qiagen |
| pTEX4577 | Plasmid vector, derivative of pBluescript KS(−); Kanr | 39 |
| pTEX5235 | Plasmid vector, derivative of pBluescript KS(−); Spcr | 43 |
| pTEX5236 | Cosmid vector, derivative of pBeloBAC11; Camr | 37 |
| 43 | ||
| pTEX10000 | pTEX5235 derivative expressing N-terminal part of SagA | This study |
| pTEX10001 | pTEX5235 derivative expressing C-terminal part of SagA | This study |
| pTEX10002 | pTEX5236 derivative expressing full-length SagA | This study |
| pTEX10003 | pAT18 derivative expressing SagA | This study |
| Strains | ||
| TX16 | E. faecium endocarditis isolate (Houston, Tex.) | 1 |
| TX1330 | E. faecium community-derived fecal isolate (Houston, Tex.) | 38 |
| TX10003 | TX1330 with pTEX10003 | This study |
| TX10004 | TX10003 with a disrupted chromosomal sagA | This study |
| 448 | E. faecium blood isolate (Poland) | This study |
| DH5α | E. coli strain | Qiagen |
| d1-27 (TX10000) | DH5α with plasmid expressing N-terminal part of SagA | This study |
| d1-29 (TX10001) | DH5α with plasmid expressing C-terminal part of SagA | This study |
| D7A3 (TX10002) | DH5α with cosmid expressing whole-length SagA | This study |
| M15 (pREP4) | Host for pQE31; derivative of E. coli K-12 with plasmid pREP4 expressing lac repressor; Kanr | Qiagen |
Spc, spectinomycin; Cam, chloramphenicol; Amp, ampicillin; Kan, kanamycin; Erm, erythromycin; superscript “r,” resistance.
Construction of an E. faecium genomic library and immunoscreening.
E. faecium strain TX16 (also known as TX0016, TEX16, and DO [see also the partial genome sequence at http://www.hgsc.bcm.tmc.edu/microbial/efaecium/]) was chosen as the source of DNA for the construction of genomic libraries. A large-insert library was made with vector pTEX5236 (43) by a method previously described (49). The plasmid pTEX5236 is a derivative of the pBeloBAC11 vector (37), with an oriT from RK2 added. This vector has been shown to stably maintain large inserts of enterococcal DNA (unpublished results). A small-insert library was made with a pBluescript SK(−)-derived vector pTEX5235 (43). About 1 mg of TX16 genomic DNA was partially digested with 10 and 20 U of Sau3AI, respectively, for 1 h at 37°C, pooled, and fractionated by size in a 10 to 40% sucrose density gradient (35). Fractions containing 0.5 to 5kb DNA fragments were selected for ligation with vector pTEX5235, which had been previously digested with BamHI and dephosphorylated with shrimp alkaline phosphatase (U.S. Biochemicals, Cleveland, Ohio). The ligation mixture was used to transform E. coli DH5α. The transformation mixture was diluted and plated with antibiotic selection to produce ca. 100 colonies per plate. Colony immunoblots were performed with patient sera by a previously described method (49). About 10,000 pTEX5235 clones were screened to recover clones that reacted with at least one of the patient sera.
DNA techniques.
PCR and DNA sequencing were performed by standard methods (35). The primers used in the present study are listed in Table 2. Distribution of sagA gene among E. faecium was determined by Southern blotting with 11 E. faecium strains and pTEX10000 and pTEX10001 (Table 1) as probes. Southern blotting with high-stringency hybridization was as described previously (2). In order to obtain the complete sequence of the sagA gene, clones from the large insert pTEX5236 library were screened with plasmids pTEX10000 and pTEX10001 as probes; one positive clone, D7A3 (TX10002), was chosen, and oligonucleotide primers, designed from plasmids pTEX10000 and pTEX10001, were used to determine the DNA sequence of this cosmid. Screening of the large-insert library with DNA probes was performed by high-stringency colony hybridization (35).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotidea | Sequence (5′-3′) | Usage |
|---|---|---|
| E54.1 | AATGGTATGTTCAATGACC | Expression |
| E54.2 | AGAAGCACGCGAACAAGCA | Expression |
| E54.3 | TCTGCTTGTTCGCGTGCTT | Expression |
| E54.4 | CACCATTCACACTTCCAGA | Expression |
| E54.5 | CATGCTGACAGCAAAGTCA | Expression |
| Eh54.1 | GTCGGAGGAATGAAGAGT | Disruption |
| Eh54.3 | GTTGACGAGCTTGTTCTG | Disruption |
| sagAup | CGGAATTCGAAATGCTAGACTTAACGC | Complementation |
| sagAdown | CGGAATTCGCAAGCAGGTATCGACATG | Complementation |
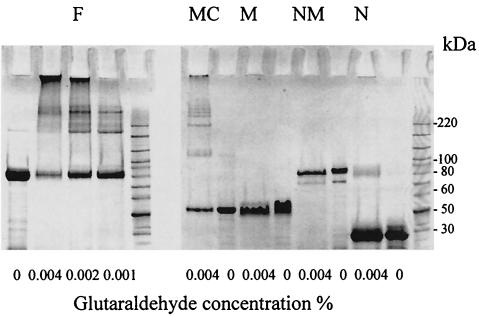
E54.1 and E54.5 were used to express the full-length SagA (F). E54.1 and E54.3 were used to express the N-terminal domain of SagA (N). E54.1 and E54.4 were used to express the N-terminal half of SagA (containing the N-terminal and middle domains [NM]). E54.2 and E54.4 were used to express the middle (repeat) domain of SagA (M). E54.2 and E54.5 were used to express the C-terminal half of SagA (containing middle and C-terminal domains [MC]). Letter codes correspond to lane designations in Fig. 5.
Computer analysis.
