Abstract
Staphylococcus aureus is a gram-positive pathogen that is capable of expressing a variety of virulence proteins in response to environmental signals. Virulence protein expression in S. aureus is controlled by a network of regulatory loci including sarA and agr. The sarA/agr network is associated with the expression of cell wall-associated adhesins during exponential growth and the expression of secreted enzymes and toxins in the transition to post-exponential growth. A number of sarA homologs, including sarT and sarS, have been identified in the S. aureus genome. Previous studies have shown that sarA influences expression of both sarT and sarS in the global regulatory network. SarS has been shown to bind to the spa promoter to induce expression of protein A. SarT, one of the SarA homologs that represses hla expression and is repressible by SarA and agr, was found to induce sarS expression in this report. Northern blot analysis of sarS and spa expression in S. aureus RN6390, and the isogenic sarT, sarT sarA, and sarT agr mutants showed that while sarA regulated spa expression directly, the agr locus used sarT as an intermediary to regulate sarS, thus leading to spa repression in agr-activated cells. Gel shift and footprinting analysis showed that SarT binds to the sarS promoter, indicating that the interaction of the sarT gene product with the upstream region of sarS is likely direct. Induction of sarS and spa by SarT in agr+ strains was confirmed by a tetracycline-inducible system to titrate sarT expression.
Throughout in vitro growth, and presumably during an infection, Staphylococcus aureus responds to environmental signals by selectively generating specific virulence proteins. During the exponential phase in vitro, S. aureus synthesizes surface proteins that mediate attachment, including protein A, fibrinogen, and fibronectin binding proteins (27, 45). In an infection, bacterial attachment would facilitate active colonization and formation of bacterial vegetations within the host (15, 27, 45). As the cells enter the post-exponential phase in vitro, proteins mediating attachment are repressed, while S. aureus synthesizes exoproteins (including hemolysins, toxins, proteases, and lipases) capable of causing host cell tissue damage (21, 47). By lysing host cells, proteins expressed in the post-exponential phase likely aid in the acquisition of additional nutrients and facilitate dissemination of S. aureus in vivo (45). A complex regulatory network, with many linked components apparently imparts precise and flexible coordination of protein expression in response to the microenvironment (1, 9, 36). The activation, repression, and interactions of the regulatory genes must be understood to develop a comprehensive picture of the infectious process and disease development.
The sarA and agr loci comprise a primary global regulatory system that coordinates synthesis of cell wall and extracellular virulence proteins during late exponential and post-exponential phases (1) (Fig. 1). Transcriptional profiling studies comparing sarA and agr mutants with wild-type cells at different growth phases indicate that in addition to the virulence proteins, sarA and agr regulate expression of a number of genes involved in metabolic processes, and activation of metabolic and virulence genes is associated with the transition from exponential to post-exponential phase of growth (21).
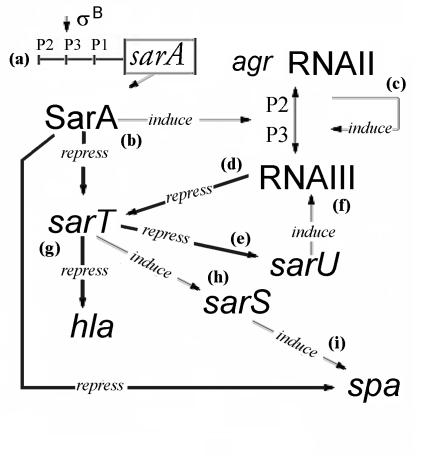
FIG. 1.
A simple overview of the predicted sarA/agr regulatory web involving SarT and SarS. (a) SarA is transcribed from one of three nested promoters (P1, P2, or P3). (b) SarA represses expression of spa (protein A) and sarT (17, 48, 49) and induces agr RNAII (19, 38). (c) agr RNAII encodes a two-component quorum-sensing system, which activates expression of agr RNAIII, a pleiotropic regulator for expression of proteins associated with virulence (23, 27, 42, 46). (d) agr RNAIII represses sarT. (e) Increased expression of sarT causes repression of sarU, particularly during exponential growth (35). (f) sarU induces expression of agr RNAIII (35). (g) SarT represses expression of hla (48). (h) SarT induces expression of sarS (this study), and sarS induces expression of spa (17). The conditions and regulators mediating induction of sarT have not yet been determined.
The agr locus (Fig. 1) is a pleiotropic regulator for the synthesis of exoproteins as well as a repressor of cell wall synthesis during the post-exponential phase (10, 23, 25, 27, 42, 46). The agr locus encodes two divergent transcripts, RNAII and RNAIII, originating from two adjacent promoters, P2 and P3, respectively (27, 41) (Fig. 1). RNAII encodes four genes, agrB, -D, -C, and -A, which comprise the genetic elements of a two-component quorum-sensing system (25, 30, 38). The RNAII gene product AgrD encodes a 46-residue peptide that is processed to a cyclic peptide (autoinducing peptide [AIP]) by AgrB and exported (24, 25). AgrC is a transmembrane sensor that is autophosphorylated when activated by a threshold concentration of AIP and, in turn, activates AgrA, the response regulator for transcriptional activation of the agr P2 and P3 promoters (30, 41). The P3 promoter mediates transcription of RNAIII, the agr regulatory molecule. RNAIII mRNA forms a complex three-dimensional structure containing multiple loops that are believed to control transcription and under certain circumstances, translation of exoprotein genes by binding to specific target sites (5, 23, 37, 42).
The agr P2-P3 interpromoter region, in addition to self-activation via AgrA, is also activated by SarA, the sarA gene product (16). The sarA locus consists of a 372-bp open reading frame driven by three sequential promoters (P2, P3, and P1) (4, 34) (Fig. 1a). Due to their overlapping nature, all three transcripts encode the sarA open reading frame. The P2 transcript encodes the entire sarA locus, including the triple promoter region (11). The P1 and P2 promoters are active during exponential and late exponential growth, while the P3 promoter is most active during the post-exponential phase and is regulated by σB, a stress-induced transcription factor (34). Protein-DNA binding studies revealed that SarA binds to a 29-bp sequence in the agr P2-P3 interpromoter region (Fig. 1c), activating agr RNAII and RNAIII transcription (19, 38).
