Abstract
Previous studies have demonstrated that the Moraxella catarrhalis surface antigen UspA1 is an adhesin for Chang human conjunctival cells. The present report demonstrates that lack of UspA1 expression does not affect the adherence of strain O35E to A549 human lung cells or primary cultures of human middle ear epithelial (HMEE) cells. These results imply that another molecule mediates the adherence of M. catarrhalis to these two cell lines. To identify this adhesin, strain O35E was mutagenized with a transposon and 1,000 mutants were screened in a microcolony formation assay using A549 cells. Nine independent isolates exhibited an 8- to 19-fold reduction in adherence and contained a transposon in the same locus. Nucleotide sequence data and PCR analysis indicated that the transposons were inserted in different locations in the gene encoding the surface protein Hag. Quantitative assays using one representative transposon mutant, O35E.TN2, showed considerably decreased binding to A549 as well as HMEE cells. However, this mutant adhered at wild-type levels to Chang conjunctival cells. These findings suggest that the M. catarrhalis Hag protein is an adhesin for cell lines derived from human lung and middle ear tissues.
Moraxella catarrhalis is a significant pathogen of the human respiratory tract. This gram-negative bacterium is now recognized as a leading cause of otitis media (middle ear infection), together with Streptococcus pneumoniae and nontypeable isolates of Haemophilus influenzae (11, 15, 28, 39). More than 80% of infants have at least one episode of ear infection by the age of 3 years, and M. catarrhalis causes 15 to 20% of all cases (9, 14, 28, 29, 31, 39, 50, 59). In the United States, ∼24 million office visits are made annually for the treatment of otitis media (29, 30, 39, 59) and of these, roughly a sixth are caused by M. catarrhalis. This organism also causes 10 to 35% of all cases of lower respiratory tract infections in adults that are suffering from chronic obstructive pulmonary disease (39, 52, 53). This disease is the fourth leading cause of death in the United States, surpassed only by heart attacks, cancer, and stroke (3, 43). Lower respiratory tract infections contribute substantially to the progression of chronic obstructive pulmonary disease; thus, infectious exacerbations due to M. catarrhalis constitute an important health problem.
M. catarrhalis has also been associated with diseases such as wound infections, bronchitis, conjunctivitis, sinusitis, bacteremia, pneumonia, meningitis, pericarditis, and endocarditis (8, 10, 11, 28, 39, 44, 55, 60, 63, 64). Patients with underlying conditions are particularly susceptible. Long considered to be a nonpathogenic inhabitant of the human nasopharynx, M. catarrhalis has clearly emerged as an important cause of infectious diseases, and under unpredictable circumstances (e.g., immunosuppressive conditions or chronic disease), the organism can be virulent and cause serious organ complications.
M. catarrhalis infections are a matter of concern due to the lack of a vaccine and the rapid emergence of antibiotic resistance observed in clinical isolates (32). Unfortunately, little is known about the pathogenesis of this organism, particularly how M. catarrhalis binds to human cells. By in vitro analyses, the proteins UspA1 and UspA2H have been directly demonstrated to be adhesins (33), and the Hag protein has been associated with the ability to bind erythrocytes (16, 17, 47) and immunoglobulin D (IgD) (21, 45, 47). The outer membrane proteins UspA2 and OMPCD have been shown to bind to the extracellular matrix protein vitronectin (36) and to human middle ear mucin (49), respectively, and antibodies binding to M. catarrhalis lipooligosaccharides can inhibit adherence to human cells (25). Recently, the protein McaP was also shown to be involved in the ability of M. catarrhalis to bind to several human cell lines in vitro (61).
In an effort to identify other M. catarrhalis gene products involved in adherence to human cells, we have developed the ability to randomly mutagenize M. catarrhalis with a transposon. The resulting mutants were screened to identify those that no longer bound to A549 human lung cells in microcolony formation assays. In this report, we present data that suggest that the M. catarrhalis Hag protein acts as an adhesin for human lung and middle ear cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. M. catarrhalis strains were cultured at 37°C in Todd-Hewitt (TH) broth (Difco) or on TH agar plates in an atmosphere of 92.5% air-7.5% CO2. M. catarrhalis transposon mutants were selected with kanamycin (KAN) at a concentration of 20 μg/ml. Escherichia coli strains were grown in Luria-Bertani (LB) broth (Difco) or on LB agar plates. For the selection of recombinant E. coli clones, the LB medium was supplemented with 15 μg of chloramphenicol/ml or 50 μg of KAN/ml.