The basic local alignment search tool (BLAST) network service at the National Center for Biotechnology Information was used to search for homologous sequences in the protein database and the nucleotide database. The pI/MW program at the ExPASy molecular biology server of the Swiss Institute of Bioinformatics was used to predict the protein pI and molecular weight. The PSORT program at the GenomeNet database service was used to predict protein sorting signals and localization sites. The coilscan program in the GCG software package (Genetics Computer Group, Madison, Wis.) was used to locate coiled-coil structures in protein sequences.
Disruption of the chromosomal sagA gene of E. faecium TX1330.
After failure to generate an insertion disruption mutation in sagA gene, complementation of sagA in E. faecium was performed as follows. The full-length sagA gene and about 200 bp upstream were amplified from E. faecium by using the primers sagAup and sagAdown (Table 2) and cloned into the EcoRI site of pAT18, a shuttle vector which has an erythromycin resistance gene and can replicate in enterococci. The pAT18-carrying sagA (pTEX10003) was then electroporated into E. faecium TX1330, and 10 μg of erythromycin/ml was used for selection.
Disruption of the chromosomal sagA gene was performed as follows. The internal fragment of sagA was amplified by using oligonucleotides Eh54.1 and Eh54.3 (Table 2) and cloned into plasmid vector pTEX4577 (Table 1), which contains a kanamycin resistance gene and does not replicate in gram-positive bacteria. The resulting construct was electroporated into E. faecium TX10003 (TX1330 with pTEX10003), and 10 mg of kanamycin/ml was used for selection. Disruption of the chromosomal sagA gene was confirmed by PCR.
The essentiality of the sagA gene for E. faecium growth was examined as follows. The TX1330 strain with a disrupted chromosomal sagA gene, and the extrachromosomal sagA gene on pAT18 (TX10004) was grown overnight without antibiotics. The overnight culture was diluted and plated without selection on five BHI agar plates. About 100 colonies were obtained on each plate. These colonies were then replica plated onto BHI-erythromycin (10 μg/ml) agar plates. The colonies that lost erythromycin resistance were streaked onto BHI-kanamycin (10 mg/ml) agar plates to test the maintenance of the disruption of chromosomal sagA without transcomplementation with pAT18-sagA. At the same time, TX10004 was grown overnight with 10 mg of kanamycin/ml, plated on BHI-kanamycin (10 mg/ml), and replica plated onto BHI-erythromycin (10 μg/ml) to determine whether any of these kanamycin resistance colonies has lost pTEX10000 (pAT18-sagA).
Production of recombinant full-length and partial SagA proteins and generation of antiserum.
The sequences encoding full-length SagA protein and different domains were amplified (expression primers, see Table 2) and cloned in frame into the BamHI site of vector pQE31 (QIAexpressionist; Qiagen, Inc., Valencia, Calif.). The His-tagged SagA protein and domains were expressed in E. coli and purified under native conditions. The purified full-length SagA protein was sent to Bethyl Laboratories (Montgomery, Tex.), where immune serum was produced. In this case, antibodies to purified SagA protein were elicited in rabbits by intravenous immunization with four 100-μg doses of protein in phosphate-buffered saline (PBS) spaced 7 days apart.
Protein analysis.
To localize SagA protein in E. faecium, cell-associated proteins were prepared by treatment with sodium dodecyl sulfate (SDS) as previously described (49), and supernatant proteins were prepared by concentration to a 1/30 volume with a Millipore centrifugal filter device (10-kDa cutoff). Proteinase K and periodate treatment were performed as described previously (49). Glycoprotein labeling was performed as described by the manufacturer (ECL Glycoprotein Detection System; Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). SDS-polyacrylamide gel electrophoresis (PAGE) and Western blots were also performed as described previously (49).
Cross-linking was performed as described by Harborth et al. (11) with minor modifications. The purified recombinant SagA proteins were dialyzed for 12 h at 4°C against PBS. Glutaraldehyde (stock concentration of 0.01, 0.02, or 0.04%) was added to 10 μl of protein sample (0.2 to 0.4 mg/ml) at a final concentration of 0.001, 0.002, or 0.004%. Samples were incubated for 30 min at 14°C. The cross-linking reactions were terminated by the addition of 5 μl of the PBS buffer containing 2 M glycine. Cross-linked samples were resolved on SDS-7.5% PAGE.
A cell wall hydrolase assay was performed as described previously (3) with minor modifications. Then, 10 μl of concentrated TX16 supernatant (as described above) was mixed with 2× SDS loading buffer, boiled for 3 min, and applied to a SDS-10% PAGE gel containing 0.2% (wt/vol) lyophilized Micrococcus lysodeikticus or SDS-treated TX16 cells (36). Renaturation of lytic enzymes was done by overnight incubation in 25 mM Tris-HCl (pH 8.0) containing 1% (vol/vol) Triton X-100 at 37°C with gentle reciprocal shaking. Lytic zones in gels appeared as clear bands within the opaque gel matrix. The proteins on the gel were then transferred to a nitrocellulose membrane, and Western blotting was performed with anti-SagA antibody to locate the SagA protein on the gel. The positions of the SagA protein and the lytic zone were then compared to determine whether the SagA protein had lytic activity.
Isolation of fibrinogen-binding protein from E. faecium.
Dot blot assay was performed with horseradish peroxidase (HRP)-labeled fibrinogen and supernatant or surface crude extract of E. faecium. Conjugates of fibrinogen with HRP were obtained by peroxide oxidation (44). To extract supernatant proteins, E. faecium was incubated (without shaking) in Todd-Hewitt broth for 24 h at 37°C, bacterial culture supernatant was subjected to precipitation with ammonium sulfate (70% of saturation), and concentrated proteins were dialyzed against 0.001 M NH4HCO3. To extract surface proteins, an overnight culture (as described above) was harvested and washed with saline, and ca. 5 g (wet weight) of bacterial pellet was subjected to a four-step extraction: (i) 1 M NaCl in 0.05 M Tris-Cl (pH 7.5), (ii) 2 M KSCN in 0.05 M sodium acetate buffer (pH 5.0), (iii) 6 M urea in 0.1 M glycine buffer (pH 2.5), and (iv) 1 M HCl. The first three steps were performed at room temperature, and the last step was performed at 95°C. The supernatant and surface crude extracts were dotted onto nitrocellulose filters, air dried, and blocked for 15 min with 2% milk in saline containing 0.05% Tween 20, followed by incubation with fresh blocking solution containing labeled fibrinogen (1 μg/ml) for 1 h at room temperature with shaking. The filters were then washed with saline containing 0.05% Tween 20. Reaction was developed in 10 mM Tris-Cl (pH 7.4) with 0.06% 4-chloro-1-naphthol and 0.01% H2O2.