While the sarA/agr system is the major controlling element for the expression of a variety of virulence proteins during the growth cycle (10, 13, 20), a series of other SarA-like proteins, discovered by promoter trap and genomic scanning, also appear to interact within the sarA/agr global regulatory network. Examples of these SarA homologs include sarR, which appears to repress sarA expression by binding to the sarA P1 and P3 promoters (33), and sarS, which induces spa (protein A) transcription (17, 49) (Fig. 1i). sarT, which is repressed by sarA and agr (Fig. 1b and d), has been shown by Northern blot, Western blot, and hemolytic assays to repress hla, the gene encoding α-hemolysin (48) (Fig. 1g). Interestingly, even though sarT is repressed by agr, it was shown both by Northern blotting and by a promoter-reporter system that repression of sarT could result in up-regulation of sarU, an activator of RNAIII, thus implying a positive-feedback control loop (35, 48) (Fig. 1e and f).
The sarS locus, adjacent to the spa gene, has been linked with spa induction (17, 49) (Fig. 1i). In this study, we showed that expression of SarT was accompanied by increased transcript levels of sarS and spa. Northern blot analysis of sarS and spa expression in S. aureus RN6390 and the isogenic sarT, sarT sarA, and sarT agr mutants showed that while sarA regulates sarS and spa expression directly, agr utilizes sarT to down-regulate sarS and thereby repress spa expression (Fig. 1h). Gel shift and DNA footprinting analysis revealed that purified SarT protein binds specifically to the sarS promoter region. Importantly, sarS and spa expression was induced in a dose-dependent manner when sarT expression was driven by a tetracycline-inducible promoter. Taken together, these data suggest that SarT is a positive regulator of sarS and the ensuing spa expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. Phage 80α (39) was used as a transducing phage. Escherichia coli strains were grown in Luria-Bertani (LB) medium (32). S. aureus strains were maintained in tryptic soy medium (Difco) and grown in CYGP or 03GL medium (39). Erythromycin (5 μg/ml), chloramphenicol (34 μg/ml in E. coli and 10 μg/ml in S. aureus), tetracycline (5 μg/ml), ampicillin (50 μg/ml), and kanamycin (50 μg/ml) were used for selection of transformants and transductants.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) |
|---|---|---|
| S. aureus | ||
| 8325-4 | Prophage-cured strain of NCT8325, harboring an 11-bp deletion in rsbU, which regulates sigB activity by activating rsbV, a factor that competitively binds to the anti-sigma factor RsbW | 39, 42 |
| RN4220 | Mutant strain of 8325-4 that accepts foreign DNA | 39 |
| RN6390 | agr+ laboratory strain related to 8325-4 | 39 |
| RN6911 | agr mutant of RN6390 (Δagr::tetM) | 42 |
| SH1000 | 8325-4 with intact rsbU | 22 |
| ALC1905 | sarT mutant of RN6390 (sarT::ermC) | 48 |
| ALC1927 | sarS mutant of RN6390 (sarS::ermC) | 15 |
| ALC2009 | ALC1927 with pSK236 containing a 1.2-kb sarS coding region fragment | 17 |
| ALC2056 | agr sarT mutant of RN6390 | 48 |
| ALC2057 | RN6390 sarA::kan | 48 |
| ALC2071 | ALC1905 with pALC2047 | 48 |
| ALC2076 | ALC2056 with pALC2047 | 48 |
| ALC2122 | ALC2057 with sarT::ermC (sarA sarT) | 48 |
| ALC2150 | ALC2122 with pALC2047 | 48 |
| ALC2221 | ALC1905 with pALC2217 | This study |
| ALC2235 | ALC2056 with pALC2217 | This study |
| ALC2236 | ALC2122 with pALC2217 | This study |
| ALC2253 | ALC2122 with pALC2073 | This study |
| ALC2254 | ALC2056 with pALC2073 | This study |
| ALC2255 | ALC1905 with pALC2073 | This study |
| ALC2305 | RN4220 with pALC2229 | This study |
| ALC2313 | RN4220 sarT::tetK | This study |
| ALC2389 | sarT mutant of SH1000 (sarT::tetK) | This study |
| ALC2394 | ALC2389 with pALC2047 | This study |
| ALC2398 | RN6390 with pALC2047 | This study |
| ALC2399 | ALC2389 with pALC2217 | This study |
| ALC2402 | ALC2389 with pACL2073 | This study |
| Plasmids | ||
| pALC1894 | pUC18 with 3.2-kb fragment containing the sarT coding region | 48 |
| pALC2047 | pSK236::sarT | 48 |
| pALC2073 | pSK236::xyl/tetO promoter::tetR | 3 |
| pALC2217 | sarT ligated into the EcoRI and SstI sites of pALC2073 | This study |
| pALC2223 | pALC1894 containing the sarT mutation (sarT::tetK) | This study |
| pALC2229 | pBT2 with the sarT mutation (sarT::tetK) | This study |
| pACL2321 | pCR2.1 Topo with the sarS promoter fragment | This study |
Generating a sarT mutant.
The tetK gene was cloned from pT181 (26) using the primers 5′-GAT AAA AGA AAT TTC GCC AGT C-3′ and 5′ ACG CGT CGA CAC TCG TTA ATA CGT GTG CTC TG-3′ and ligated into pCR2.1 (Invitrogen, San Diego, Calif.). tetK was ligated into a blunted NdeI site within the sarT coding region of pALC1894 (48), the insertion was confirmed by DNA sequencing, and the resulting 4.9-kb fragment containing tetK inserted into the sarT region was ligated into pBT2 (7). The resulting plasmid, pALC2229, was electroporated into S. aureus RN4220 and propagated through several cycles of alternating the temperature between 30 and 42°C as described elsewhere (8). Chloramphenicol-sensitive, tetracycline-resistant colonies, representing possible double-crossover events, were selected (14) and screened for tetK insertion into sarT by Southern blotting and by sequencing of the PCR fragment containing the junctional fragment. The S. aureus RN4220 sarT::tetK mutant (ALC2313) was used as the source for transduction of the sarT::tetK mutation to other S. aureus strains.