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| M. catarrhalis | ||
| O35E | Wild-type isolate | 22 |
| O35E.1 | uspA1 isogenic mutant of strain O35E | 1 |
| O35E.TN2 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN19 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN34 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN107 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN114 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN115 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN184 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN246 | Adherence negative hag transposon mutant of strain O35E | This study |
| O35E.TN532 | Adherence negative hag transposon mutant of strain O35E | This study |
| E. coli EPI300 | Cloning strain | Epicentre |
| Plasmids | ||
| pCC1 | Cloning vector | Epicentre |
| pUC4K | Source of the Kanr cassette | AmershamBiosciences |
| pMHO35ETN2 | pCC1 containing the transposon and flanking M. catarrhalis DNA of strain O35E.TN2 | This study |
| pMHO35ETN114 | pCC1 containing the transposon and flanking M. catarrhalis DNA of strain O35E.TN114 | This study |
| pMHO35ETN532 | pCC1 containing the transposon and flanking M. catarrhalis DNA of strain O35E.TN532 | This study |
Chang (human conjunctival epithelium; ATCC CCL20.2) and A549 (human type II alveolar lung epithelium; ATCC CCL85) tissue culture cell lines were cultured in Ham's F12 medium (Cellgro) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (GIBCO), 0.15% (vol/vol) sodium bicarbonate (Cellgro), and 4 mM l-glutamine (Cellgro) at a temperature of 37°C in an atmosphere of 92.5% air-7.5% CO2. Primary cultures of human middle ear epithelial (HMEE) cells were cultured in modified minimal essential medium α (GIBCO) as previously described (62).
Recombinant DNA techniques.
Standard molecular biology methods were performed as described elsewhere (51). M. catarrhalis genomic DNA was purified using an Invitrogen Easy-DNA kit under the conditions recommended by the manufacturer. The libraries of M. catarrhalis O35E.TN2, O35E.TN114, and O35E.TN532 DNA fragments were generated with the CopyControl Fosmid library production kit (Epicentre) following the manufacturer's instructions. Plasmid DNA was purified with a QIAprep Spin Miniprep system (Qiagen). Southern hybridization experiments were performed with a North2South chemiluminescent nucleic acid hybridization and detection system (Pierce) following the manufacturer's recommended procedure. The 1.2-kb KAN resistance cartridge from the plasmid pUC4K was used as a probe in these experiments.
Transposon mutagenesis of M. catarrhalis and screening for adherence-negative mutants.
M. catarrhalis O35E cells were grown to a density of 108 CFU/ml in 10 ml of TH broth, harvested by centrifugation, washed once with a 1 mM HEPES (pH 7.0) solution, and then washed twice with 10% (vol/vol) glycerol in distilled water. The final cell pellet was resuspended in 100 μl of 10% glycerol, and a 20-μl portion of this cell suspension was mixed with 20 ng of EZ::TN <KAN-2> Transposome (Epicentre). This mixture was electroporated immediately using a BTX Transporator Plus apparatus, mixed subsequently with 1 ml of sterile TH broth, and incubated at 37°C with shaking for 3 h. The electroporated cells were then spread onto TH agar plates supplemented with KAN (the antibiotic marker associated with the transposon) and incubated overnight at 37°C.
Kanr colonies were screened in a microcolony formation assay to identify M. catarrhalis transposon mutants that were substantially reduced in adherence to A549 human lung cells. In this assay, each Kanr colony obtained by electroporation with the EZ::TN <KAN-2> Transposome was inoculated into one well of a 48-well tissue culture plate (CELLSTAR) containing 1.2 × 105 A549 cells. After incubation at 37°C for 20 h, the plate was centrifuged for 5 min at 165 × g, incubated for an additional 20 h at 37°C, centrifuged again, and finally rinsed five times with phosphate-buffered saline (PBS) to remove unbound M. catarrhalis cells. The content of each well was fixed in methanol for 5 min and stained with Giemsa Accustain (Sigma), and the adherence of each mutant was evaluated by visual inspection of wells under the microscope. These microcolony formation assays were performed at least twice more to verify the reduced binding of M. catarrhalis transposon mutants.
Adherence assay.