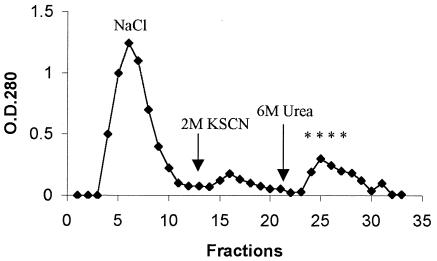
The supernatant crude extract (as described above) was subjected to affinity chromatography as follows. Columns packed with fibrinogen-Sepharose (Pharmacia Biotech) were equilibrated with the buffer 0.05 M Tris-Cl (pH 7.5)-0.25 M NaCl. Elution of proteins bound to the column was performed with (i) NaCl gradient (0.25 to 1 M) in 0.05 M Tris-Cl (pH 7.5) buffer, (ii) 2 M KSCN in 0.1 M sodium acetate buffer (pH 5.0), and (iii) 6 M urea in 0.1 M glycine buffer (pH 2.5). The flow rate was 40 ml/h, and the fraction sizes were 10 ml. The binding activity to fibrinogen was assayed in protein dot blot technique (described above) by using 5 μl of each collected fraction. Fractions positive in the assay were dialyzed against 0.05 M Tris-Cl (pH 7.5), concentrated on Centricon filters (30-kDa cutoff [Amicon]) and stored at −20°C. The purified fibrinogen-binding protein was subjected to preparative SDS-PAGE and blotted onto Immobilon membranes (Millipore, Italy), followed by N-terminal sequencing by Edman degradation in a gas-liquid sequenator 473A (Applied Biosystems).
Binding of recombinant SagA to ECM proteins.
Polyvinyl chloride plates (Becton Dickinson Labware, Franklin Lakes, N.J.) were coated with 1 μg of ECM proteins or BSA in 0.05 M carbonate buffer (pH 9.6)/well and then incubated at 4°C overnight. Wells were washed five times with PBST (PBS with 0.01% Tween 20) and blocked with 5% bovine serum albumin (BSA) at 37°C for 1 h. After the wells were washed, various concentrations of recombinant SagA proteins (0 to 20 μg/100 μl) in PBS with 1% BSA were added to the wells, followed by incubation at 37°C for 2 h. Wells were washed with PBST to remove the unbound proteins, and bound recombinant SagA proteins were detected by peta-His monoclonal antibodies (Qiagen), followed by the addition of HRP-conjugated goat anti-mouse immunoglobulin G antibodies (Life Technologies, Inc.). Relative binding was measured by monitoring A450 after the addition of 3,3′,5,5′-tetramethylbenzidine and H2O2.
GenBank accession number.
The GenBank accession number for E. faecium sagA and its flanking regions is AF242196.
RESULTS
Identification of the sagA gene of E. faecium.
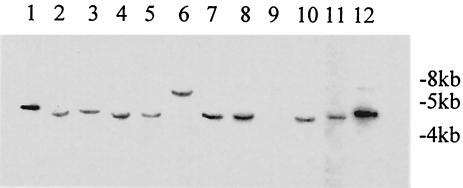
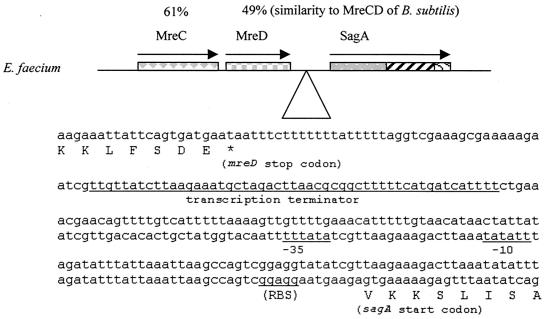
After we immunoscreened an E. faecium genomic expression library in plasmid pTEX5235, we obtained 36 immunopositive clones. Some of the immunopositive clones and the BLAST search results with the corresponding antigen-encoding genes are shown in Table 3. Among the 36 immunopositive clones, clone d1-27 showed one of the strongest reactions, suggesting that it is a prominent antigen expressed by E. faecium. The deduced peptide sequence of clone d1-27 showed 68% similarity to the N-terminal part of the Enterococcus hirae (previously misidentified as E. faecium [Marc Solioz, personal communication]) P54 protein (10). Another clone, d2-29, that showed weak reactivity with the sera showed 89% similarity (at amino acid level) to the C-terminal part of the E. hirae P54 protein. The gene in clones d1-27 and d2-29 was named sagA (for secreted antigen [see below]) and was found in all 11 E. faecium strains from different clinical and/or community and geographic sources (Fig. 1). The complete sequence of the sagA gene and its flanking genes from TX16 was obtained. The open reading frames upstream of sagA showed 61 and 49% similarity (in amino acid level) to the mreCD genes of Bacillus subtilis (Fig. 2), coding for cell shape determinants (21, 46). Between the putative mreCD and the sagA genes, there is a 258-bp region containing a putative transcription terminator and a putative promoter (Fig. 2), suggesting that the putative mreCD and the sagA genes are not in an operon. The gene downstream of sagA showed 66% similarity to a putative oligoendopeptidase gene of B. subtilis and is transcribed in the opposite orientation.
TABLE 3.