The sarT mutant of S. aureus SH1000 was generated by transducing strain SH1000 with an 80α phage lysate of S. aureus ALC2313 as previously described (14, 50). To confirm the genotype, chromosomal DNA from S. aureus cultures grown overnight was isolated by lysostaphin lysis and phenol extraction as described previously (14). DNA extracts of the putative transductants were digested with restriction enzymes and screened by Southern blotting for the presence of antibiotic resistance genes and shifts in the sizes of restriction digest fragments as previously described (48). Northern blot analysis with relevant DNA probes confirmed the loss of RNA message.
RNA analysis.
Cells were grown to exponential (optical density at 650 nm [OD650] of 0.7), late exponential (OD650 of 1.1), and postexponential (OD650 of 1.7) phases, and RNA was extracted using the Trizol isolation procedure (Gibco BRL, Gaithersburg, Md.) as previously described (18, 28, 48). RNA concentrations in the extracts were determined by absorbance at 260 nm using an Eppendorf BioPhotometer (Brinkmann, Westbury, N.Y.). Part (20 or 30 μg) of each sample was electrophoresed through a 1.5% agarose-0.66 M formaldehyde gel in 4-morpholinepropanesulfonic acid (MOPS) (Roche Diagnostics, Indianapolis, Ind.) buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA [pH 7.0]) and blotted onto Hybond-N+ membranes (Amersham, Arlington Heights, Ill.) as previously described (16). Prior to blotting, the gel was viewed under UV light to ensure that equivalent amounts of ethidium bromide-stained rRNA bands were present for each sample. After blotting, the gel was viewed under UV light to confirm complete RNA transfer. Northern blots were probed with radiolabeled DNA probes for the detection of sarT, sarS, RNAIII, hla, spa and HU (a housekeeping transcript) transcripts as previously described (48). The HU transcript served as an internal gel loading control (20, 27).
Cell wall-associated proteins.
Staphylococcal strains were grown overnight in CYGP broth and washed, and the cell density was adjusted to an OD650 of 1.0 in PBS. Cell wall-associated proteins were extracted after lysostaphin digestion in a hypertonic buffer (0.05 M Tris, 0.145 M NaCl, 30% raffinose [pH 8.0]) with protease inhibitors as previously described (13). Equivalent amounts of cell wall extracts were separated by electrophoresis on sodium dodecyl sulfate-10% polyacrylamide gels and transferred to nitrocellulose membranes by electroblotting. To detect protein A, nitrocellulose blots that had been incubated overnight in bovine serum albumin (BSA) blocking buffer (19) were incubated with affinity-purified chicken anti-protein A antibody (1:3,000 dilution) (Accurate Chemicals, Westbury, N.J.) and then with an alkaline phosphatase-conjugated F(ab′)2 fragment of sheep anti-chicken immunoglobulin G (Jackson Immunoresearch, West Grove, Pa.). Reactive bands were detected by incubating the blot with Nitro Blue Tetrazolium (NBT)/5-bromo-4-chloro-3-indolylphosphate (BCIP) substrate (Sigma Chemical Co., St. Louis, Mo.). Band intensities were determined by densitometric scanning using SigmaGel software (Jandel Scientific, San Rafael, Calif.), with the data presented as integrated area units (IAU).
Expression and purification of SarT.
The 420-bp DNA fragment containing the sarT coding region was cloned from S. aureus RN6390 chromosomal DNA by PCR with the primers 5′-GTA AGG GAT GAA CTC GAG ATG AAT GAT TTG AA-3′ and 5′-ACG GGG ATC CAA AAA TAC ATT TAA CTG CAC CAA-3′, ligated into the XhoI and BamHI sites of pET14b (Novagen, Madison, Wis.), and transformed into BL21 (Novagen). Proper insertion of the sarT gene into the recombinant plasmid was confirmed by restriction digestion and by sequencing (Novagen). Protein expression was induced in broth culture with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the cells were harvested and lysed, and the protein was purified on a His-Tag column following the manufacturer's protocols (Novagen). The purity of SarT was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the authenticity was verified by microsequencing the first 10 amino acids of the purified, thrombin-cleaved protein (48).
Doxycycline induction.
The effects of SarT on target gene expression were evaluated with a tetracycline-inducible promoter system to yield a gradient of sarT expression. The sarT coding region, together with the upstream ribosomal binding site, was ligated downstream of the xyl/tetO promoter in pALC2073, a derivative of pSK236, containing the tetR repressor (3). Insertion was confirmed by restriction mapping and DNA sequencing. sarT mutant strains were transformed with the resulting tetracycline-inducible sarT-expressing plasmid (pALC2217) or with pALC2073 as a control. Cultures of the sarT mutant strains harboring the plasmids (cultures had been grown overnight in broth) were diluted 1:100 in CYGP medium and incubated with shaking at 37°C. To capture differential expression during the transition from the late exponential phase (OD650 of ≅1.4) to the post-exponential phase (OD650 of >1.7), the cultures were grown to an OD650 of 1.1 and doxycycline was added at concentrations of 0, 35, and 50 ng/ml to induce gene expression. Cultures were harvested for RNA analysis 1 and 2 h after the addition of doxycycline (48). Northern blots of RNA from the above strains were probed with 32P-labeled specific gene fragments as previously described (16) for the detection of sarT, sarS, and spa transcripts.
DNA gel shift assay of SarT with the sarS promoter.
The binding of SarT to the sarS promoter was confirmed by gel shift assays. A DNA fragment encompassing the sarS promoter region (49) was cloned using the primers 5′ CCC GGT ACC TAT TAC GCT TAC CTC GCT TTA-3′ and 5′-CCC GGA TCC TTT CAT TGT TTT ATC TC-3′, ligated into pCR2.1 Topo (Invitrogen, Carlsbad, Calif.), and confirmed by sequencing. A plasmid harboring the sarS promoter region fragment (pALC2321) was digested with SalI and EcoRI to yield a 264-bp fragment (nucleotides [nt] 125169 to 125432) (29) for use in gel shift experiments. The gel-purified fragment was dephosphorylated and then end labeled with [γ-32P]ATP using T4 polynucleotide kinase (U.S. Biochemical Corp, Cleveland, Ohio). The labeled fragment was incubated with various concentrations of purified SarT protein and analyzed by nondenaturing polyacrylamide gel electrophoresis as previously described (33). Controls were BSA, the sarA P3 promoter fragment as the nonspecific competitor, and unlabeled sarS promoter fragment as the specific competitor.