Results of BLAST searches with immunopositive clones containing a single gene
| Clone | BLAST hit | Organism | Expect valuea |
|---|---|---|---|
| d2-1 | ABC transporter | Enterococcus gallinarum | 3e-14 |
| d2-34 | Cation-transporting ATPase | Bacillus subtilis | 1.0e-45 |
| d1-39 | Cation-transporting ATPase | Bacillus subtilis | 1.0e-45 |
| d2-55 | Lactose-binding protein | Agrobacterium radiobacter | 2.6e-63 |
| d1-27 | P54 | Enterococcus hirae | 5e-46 |
| d2-29 | P54 | Enterococcus hirae | 2e-64 |
| d2-61 | P54 | Enterococcus hirae | 1e-36 |
| d1-48 | GroEL | Lactobacillus | 9.8e-129 |
| d1-47 | Dihydrolipoamid S-acetyltransferase | Enterococcus faecalis | 2.4e-59 |
| d2-2 | Hypothetical protein | Enterococcus faecium | 5e-82 |
| d2-46 | Thiophene and furan oxidation protein | Bacillus subtilis | 1.7e-63 |
| d2-44 | SocE (stringent response regulator) | Myxococcus xanthus | 3e-16 |
| d1-17 | Lmo0950 (hypothetical) | Listeria monocytogenes | 6e-16 |
| d2-37 | Orf122 | Chlorobium tepidum | 5e-13 |
| d1-31 | Glycerol uptake facilitator protein | Streptococcus pneumoniae | 3.3e-11 |
| d1-28 | Glycerol uptake facilitator protein | Streptococcus pneumoniae | 3.3e-11 |
| d1-13 | Low-affinity penicillin-binding protein | Enterococcus hirae | 1.3e-96 |
| d1-46 | No match |
The expert value is a parameter that describes the probability that a match occurs by chance when a search is done in a database of a particular size and composition.
FIG. 1.
Southern blots of genomic DNA digested with EcoRI by using pTEX10000 as probe (the same pattern is obtained with pTEX10001). Lanes 1 to 8 and lanes 10 to 12 are E. faecium (lane 1, TX16; lane 6, TX1330), and lane 9 is E. faecalis (used as a negative control).
FIG. 2.
Map of the sagA gene and its genomic environment. The percent similarities to MreC and MreD of B. subtilis are given. An insert with a sequence upstream of the sagA gene is shown with putative transcription terminator, −35, −10, and ribosome-binding site (RBS) sequences underlined.
Characterization of SagA protein.
The gene product of sagA has a predicted size of 524 amino acids and was predicted by PSORT to be secreted, with a predicted N-terminal signal peptide of 27 amino acids. The predicted mature gene product (without signal peptide) has a molecular mass of 53 kDa and a pI of 4.36. The gene product can be divided into three domains. The N-terminal domain (251 amino acids, ca. 27 kDa) was predicted by Coilscan to form a coiled-coil structure that is similar to the N-terminal part of an extracellular 45-kDa protein (41% similarity and 32% identity) of Listeria monocytogenes (36), as well as to the N-terminal part of the Streptococcus mutans SagA protein (42% similarity and 35% identity), which was named after our E. faecium SagA protein based on the sequence homology (7). The C-terminal domain (126 amino acids, ca. 13 kDa) of the sagA gene product has a motif of unknown function that is present in other proteins, including the P45 (50% similarity and 38% identity) and P60 (52% similarity and 43% identity) proteins of L. monocytogenes (5, 12, 13), but is not in the S. mutans SagA protein. The central domain (147 amino acids, ∼15 kDa) of the sagA gene product contains two kinds of similar direct repeats, repeats A (repeated three times) and B (repeated four times), each containing 12 amino acids (repeat A, S S A/TT/AQ S S A T/ME E S; repeat B, T A/VP E S S A/TT E E S T), which was not found in other known proteins.
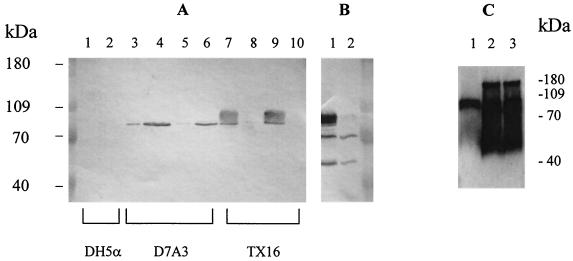
Using antiserum raised against recombinant SagA, the SagA protein was found in E. faecium supernatant from both exponential and stationary phases (Fig. 3). Anti-SagA antibody mainly reacted with TX16 supernatant and not with whole-cell lysates (Fig. 3A, lanes 7 to 10), indicating that the SagA protein was secreted into the medium. The reactive species appeared as a smear at a position from 80 to 90 kDa (Fig. 3A), which was susceptible to proteinase K treatment, but resistant to periodate treatment and glycoprotein labeling, suggesting that it was not modified by carbohydrates. Patient serum S0111 (along with a pooled human serum from healthy volunteers) was also used to perform Western blotting with the E. faecium cell lysate and the supernatant (Fig. 3B). Three major bands appeared on the supernatant blot (as opposed to negative result with normal human serum [data not shown]), while the broad band at the position of SagA protein was the only one missing from the cell lysate blot, suggesting that the SagA protein was the major secreted antigen. With the E. coli clone D7A3, which contains the complete sagA gene, the SagA protein was mostly found in the whole-cell lysate (Fig. 3A, lanes 3 to 6). It appeared as a single homogeneous band of ca. 80 kDa, corresponding to the bottom of the smear seen with E. faecium strains. When overexpressed in E. coli M15 from pQE31 (Table 1), a stable oligomer of SagA protein was seen (Fig. 4B). To further study the involvement of different domains in oligomerization of SagA protein, a cross-linking experiment was performed with recombinant full-length and partial SagA proteins (Fig. 5). The full-length and the 27-kDa C-terminal half of the SagA protein showed a series of high-molecular-weight forms in the presence of the cross-linker, the N-terminal domain showed a weak band at a high-molecular-weight position, and the middle repeat containing domain did not show any high-molecular-weight forms. These results suggest that the C-terminal domain is important for formation of higher oligomeric forms of the SagA protein.
FIG. 3.
(A) Western blot of different cell culture fractions with anti-SagA antibody. Lanes 1, 3, 5, 7, and 9, supernatants; lanes 2, 4, 6, 8, and 10, whole-cell lysates. Lanes 3, 4, 7, and 8 show overnight cultures; lanes 5, 6, 9, and 10 show 4-h cultures (optical density at 600 nm of 0.5). Lanes 1 and 2, E. coli; lanes 3, 4, 5, and 6, E. coli clone D7A3; lanes 7, 8, 9, and 10, E. faecium TX16. (B) Western blot with patient serum and E. faecium TX16. Lane 1, supernatant; lane 2, whole-cell lysate. (C) Western blot with anti-SagA antibody. Lane 1, E. faecium TX1330 supernatant; lane 2, TX10003 (TX1330 with sagA gene expressed in trans) supernatant; lane 3, TX10004 (TX1330 with sagA gene expressed in trans, chromosomal sagA disrupted) supernatant.
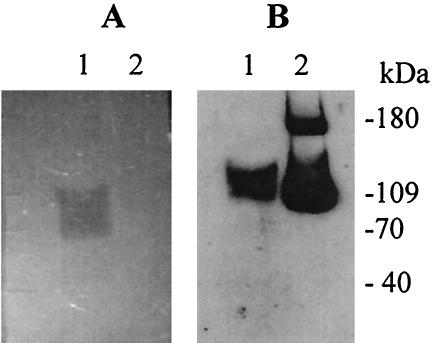
FIG. 4.
Cell wall hydrolase assay and Western blot. E. faecium supernatant and recombinant SagA protein were run on an SDS-PAGE gel containing E. faecium cell wall extract, renatured to detect lytic activity (A), and then blotted with anti-SagA antiserum to localize SagA on the gel (B). Lane 1, TX16 supernatant; lane 2, recombinant SagA protein from E. coli M15 and formation of a 180-kDa SagA oligomer possibly due to large amount of SagA.
FIG. 5.
Cross-linking of recombinant full-length and partial SagA. Different concentrations of glutaradehyde were used (given at the bottom of panels) and products were run on SDS-PAGE. Lanes: F, full-length SagA; N, N-terminal domain of SagA; NM, N-terminal half of SagA (containing the N-terminal and middle domains); M, middle (repeat) domain of SagA; MC, C-terminal half of SagA (containing the middle and C-terminal domains).
Since the listerial P60 and P45 proteins, which have homology to SagA, both exhibit peptidoglycan hydrolytic activity (5, 36) and the S. mutans SagA protein was shown to be involved in cell wall maintenance (7), we suspected that the E. faecium SagA might also have similar cell wall lytic activity. However, under our testing conditions, lytic activity could not be detected on M. lysodeikticus or E. faecium cells for either native SagA protein in the supernatant of E. faecium TX16 or the purified His-tagged SagA protein expressed in E. coli, although a lytic band was present at a position slightly lower than the location of the SagA protein on the gel, when we compared the cell wall gel (Fig. 4A) and the Western blot with anti-SagA antiserum (Fig. 4B). The SagA protein ran more slowly in the cell wall gel than in the regular SDS-PAGE gel (Fig. 3 and 4B), suggesting that the SagA protein may be retarded by interaction with cell wall, whereas the protein marker was not.
The sagA gene is essential for E. faecium growth.
Disruption of the sagA gene in E. faecium TX1330 was not successful, although the same method has been successfully applied to obtain other mutants of E. faecium TX1330 (38). Disruption of chromosomal sagA gene was possible, however, after transcomplementation with pAT18-sagA. Overexpression of SagA in the complemented E. faecium strain TX1330 was detected by Western blotting (Fig. 3C); overexpressed SagA protein was found mainly in culture supernatant and formed a stable oligomer that was not disrupted in SDS-PAGE gels (Fig. 3C); while, in this experiment, an oligomer was not seen with the wild-type strain, on a few occasions we did observe a very faint 180-kDa anti-SagA reactive band in the supernatant from the wild-type strain (data not shown). When TX10004 (TX1330 strain with a disrupted chromosomal sagA gene and an extrachromosomal sagA gene on pAT18) was grown overnight without antibiotics, about one-third of the colonies lost the pAT18-sagA plasmid, suggesting that this plasmid was not stable in TX1330. At the same time, the colonies that lost the pAT18-sagA had lost the high-level kanamycin resistance, indicating that these colonies had reverted to the wild type. When TX10004 was grown overnight with 10 mg of kanamycin/ml and plated on BHI-kanamycin (10 mg/ml), all colonies (ca. 500 colonies were tested) on BHI-kanamycin (10 mg/ml) agar plates also grew on BHI-erythromycin (10 μg/ml) agar plates, suggesting that the extrachromosomal sagA has to be present for E. faecium with a disrupted chromosomal sagA to grow under the testing condition.
Identification of SagA protein as fibrinogen-binding protein.
In earlier attempts to screen for enterococcal clinical isolates that bound to fibrinogen, we identified E. faecium isolate 448, which was one of the strongest fibrinogen-binding E. faecium isolates in a dot blot assay with HRP-labeled fibrinogen. Strong fibrinogen-binding activity was detected in supernatants of E. faecium 448 and rather weakly from the surface protein extracts, leading to our use of E. faecium 448 supernatant for the isolation of a fibrinogen-binding protein (Fig. 6). The fractions eluted from the fibrinogen-Sepharose affinity column with 6 M urea were positive in dot blots with HRP-labeled fibrinogen. N-terminal sequencing of the protein identified a 20-amino-acid sequence, DFDSQIQQQDQKIADLKNQQ, identical to the predicted N-terminal sequence of the mature SagA. After we subsequently identified sagA and had raised antibodies aginst recombinant SagA, we reexamined the protein purified from E. faecium 448 and found that it reacted with the anti-SagA antibody and, on the Western blot, it appeared as a smear at a position from 80 to 90 kDa, which was the same pattern seen with the TX16 supernatant, thus further confirming that the fibrinogen-binding protein isolated from strain 448 is SagA.