DNase I footprinting.
DNase I footprinting assays with a 287-bp DNA fragment encompassing the promoter region upstream of sarS were performed as previously described (20). The top-strand primer 5′-ACA TCT AGA TGT TGT TAT TGT TAA CAA GCG −3′ at positions −288 to −259 from the translational start site (ATG) and the bottom-strand primer 5′-CTG TCC ATG GTT TTA TCT CCT TGT ATA TGC AC-3′ at positions −2 to −33 from the translational start site were used to amplify the sarS promoter region. To label the PCR product, only one of the primers was end labeled prior to the PCR. For the assay, the binding reactions were performed in the 100-μl reaction volume containing 20 mM Tris-Cl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 2 mM dithiothreitol, 10 μg of BSA, 0.4 μg of calf thymus DNA, radiolabeled template DNA (10,000 cpm for each reaction), and various amounts of purified SarT at room temperature for 30 min. DNase I (0.02 U) (Boehringer GmbH, Mannheim, Germany) was added and allowed to incubate for 1 min at room temperature. The reaction mixtures were extracted with phenol-chloroform, ethanol precipitated, washed with 70% ethanol, dried, and resuspended in loading buffer (98% deionized formamide, 10 mM EDTA [pH 8.0], 0.025% [wt/vol] xylene cyanol FF, 0.025% [wt/vol] bromophenol blue). The samples were denatured at 95°C for 5 min and analyzed on a 6% denaturing polyacrylamide sequence gel. Positions of the protected region were derived by comparing the footprint with the A+G sequencing ladder of the same fragment (34).
Densitometry.
Blots were scanned with an Hewlett-Packard desktop scanner set at 200 dots per inch. Band density was determined from the scan image using the SigmaGel (Jandel Scientific) software. Graphs were drawn using Excel (Microsoft). Blot scans were processed into figures using Adobe Photoshop LE (Adobe Systems Inc., San Jose, Calif.).
RESULTS
Effects of sarT on sarS and spa expression.
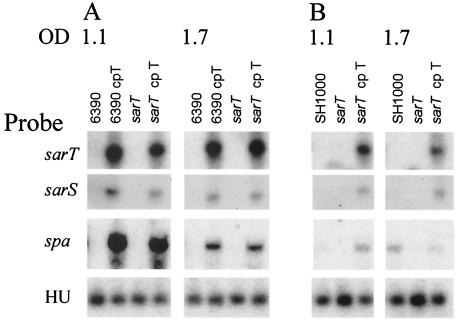
In a previous study (48), we generated a sarT mutant harboring a plasmid (pALC2047) bearing the sarT gene (sarT in trans) that expressed a high level of SarT. We noted that on a Northern blot, the complemented sarT mutant strain (S. aureus ALC2071 [sarT cpT]) expressed high levels of sarS and spa messages during late exponential and post-exponential growth phases, concomitant with increased sarT expression in these strains (Fig. 2A). The same pattern was obtained when parental strain RN6390 was transformed with pALC2047 (6390 cp T [Fig. 2A]). As has been found previously (12), strain RN6390 was a low protein A producer, as evidenced by our failure to detect a significant level of spa transcription in this strain at both growth phases (OD650s of 1.1 and 1.7 [Fig. 2A]).
FIG. 2.
Effects of sarT mutation on expression of sarS and spa. Northern blots of RNA extracted from S. aureus RN6390 (6390) (A) and SH1000 (B) and their isogenic sarT mutant and sarT-complemented strains were hybridized with sarT, sarS, spa, or HU probes. RNA was extracted during late exponential (OD650 of 1.1) and postexponential (OD650 of 1.7) phases of growth. Results are representative of multiple Northern blots, utilizing RNA extracts from multiple experimental sets as previously described (48). HU is constitutively expressed and served as an internal loading control (20, 27). Band density measurement for HU had a mean IAU of 3,455 (standard deviation, 498). The presence of pALC2047, a plasmid bearing an intact sarT, is indicated by cp T (for complementation in trans).
S. aureus RN6390 harbors an 11-bp deletion in rsbU, a gene encoding an anti-sigma factor that, when activated, increases sigB expression approximately 50% (43). σB has been implicated in regulating expression of sarA and agr (Fig. 1a) (6, 22). Since sarA and agr expression influence expression of sarT and sarS (17, 48) (Fig. 1b and d), we wanted to confirm that the effect of sarT on sarS in strain RN6390 was not due to the mutation in rsbU. Accordingly, a sarT mutant was generated by transducing the sarT mutation into S. aureus SH1000 (S. aureus 8325-4 rsbU+ [22], kindly provided by S. J. Foster), using phage 80α.
In Northern blots of late exponential and post-exponential RNA of S. aureus SH1000, expression of sarT and sarS was not detected (Fig. 2B). Interestingly, spa expression was detected during the post-exponential phase (OD650 of 1.7), but not at the late exponential phase (OD650 of 1.1) (Fig. 2B). In the SH1000 sarT mutant, neither sarS nor spa expression was detected (Fig. 2B). Complementation of the sarT mutant of SH1000 in trans resulted in increased expression of sarS and spa, particularly during the late exponential phase at an OD650 of 1.1 (Fig. 2B). Since sarS was expressed at equivalent levels in both the RN6390 and SH1000 sarT-complemented mutant strains (the sarS band density for RN6390 sarT-complemented mutant was 869 IAU versus 898 IAU for the SH1000 sarT-complemented mutant at the post-exponential phase) (Fig. 2), σB activity does not appear to directly affect the relationship between sarT and sarS. However, in the complemented sarT mutant, the expression of spa was lower than in the RN6390 counterpart. Although SarA has been shown to be a direct effector of spa expression (Fig. 1b) (12), the discrepancy in spa expression between RN6390 and SH1000 cannot be attributed to the effect of σB on sarA expression alone.
Effect of sarS on sarT.
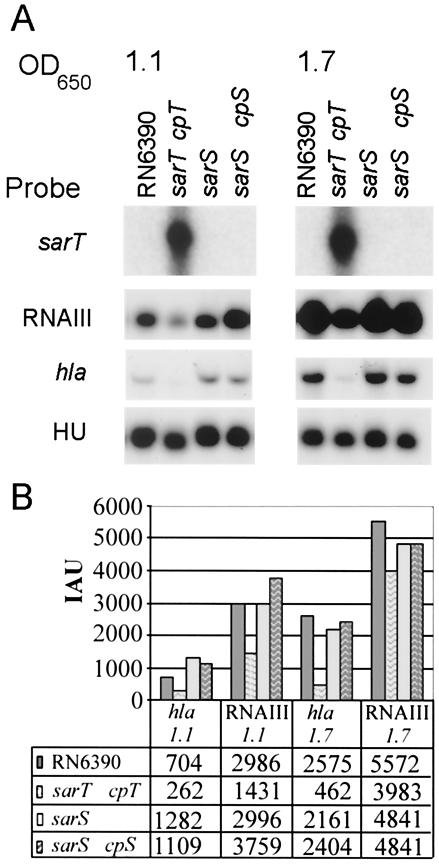
While sarT appears to influence sarS expression, we wondered if sarS, in turn, modulates sarT. We thus examined the expression levels of sarT and of agr RNAIII and hla, two targets of sarT repression (48), in a sarS mutant and its sarS-complemented counterpart (S. aureus ALC2009) (17). As with S. aureus RN6390, sarT expression was not detected in either the sarS mutant or its isogenic sarS-complemented strain (Fig. 3A). Further, the expression levels of hla and RNAIII, target genes of sarT, did not vary substantially from the RN6390 levels in either the sarS mutant or its isogenic complement (Fig. 3). In contrast to RN6390, the parental strain (which has a single copy of sarT), the sarT mutant expressing sarT from a multicopy plasmid in trans expressed considerably lower levels of hla and RNAIII during the late exponential and post-exponential phases (Fig. 3). These results are consistent with the previous observation that sarT is a repressor of hla (48) and RNAIII by virtue of a feedback loop involving sarU (35) (Fig. 1).
FIG. 3.
Effects of sarS mutation on sarT, RNAIII, and hla expression. (A) Northern blots of RNA extracted from S. aureus RN6390 and isogenic mutants at late exponential (OD650 of 1.1) and postexponential (OD650 of 1.7) phases of growth and probed with sarT, RNAIII, or hla probe. Results are representative of Northern blots made from three independently isolated sets of RNA extracts. cpT, complemented with sarT in trans; cpS, complemented with sarS in trans. (B) RNAIII and spa signals quantified by densitometry. The mean IAU for the HU bands was 3,849 (standard deviation, 306).
In a previous study (48), expression of hla and RNAIII in a sarT mutant of S. aureus RN6390 was approximately double the levels seen for RN6390. As agr RNAIII and hla expression levels in the sarS mutant and its isogenic complemented strain did not differ substantially from those seen in RN6390 (Fig. 3B) and since any significant alterations in sarT expression would be reflected in changes in RNAIII (sarT ↑ → RNAIII↓) and hla (sarT↓ → hla↑) expression, these data are consistent with the hypothesis that sarS does not exert any great inductive or repressive effect on sarT.
Effects of sarA and agr mutations.
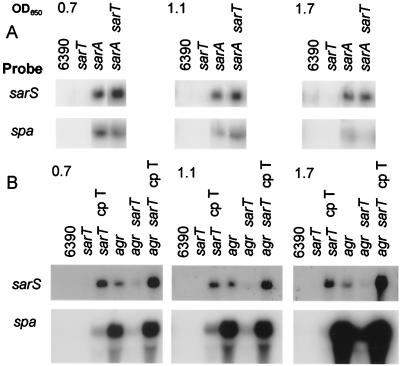
In the regulatory pathway (Fig. 1), SarA has been found to repress sarT (48), thus accounting for increased transcript levels of sarT in the sarA mutant compared to the parent (Fig. 4A). To determine whether SarA represses spa directly (SarA↑→ spa↓) or indirectly via sarT (SarA↑→ sarT↓ → sarS↓ → spa↓), a double sarT sarA mutant was generated and tested for sarS and spa expression in S. aureus RN6390. Expression levels of sarS and spa remained high in the sarA sarT double mutant compared with the single sarA mutant (Fig. 4A). As expected, sarS and spa expression was not readily detected in the sarT mutant. This suggests that sarA is repressing spa directly, rather than indirectly via sarT and sarS (Fig. 1), since a sarA sarS double mutant has previously been found to express a high level of spa despite the absence of sarS (17, 49).
FIG. 4.
Expression of sarS and spa in agr, sarA, agr sarT, and sarA sarT mutant strains. RNA extracts from S. aureus taken at exponential (OD650 of 0.7), late exponential (OD650 of 1.1), and postexponential (OD650 of 1.7) phases of growth were hybridized with sarS and spa probes. sarT and sarA mutant strains (A) and sarT and agr mutant strains (B) are shown. Results are representative of multiple Northern blots utilizing RNA extracts from multiple experimental sets as previously described. 6390, RN6390; cp T, complemented with sarT in trans.
Previous studies also showed that transcription levels of sarS (17) and spa are increased in an agr mutant (12, 23). The transcription of spa is also reduced in the agr sarS double mutant, demonstrating that agr represses spa expression probably by repressing sarS, an activator of spa (17). To ascertain whether sarT induces sarS directly in an agr mutant (agr↓ → sarT↑ → sarS↑) (Fig. 1), we generated an agr sarT double mutant and compared sarS and spa expression with expression in the isogenic single agr mutant (Fig. 4B). In the agr sarT double mutant, sarS and spa expression levels were reduced from those of the agr mutant during both exponential and post-exponential growth. Importantly, the absence of sarS in the double mutant correlated with notably reduced spa expression. Complementing the double agr sarT mutant with sarT in trans resulted in increased levels of sarS and spa expression compared to those of the double agr sarT mutant (Fig. 4B), particularly in the post-exponential growth phase. The same expression pattern was also seen in agr sarT and sarT-complemented double mutants of SH1000 (data not shown). Collectively, these data suggest that the increase in spa expression seen in an agr mutant is likely the result of sarT induction of sarS. As expression of sarT also has a mild repressive effect on agr (48), the decreased sarS message in an agr sarT double mutant also indicated that the high levels of sarS message in an agr mutant was not due to repression of agr RNAIII by SarT (i.e., SarT↑ → agr↓ → sarS↑).
Cell wall-associated protein A.