FIG. 6.
Isolation of a fibrinogen-binding protein from E. faecium 448 supernatant. Asterisks indicate fibrinogen-binding fractions in a dot blot; arrows indicate when the column was washed with elution buffers.
Binding of recombinant SagA protein to ECM proteins.
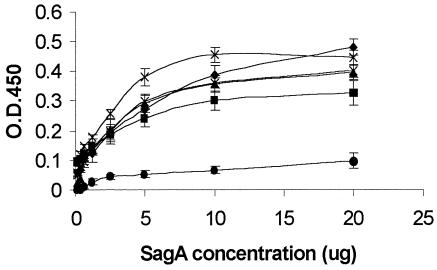
Binding of recombinant SagA protein to immobilized fibrinogen, collagen type I, collagen type IV, fibronectin, and laminin was demonstrated in an enzyme-linked immunosorbent assay (ELISA) (Fig. 7), with BSA as a control. Binding of recombinant SagA protein to fibrinogen, collagen type I, collagen type IV, fibronectin and laminin was found to be concentration dependent and to exhibit saturation kinetics (Fig. 7).
FIG. 7.
Binding of recombinant SagA protein to ECM proteins. The results from two independent experiments are shown. Error bars show the standard deviation. Symbols: ♦, fibrinogen; ▪, collagen type I; ▴, collagen type IV; ×, laminin; ✠, fibronectin; •, BSA.
DISCUSSION
In our effort to identify E. faecium proteins that were expressed and were antigenic during human infections, a wide variety of antigen-encoding genes were sequenced. Analysis of these sequences revealed similarity to proteins involved in transport, chaperone activity, regulation, and metabolism. Study of these in vivo expressed antigens should improve our understanding of the biology of E. faecium in infection and could lead to the discovery of virulence factors and to the development of serodiagnostic and therapeutic tools. One of the E. faecium antigen-encoding genes, sagA, as well as its protein product, was characterized here. The SagA protein is remarkable as a prominent antigen that is quantitatively secreted to the medium, that binds to ECM proteins, and that, at the same time, appears to be essential for E. faecium growth.
The sagA gene is located in a cluster of genes encoding proteins (MreCD) involved in cell wall metabolism and encodes a protein with sequence homology to cell wall metabolism-related proteins such as SagA of S. mutans and P60 and P45 of L. monocytogenes, suggesting that the SagA protein may be involved in cell wall metabolism. The murein hydrolase activity of P60 protein has been shown to be dependent on the conserved single cysteine residue, which was also present in E. faecium SagA protein at the C-terminal domain (5). However, when we performed a cell wall hydrolase assay with M. lysodeikticus and E. faecium cells, no lytic activity was detected for the native SagA protein in TX16 supernatant or the purified recombinant SagA protein. Thus, if SagA is a cell wall hydrolase, it may act on a different substrate or just be inactive in this assay. Putative mreCD genes were found upstream of both E. faecium and S. mutans sagA genes (7), suggesting the two genes have similar gene organizations and may have related functions. However, the S. mutans SagA protein does not share the repeat region nor the C-terminal domain with E. faecium SagA and has been shown to be a glycoprotein associated with cell wall, whereas the E. faecium SagA protein appeared to be quantitatively secreted to the medium and was resistant to periodate treatment and glycoprotein labeling, suggesting possibly different structures and/or functions for the two proteins. Possible explanations for the smear pattern of native SagA protein on SDS-PAGE gel include that the protein may indeed be modified by carbohydrate, but undetectable under our testing conditions, or that the protein may undergo other modifications or interact with other protein(s).
Proteins with a role in cell growth may also have other functions, such as the P60 protein of L. monocytogenes. On the one hand, P60 protein is a murein hydrolase that has an essential role in cell division and, on the other hand, it is involved in the invasion by L. monocytogenes of some selected mammalian cells, such as fibroblasts, macrophages, and hepatocytes (5, 12, 13). In the present study, various findings indicate that the SagA protein is an adhesin with broad-spectrum binding to ECM proteins. This conclusion was supported by the isolation of a fibrinogen-binding protein from an E. faecium strain, whose N-terminal 20 amino acids matched the SagA protein sequence. The fibrinogen-binding protein was also confirmed to be the SagA protein by its size, by its reactivity with anti-SagA antibody, and by the pattern it shows on the Western blot. In the procedure for isolation of a fibrinogen-binding moiety, this protein was eluted from the fibrinogen-Sepharose column with 6 M urea, suggesting a specific interaction between SagA protein and fibrinogen. Additional evidence for the binding ability of SagA derives from enzyme-linked immunosorbent assays with recombinant SagA protein; this evidence not only confirmed the interaction between SagA and fibrinogen but also demonstrated the binding of SagA to other ECM proteins, including collagen type I, collagen type IV, fibronectin, and laminin.