We also confirmed the Northern blot results by analyzing protein A levels in post-exponential-phase cultures of S. aureus RN6390 (cultures grown overnight) with immunoblots (Fig. 5). Consistent with the spa message detected in the Northern blots, levels of cell wall-associated protein A were not detected in the parental RN6390 and sarT and sarS mutant strains. The intensity of the protein A band in the sarT mutant strain harboring sarT in trans (870 IAU) was consistent with the light spa band seen on the Northern blot (Fig. 4B). The intensity of the protein A band in the agr sarT mutant was lower (1,038 IAU) than that seen in the agr mutant strain (1,350 IAU). However, the protein A band of the agr sarT mutant on the immunoblot seemed higher than what one would expect from the level of spa transcription on the Northern blot (Fig. 4B). This could conceivably be explained by technical differences, since cell wall protein A tends to accumulate throughout the growth cycle, while the Northern blot data are expected to reflect spa expression at a single time point during growth (Fig. 4).
FIG. 5.
Western blot of S. aureus cell wall-associated protein extracts probed with chicken anti-protein A antibody. Equivalent amounts of cell wall extracts were applied to the lanes, electroblotted, and probed with chicken anti-protein A antibody as described in the text (17, 48). The positions of standard protein molecular mass standards (Std) (in kilodaltons) are shown to the left of the blot. The positions of intact protein A (large arrowhead) and degraded protein A bands (small arrowheads) are shown to the right of the blot. Results are representative of two independent experiments. 6390, RN6390; cpT, complemented with sarT in trans.
As has been noted previously (17, 49), additional lower-molecular-weight protein A-specific reactive bands were present in the sarA mutants (Fig. 5). This is presumably due to degradation of protein A as a result of increased proteolytic activity in a sarA mutant (17, 49). Interestingly, protein A showed less apparent degradation in the sarA sarT double mutant than in the sarA mutant. However, the intensities of the four bands seen in the sarA mutant (138, 134, 795, and 307 AIU) and the two major bands seen in the sarA sarT mutant (1,031 and 188 AIU) added up to 1,374 and 1,219 AIU, respectively, suggesting that protein A expression was likely to be similar in the two strains.
Gel shift and DNase I footprinting.
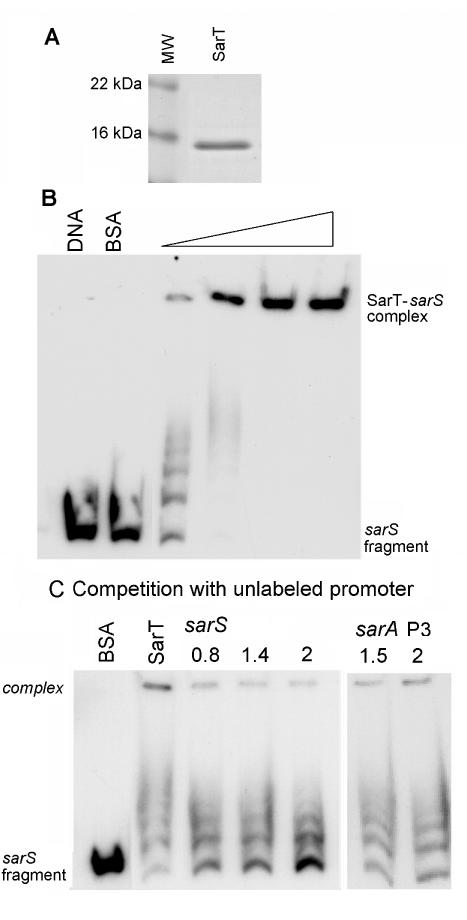
Northern and Western blots of sarS and spa RNA levels in various mutants led to the hypothesis that sarT regulates sarS expression. We evaluated the binding of the SarT protein to the region upstream of the sarS promoter. A 264-bp region of the sarS promoter was cloned and purified. Various concentrations of purified SarT protein (Fig. 6A) were incubated with the 32P-end-labeled fragment encompassing the sarS promoter region, and the reaction mixtures were analyzed by gel shift assays (Fig. 6B). The dose-dependent retardation in the mobility of the sarS promoter fragment with increasing amounts of SarT (Fig. 6B) indicated that SarT binds to the sarS promoter region. As SarA, SarR, and possibly other SarA homologs are believed to form dimeric structures (31), it is likely that the multiple banding pattern (Fig. 6B, third lane from the left) may result from SarT forming dimers and tetramers on the DNA strand, since there were only two binding sites for SarT on the sarS promoter (see below).
FIG. 6.
Gel shift assay of the sarS promoter. (A) Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis of SarT purified from the pET14b expression vector. MW, molecular mass standards. (B) Gel shift assay showing the effect of increasing concentrations of SarT protein on the mobility of a 264-bp fragment (nt 125169 to 125432) (29). The sarS promoter fragment was end labeled with [γ-32P] ATP, and approximately 11,000 cpm was used in each lane. Purified SarT at 0.19, 0.38, 0.56, and 0.75 μg (indicated by the thickness of the triangle over the lanes) were added to the lanes. (C) Competition assays. Increasing amounts of unlabeled sarS fragment (specific competitor [in micrograms]) and sarA P3 promoter fragment (nonspecific competitor [in micrograms]) (161 bp, nt 365 to 525) (4) were added to a mixture containing sarS promoter (10,000 cpm) and 0.45 μg of SarT protein.
To confirm that binding was specific, the assay was repeated with unlabeled sarS promoter as a competitor (Fig. 6C). The gel shift pattern showed a dose-dependent reduction in the intensity of the SarT-sarS promoter complex, while the intensity of the unbound sarS promoter fragment increased, indicating that the unlabeled DNA fragment containing the sarS promoter is able to bind SarT competitively. No comparable shift in the labeled sarS promoter region fragment was noted when the noncompetitive sarA P3 promoter was added to the SarT-sarS promoter DNA mix.