Interaction of pathogenic bacteria with mammalian ECM and serum proteins has been widely studied and been shown to be involved in bacterial virulence (9, 19, 30, 47). Although most of these studies are focused on bacterial-surface-located adhesins, secreted proteins that bind to ECM proteins have also been reported. Such examples include the Efb and Eap/Map proteins of Staphylococcus aureus (4, 6, 8, 14-16, 18, 27-29). Efb (for extracellular fibrinogen-binding protein) is a potential virulence factor of S. aureus in animal models and very immunogenic in the course of human infection (8, 20, 28). The secreted Efb protein, by binding to fibrinogen, inhibits ADP-induced, fibrinogen-dependent platelet aggregation, probably by blocking platelet integrin GPIIb/IIIa-binding to fibrinogen; this inhibition is thought to be responsible for delay in wound healing (29). Another secreted protein, Eap (for extracellular adherence protein)/Map, has been shown to have broad-spectrum binding characteristics, to promote adherence of S. aureus to eukaryotic cells (probably via its interaction with staphylococcal surface receptors), to serve as an anti-inflammatory factor via its specific interactions with ICAM-1 and ECM proteins, to produce oligomeric forms, and to mediate staphylococcal agglutination (6, 14-16, 18, 27). The E. faecium SagA protein, like the Eap protein, was shown to produce oligomeric forms, and the C-terminal domain of SagA may be responsible for the oligomerization. A highly similar C-terminal domain (70% identity) was found in at least one other E. faecium protein (unpublished data); whether these proteins can interact with each other, as the C-terminal domain of SagA can do, is not known. Since the C-terminal domain of SagA is not present in Eap, these two proteins may use different mechanisms to form oligomers. The E. faecium SagA protein does not show significant sequence homology or structural similarity (such as the major histocompatibility complex-like conformation of Eap/Map protein) to known ECM binding proteins, and attempts to search for a known binding motif in SagA have not been successful, suggesting a possible novel binding mechanism for SagA.
In conclusion, a major secreted antigen, SagA, as well as its encoding gene, was identified in E. faecium. The SagA protein appears to be essential for E. faecium growth, perhaps due to an as-yet-unknown function in cell wall metabolism and, when overexpressed, forms oligomers. The native SagA protein was isolated as a fibrinogen-binding protein, and the recombinant SagA protein shows binding activity to a number of ECM proteins. The SagA protein is composed of three domains with distinct sequence and/or structural characteristics. The involvement of different domains of SagA protein in its structure or function and the physiological and pathogenic relevance of the protein will be the subject of future studies.
Acknowledgments
This work was supported by NIH grant AI42399 from the Division of Microbiology and Infectious Diseases to Barbara E. Murray and by FEMS and ESCMID scholarships to Magdalena Kawalec.
We thank Yi Xu for technical support in constructing the genomic libraries and immunoscreening; Kavindra V. Singh and Monjula Chidambaram for general technical support; Karl-Hermann Schmidt, David Šmajs, and Shuguang Liang for technical support in protein purification; and Marc Solioz at the University of Berne, Berne, Switzerland, for information about the previous misidentification of the E. hirae strain as E. faecium.
Editor: J. N. Weiser
REFERENCES
- 1.Arduino, R. C., K. Jacques-Palaz, B. E. Murray, and R. M. Rakita. 1994. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 62:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. F. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. Wiley, Boston, Mass.
- 3.Beliveau, C., C. Potvin, J. Trudel, A. Asselin, and G. Bellemare. 1991. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J. Bacteriol. 173:5619-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boden, M. K., and J. I. Flock. 1994. Cloning and characterization of a gene for a 19-kDa fibrinogen-binding protein from Staphylococcus aureus. Mol. Microbiol. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 5.Bubert, A., M. Kuhn, W. Goebel, and S. Kohler. 1992. Structural and functional properties of the p60 proteins from different Listeria species. J. Bacteriol. 174:8166-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavakis, T., M. Hussain, S. M. Kanse, G. Peters, R. G. Bretzel, J. I. Flock, M. Herrmann, and K. T. Preissner. 2002. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 8:687-693. [DOI] [PubMed] [Google Scholar]
- 7.Chia, J. S., L. Y. Chang, C. T. Shun, Y. Y. Chang, and J. Y. Chen. 2001. A 60-kilodalton immunodominant glycoprotein is essential for cell wall integrity and the maintenance of cell shape in Streptococcus mutans. Infect. Immun. 69:6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colque-Navarro, P., M. Palma, B. Soderquist, J. I. Flock, and R. Mollby. 2000. Antibody responses in patients with staphylococcal septicemia against two Staphylococcus aureus fibrinogen binding proteins: clumping factor and an extracellular fibrinogen binding protein. Clin. Diagn. Lab. Immunol. 7:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 10.Furst, P., H. U. Mosch, and M. Solioz. 1989. A protein of unusual composition from Enterococcus faecium. Nucleic Acids Res. 17:6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harborth, J., K. Weber, and M. Osborn. 1995. Epitope mapping and direct visualization of the parallel, in-register arrangement of the double-stranded coiled-coil in the NumA protein. EMBO J. 14:2447-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess, J., A. Dreher, I. Gentschev, W. Goebel, C. ladel, D. Mido, and S. H. Kaufmann. 1996. Protein p60 participates in intestinal host invasion by Listeria monocytogenes. Zentbl. Bakteriol. 284:263-272. [DOI] [PubMed] [Google Scholar]
- 13.Hess, J., I. Gentschev, G. Szalay, C. Ladel, A. Bubert, W. Goebel, and S. H. Kaufmann. 1995. Listeria monocytogenes p60 supports host cell invasion by and in vivo survival of attenuated Salmonella typhimurium. Infect. Immun. 63:2047-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain, M., K. Becker, C. von Eiff, G. Peters, and M. Herrmann. 2001. Analogs of Eap protein are conserved and prevalent in clinical Staphylococcus aureus isolates. Clin. Diagn. Lab. Immunol. 8:1271-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain, M., A. Haggar, C. Heilmann, G. Peters, J. I. Flock, and M. Herrmann. 2002. Insertional inactivation of Eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect. Immun. 70:2933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson, K., D. McDevitt, M. H. McGavin, J. M. Patti, and M. Hook. 1995. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J. Biol. Chem. 270:21457-21460. [DOI] [PubMed] [Google Scholar]