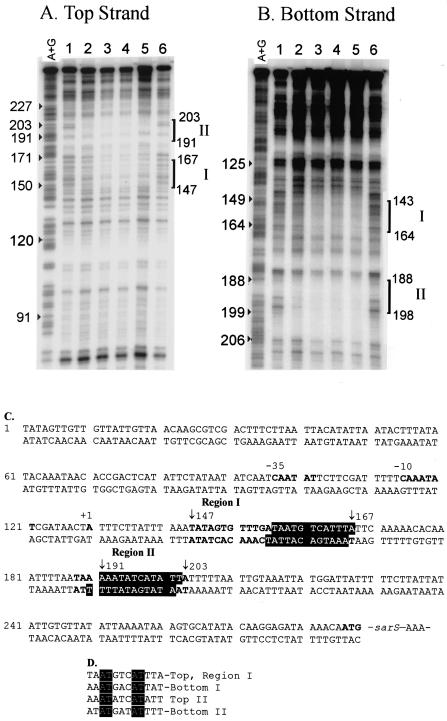
To verify the specificity and map the probable binding site of SarT on the sarS promoter region, DNase I footprinting was performed, using a 287-bp DNA fragment encompassing the sarS promoter region (49). DNase I footprinting of the top strand (Fig. 7A and C) showed two major protected regions, one region corresponding to positions 147 to 167 (region I [Fig. 7C]) and one region corresponding to positions 191 to 203 (region II [Fig. 7C]) (139 to 119 bp and 95 to 83 bp upstream of the translation start, ATG, respectively). With the bottom strand, we found binding sites at positions 144 to 164 and 188 to 199 (Fig. 7C) (142 to 121 bp and 98 to 87 bp upstream of the translation start, ATG, respectively) (Fig. 7B and C). The two protected regions occur in corresponding locations on the top and bottom strands and are separated by approximately 23 bp of AT-rich sequence.
FIG. 7.
DNase I footprinting assays of the interaction of SarT with the sarS promoter. (A and B) Top-strand (A) and bottom-strand (B) promoter fragments. Lanes: 1 to 6, labeled DNA fragments reacted with 0, 1, 2, 4, 6, and 0 μg of purified SarT protein, respectively, prior to DNase I digestion as described in the text. An A+G ladder was run in parallel to identify the positions of the protected regions. The positions of protected regions I and II are marked with brackets. (C) Sequence of the promoter region upstream of sarS, showing the two protected regions (positions 147 to 167 and positions 191 to 203 [shown in bold type]) and showing the SarT binding sites (white letters on a black background) as deduced from the footprinting analysis. (D) Putative 12-bp sarT binding sequence deduced from the protected regions on the top and bottom strands. Identical nucleotides are indicated by white lettering on a black background.
A close analysis of the four protected sequences in two regions suggested a putative conserved 12-bp SarT binding sequence with a consensus sequence of AAATG(A/T)CATTTT, present in both the top and bottom strands of region I and region II (Fig. 7D).
Doxycycline-dependent induction of sarT.
Dose-dependent induction of sarS and spa was used to confirm the regulatory control of sarT on sarS. A shuttle plasmid (pALC2217) was constructed such that sarT was placed downstream of the tetracycline-induced xyl/tetO promoter (3). sarT mutant strains of RN6390 and SH1000 were transformed with pALC2217 or with vector plasmid pALC2073 as a control. Tetracycline was used in initial experiments, but the dose range required for a suitable sarT response from the inducible promoter was too high (data not shown). Therefore, we used doxycycline as an alternative to induce a sarT response at a low enough dose level to avoid an untoward response from the control strain. Northern blots showed that sarT expression from pALC2217 occurred even in the absence of added doxycycline (Fig. 8), but the band intensity for sarT increased with increasing amounts of doxycycline, particularly in strain SH1000. sarS and spa induction roughly mirrored the sarT band intensity. Collectively, these data with the tetracycline-inducible promoter system indicated that sarT induced sarS and the ensuing spa expression.
FIG. 8.
Doxycycline induction of sarT. Northern blots of RNA extracted from S. aureus sarT mutants of strain RN6390 (A) or SH1000 (B) harboring either pALC2073 (control; tet-inducible promoter) or pALC2217 (tet-inducible promoter driving sarT) and grown in the presence of 0, 35, or 50 ng of doxycycline per ml. Doxycycline was added to late-exponential-phase cells (OD650 of 1.1), and cells were harvested for RNA 1 and 2 h after induction. Strains harboring the control plasmid were harvested only 2 h after the addition of doxycycline. Results are representative of multiple experiments.
DISCUSSION
S. aureus exhibits complex patterns of protein expression in response to different environmental conditions (1, 36). It is therefore not surprising that S. aureus harbors a complex regulatory network to coordinate expression of its array of virulence proteins. The sarA/agr interface within the regulatory system controls growth phase-mediated repression of cell wall-associated proteins and induction of late exponential to post-exponential expression of a variety of extracellular toxins and enzymes (1, 10, 14, 17, 19, 21, 23, 27, 39, 40, 46).
SarT (48) and SarS (17, 49), two recently described SarA homologs, have been shown to be modulated by sarA and agr in the regulatory pathway (Fig. 1). In previous studies, we have shown that agr likely represses spa expression by down-regulating sarS (17), while the sarA locus probably activates hla expression by down-regulating sarT (48). The above findings are of interest because sarS and sarT, as an activator of spa and repressor of hla, respectively, participate in divergent pathways in regulating spa (agr↓ → sarS↑→ spa↑) and hla (sarA↓ → sarT↑ → hla↓), despite the ability of both sarA and agr to repress sarS and sarT. In discovering that sarS expression could be enhanced in a sarT-complemented strain (Fig. 2), we wanted to investigate the broader relationships of sarT with sarS within the sarA/agr regulatory network. S. aureus RN6390 was initially chosen for these studies because of its low basal spa expression level; hence, any alteration in sarS expression attributable to possible sarT-mediated induction can be readily detected by examining spa expression, the target gene of sarS (12). We found that the complemented sarT mutant of strain RN6390 had elevated sarS expression and was able to express spa at a high level (Fig. 2A). Parallel experiments with S. aureus SH1000 (22) and its complemented sarT mutant also showed expression patterns similar to those of RN6390 (Fig. 2B).
In earlier studies (17, 48), we found that sarT, sarS, and spa expression was elevated in a sarA mutant. In this study, we found that sarS and spa expression was elevated in the sarA sarT double mutant (Fig. 4A), suggesting that SarA may repress sarS rather than spa. However, in previous studies, spa was found to be repressed in a sarS mutant, relative to the sarS sarA double mutant, indicating that SarA repression of spa is likely independent of sarS (17, 49). Taken together, these results indicate that sarA repression of spa is independent of sarT and sarS.