- 17.Kreft, B., R. Marre, U. Schramm, and R. Wirth. 1992. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect. Immun. 60:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreikemeyer, B., D. McDevitt, and A. Podbielski. 2002. The role of the map protein in Staphylococcus aureus matrix protein and eukaryotic cell adherence. Int. J. Med. Microbiol. 292:283-295. [DOI] [PubMed] [Google Scholar]
- 19.Ljungh, A., A. P. Moran, and T. Wadstrom. 1996. Interactions of bacterial adhesins with extracellular matrix and plasma proteins: pathogenic implications and therapeutic possibilities. FEMS Immunol. Med. Microbiol. 16:117-126. [DOI] [PubMed] [Google Scholar]
- 20.Mamo, W., M. Boden, and J. I. Flock. 1994. Vaccination with Staphylococcus aureus fibrinogen binding proteins (FgBPs) reduces colonisation of S. aureus in a mouse mastitis model. FEMS Immunol. Med. Microbiol. 10:47-53. [DOI] [PubMed] [Google Scholar]
- 21.Margolis, P. S., A. Driks, and R. Losick. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175:528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 24.Nallapareddy, S. R., X. Qin, G. M. Weinstock, M. Hook, and B. E. Murray. 2000. Enterococcus faecalis adhesin, Ace, mediates attachment to proteins collagen type IV and laminin as well as collagen extracellular matrix type I. Infect. Immun. 68:5218-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nallapareddy, S. R., K. V. Singh, R. Duh, G. M. Weinstock, and B. E. Murray. 2000. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of Ace during human infections. Infect. Immun. 68:5210-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nallapareddy, S. R., G. M. Weinstock, and B. E. Murray. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 47:1733-1747. [DOI] [PubMed] [Google Scholar]
- 27.Palma, M., A. Haggar, and J. I. Flock. 1999. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 181:2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palma, M., S. Nozohoor, T. Schennings, A. Heimdahl, and J. I. Flock. 1996. Lack of the extracellular 19-kilodalton fibrinogen-binding protein from Staphylococcus aureus decreases virulence in experimental wound infection. Infect. Immun. 64:5284-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palma, M., O. Shannon, H. C. Quezada, A. Berg, and J. I. Flock. 2001. Extracellular fibrinogen-binding protein, Efb, from Staphylococcus aureus blocks platelet aggregation due to its binding to the alpha-chain. J. Biol. Chem. 276:31691-31697. [DOI] [PubMed] [Google Scholar]
- 30.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 31.Ponnuraj, K., Y. Xu, D. Moore, C. C. Deivanayagam, L. Boque, M. Hook, and S. V. Narayana. 2002. Crystallization and preliminary X-ray crystallographic analysis of Ace: a collagen-binding MSCRAMM from Enterococcus faecalis. Biochim. Biophys. Acta 1596:173-176. [DOI] [PubMed] [Google Scholar]
- 32.Rice, L. B., L. Carias, S. Rudin, C. Vael, H. Goossens, C. Konstabel, I. Klare, S. R. Nallapareddy, W. Huang, and B. E. Murray. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 187:508-512. [DOI] [PubMed] [Google Scholar]
- 33.Rich, R. L., B. Kreikemeyer, R. T. Owens, S. LaBrenz, S. V. Narayana, G. M. Weinstock, B. E. Murray, and M. Hook. 1999. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J. Biol. Chem. 274:26939-26945. [DOI] [PubMed] [Google Scholar]
- 34.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schubert, K., A. M. Bichlmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. P45, an extracellular 45-kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 37.Shiyuza, H., B. Birren, U.-J. Kim, S. T. Mancino, Y. Slepak, Y. Tacchiiri, and M. Simon. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89:8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, K. V., K. Malathum, and B. E. Murray. 2001. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob. Agents Chemother. 45:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 40.Speciale, P., G. Raucci, S. Meloni, M. L. Meloni, and T. Wadstrom. 1987. Binding of collagen to group A, B, C, D, and G streptococci. FEMS Microbiol. Lett. 48:47-51. [Google Scholar]
- 41.Switalski, L. M., A. Ljungh, C. Ryden, K. Rubin, M. Hook, and T. Wadstrom. 1982. Binding of fibronectin to the surface of group A, C, and G streptococci isolated from human infections. Eur. J. Clin. Microbiol. 1:381-387. [DOI] [PubMed] [Google Scholar]
- 42.Teng, F., K. D. Jacques-Palaz, G. M. Weinstock, and B. E. Murray. 2002. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect. Immun. 70:2010-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teng, F., B. E. Murray, and G. M. Weinstock. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid 39:182-186. [DOI] [PubMed] [Google Scholar]
- 44.Tijeesen, P. 1985. Practice and theory of enzyme immunoassays, p. 221-276. In R. H. Burdon and A. van Knippenberg (ed.), Laboratory techniques in biochemistry and molecular biology. Elsevier, Amsterdam, The Netherlands.
- 45.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 46.Varley, A. W., and G. C. Stewart. 1992. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (minCD) and cell shape (mreBCD) determinants. J. Bacteriol. 174:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westerlund, B., and T. K. Korhonen. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9:687-694. [DOI] [PubMed] [Google Scholar]
- 48.Xiao, J., M. Hook, G. M. Weinstock, and B. E. Murray. 1998. Conditional adherence of Enterococcus faecalis to extracellular matrix proteins. FEMS Immunol. Med. Microbiol. 21:287-295. [DOI] [PubMed] [Google Scholar]
- 49.Xu, Y., L. Jiang, B. E. Murray, and G. M. Weinstock. 1997. Enterococcus faecalis antigens in human infections. Infect. Immun. 65:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, Y., B. E. Murray, and G. M. Weinstock. 1998. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect. Immun. 66:4313-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, Y., K. V. Singh, X. Qin, B. E. Murray, and G. M. Weinstock. 2000. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect. Immun. 68:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zareba, T. W., C. Pascu, W. Hryniewicz, and T. Wadstrom. 1997. Binding of extracellular matrix proteins by enterococci. Curr. Microbiol. 34:6-11. [DOI] [PubMed] [Google Scholar]