We have previously shown that agr RNAIII likely regulates spa by repressing sarS, an activator of spa expression (17). However, the positive modulation of sarS by SarT (Fig. 2) leads to the question whether RNAIII represses sarS transcription directly or indirectly by suppressing sarT. A comparison of the agr and sarT mutants (Fig. 4B) revealed that sarS and spa expression was induced in an agr mutant strain but was almost totally repressed in the sarT mutant and sarT agr double mutant. Further, when sarT was returned in trans to the agr sarT mutant via a plasmid harboring the sarT gene, sarS and spa expression returned to levels seen in the agr mutant strain, suggesting that sarT induction of sarS may be important for spa expression in the agr mutant strain.
To further confirm the activation of sarS by sarT, we took advantage of an inducible promoter system that can be activated by low doses of doxycycline to express sarT in a dose-dependent manner. This plasmid, upon introduction into sarT mutants of S. aureus RN6390 and SH1000, expressed sarT, in proportion (approximately) to the doxycycline dose, in particular during the 2-h time points (Fig. 8). At the 1-h time point, the basal level of sarT expression (i.e., no doxycycline) remained quite high, especially in RN6390, so the induction phenomenon was less apparent under this condition. Nevertheless, the data on the induction time point at 2 h for both RN6390 and SH1000 indicated that SigB does not play a significant role in this sarT-sarS induction process.
Gel shift (Fig. 6) and footprinting assays (Fig. 7) confirmed that SarT binds specifically to the sarS promoter, presumably leading to sarS activation. The presence of multiple bands in gel shift assays indicated that multiple SarT dimers likely bind to the sarS promoter. Interestingly, SarT binds to two regions on the sarS promoter, separated by an extremely AT-rich 23-bp region (20 of 23 bp or 87% AT). Within the AT-rich region is a short stretch of five adenines on the top strand and four adenines on the bottom strand which potentially may play a role in DNA bending (2, 44), a process that has been recognized to play a role in growth phase-mediated transcriptional regulation (51). Additionally, the binding sites covered in both the plus and minus strands divulged a 12-bp sequence in region I that was repeated in region II (Fig. 7C). We speculate that this 12-bp sequence may represent a conserved binding site for SarT. Nevertheless, additional verification with other SarT target genes should be made to confirm this hypothesis.
In sarA or agr mutant strains where repression of sarT is relieved, sarT expression occurs during exponential growth but is highest during the post-exponential phase (48). The repression of sarT by sarA or agr, as demonstrated by the extremely low sarT transcriptional levels in S. aureus RN6390 (48), suggests that sarT induction may require specific environmental activators that in vitro growth conditions replicate poorly or that it is transitory and therefore less likely to be readily detected.
In a transcription profiling study, Dunman et al. (21) have pointed out that the pathogenic process is a complex progression of bacterial and host interactions in a dynamically shifting environment. We have characterized in sarT what may be a regulatory subroutine that is activated under a specific environmental stimulus. Further studies are required to discover the specific environmental conditions that induce expression of sarT and subsequently sarS.
Acknowledgments
This work was supported in part by PHS grant AI07519-14 and NIH grants AI43968 and AI37142.
We thank Steven Bobin of the molecular biology core facility for his assistance and sequencing advice. We thank Simon Foster of the University of Sheffield (United Kingdom) for providing the SH1000 strain.
Editor: J. T. Barbieri
REFERENCES
- 1.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Barbic, A., D. P. Zimmer, and D. M. Crotheres. 2003. Structural origins of adenine-tract bending. Proc. Natl. Acad. Sci. USA 5:2369-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benito, Y., F. A. Kolb, P. Romby, G. Lina, J. Etienne, and F. Vandenesch. 2000. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6:668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Camilli, A., D. A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, P. F., and S. J. Foster. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469-2479. [DOI] [PubMed] [Google Scholar]
- 10.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, A. L., K. Eberhardt, and J. H. Heinrichs. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 65:2243-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung, A. L., and V. Fischetti. 1988. Variation in the expression of cell wall proteins of Staphylococcus aureus grown on solid and liquid media. Infect. Immun. 56:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung, A. L., C. C. Nast, and A. S. Bayer. 1998. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect. Immun. 66:5988-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung, A. L., C. Wolz, M. R. Yeaman, and A. S. Bayer. 1995. Insertional inactivation of a chromosomal locus that modulates expression of potential virulence determinants in Staphylococcus aureus. J. Bacteriol. 177:3220-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien, Y.-T., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 20.Chien, Y.-T., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 21.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janzon, L., and S. Arvidson. 1990. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 25.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan, S. A., and R. P. Novick. 1983. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10:251-259. [DOI] [PubMed] [Google Scholar]
- 27.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 28.Kornblum, J., S. J. Projan, S. L. Moghazeh, and R. P. Novick. 1988. A rapid method to quantitate non-labeled RNA species in bacterial cells. Gene 63:75-85. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K.-I. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R.-I. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 30.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., A. Manna, R. Li, W. E. Martin, R. C. Murphy, A. L. Cheung, and G. Zhang. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6877-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morfeldt, E., D. Taylor, A. V. Gabain, and S. Arvidson. 1995. Activation of alpha toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 39.Novick, R. P. 1990. The staphylococcus as a molecular genetic system, p. 1-40. In R. P. Novick (ed.), Molecular biology of the staphylococcus. VCH Publishers, New York, N.Y.
- 40.Novick, R. P., and T. W. Muir. 1999. Virulence gene regulation by peptides in staphylococci and other Gram-positive bacteria. Curr. Opin. Microbiol. 2:40-45. [DOI] [PubMed] [Google Scholar]
- 41.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 42.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Martin, J., F. Rojo, and V. de Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58:268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingston, New York, N.Y.
- 46.Recsei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 47.Saïd-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of α-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, W. D., Jr., and G. L. Archer. 1989. Identification and cloning of the conjugative transfer region of Staphylococcus aureus plasmid pGO1. J. Bacteriol. 171:684-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vicente, M., K. F. Chater, and V. de Lorenzo. 1999. Bacterial transcription factors involved in global regulation. Mol. Microbiol. 33:8-17. [DOI] [PubMed] [Google Scholar]