Quantitative adherence assays were performed as previously described (33). Briefly, 2.5 × 105 human cells were seeded into each well of a 24-well tissue culture plate (CELLSTAR) and incubated overnight before use; the bacterial strains to be tested were also streaked onto appropriate agar plates and grown overnight. The next day, bacterial cells were scraped off of the agar plates and aseptically resuspended to an optical density of 230 Klett units into 5 ml of sterile PBS supplemented with 0.15% (wt/vol) gelatin (PBSG buffer). Portions of these bacterial suspensions (25 μl containing 107 CFU) were inoculated in duplicate into the wells of the 24-well tissue culture plate containing monolayers of human cells. The tissue culture plate was centrifuged at 165 × g to facilitate contact between bacteria and human cells and then incubated for 15 min (Chang cells and HMEE monolayers) or 3 h (A549 monolayers) at 37°C. Nonadherent bacteria were removed by gently rinsing the wells five times with PBSG buffer, and the human cells were released from the plastic support of the tissue culture plate by adding 100 μl of a solution containing 0.05% trypsin and 0.02% EDTA (Cellgro). The released human cells were suspended in 500 μl of PBSG buffer, serially diluted in PBSG buffer, and spread onto agar plates to determine the number of viable bacterial cells attached to human cells. Adherence is expressed as the percentage (± standard deviation) of bacteria attached to the human cells relative to the original inoculum added to the well. Adherence assays were repeated at least four times for each bacterial strain.
Autoagglutination assay.
The ability of M. catarrhalis strains to autoagglutinate was measured as previously reported (47). Briefly, plate-grown bacteria were suspended to a density of 300 Klett units in 5 ml of PBS. The decrease in this optical density was then monitored at regular time intervals.
PCR.
Amplification of DNA fragments was performed with Pfx DNA polymerase (Invitrogen) following the manufacturer's recommended procedure. The hag-specific oligonucleotide primers F1 (5′-GGCTTTTTGTAAAAATCACATCG-3′) and R1 (5′-TGAGCGGTAAATGGTTTAAGTG-3′) were used.
Monoclonal antibodies and characterization of selected protein antigens.
UspA1-specific monoclonal antibody (MAb) 24B5 (12), UspA1- and UspA2-reactive MAb 17C7 (2, 3, 22), and MAb 5D2 (47), which binds to the 200-kDa Hag protein, have been described elsewhere. Note that MAb 17C7 binds to an epitope that is shared by UspA1 and UspA2 and therefore cross-reacts with both proteins. Outer-membrane vesicles (46) and whole-cell lysates (12, 46) were prepared as previously reported. These antigen preparations were heated at 100°C for 15 min, separated by electrophoresis into sodium dodecyl sulfate (SDS)-7.5% polyacrylamide gels, electrophoretically transferred to polyvinylidene difluoride membranes (Millipore), and blocked for 1 h at room temperature in PBS supplemented with 0.05% (vol/vol) Tween 20 and 3% (wt/vol) skim milk (blocking buffer). The membranes were incubated overnight at 4°C with MAb 24B5, MAb 17C7, or MAb 5D2 hybridoma culture supernatants diluted in blocking buffer (1/20 [vol/vol]) and then washed five times (5 min each time) at room temperature with PBS supplemented with 0.05% (vol/vol) Tween 20. The polyvinylidene difluoride membranes were subsequently incubated for 1 h at room temperature with goat anti-mouse Ig (heavy and light drain)-horseradish peroxidase antibodies (Southern Biotechnology Associates) diluted in blocking buffer (1/3,000 [vol/vol]). The membranes were washed with PBS supplemented with 0.05% (vol/vol) Tween 20 as described above and incubated for 5 min at room temperature with a SuperSignal West Pico chemiluminescent substrate (Pierce). Selected antigens were visualized by autoradiography.
RNA purification.
RNA was purified using Trizol reagent (Gibco/BRL) and Phase Lock gel (Fischer) following the manufacturer's instructions with some modifications (54). Briefly, M. catarrhalis was grown to the logarithmic phase in broth. Bacterial cells (5 ml) were pelleted by centrifugation (30 s at 21,000 × g), resuspended in Tris-EDTA buffer (0.5 ml), and lysed by passage through a Qiashredder apparatus following the manufacturer's protocol (Qiagen). Trizol (0.5 ml) was immediately added to the lysed bacteria. These lysates were transferred to prespun Phase Lock Gel-Heavy tubes (Fischer) and allowed to incubate at room temperature for 5 min. Chloroform (0.2 ml) was added, and the samples were shaken vigorously for 15 s and then centrifuged at 12,000 × g for 10 min at 4°C. The upper aqueous phase was transferred to a microcentrifuge tube, and RNA was precipitated by the addition of 0.6 ml of isopropyl alcohol and incubation for 10 min at room temperature. The RNA pellet was collected by centrifugation at 12,000 × g for 30 min at 4°C and washed with 70% ethanol. RNA was dissolved in 100 μl of 1× DNase buffer and treated with 30 U of RNase-free DNase (Promega) for 1 h at 37°C. RNA was precipitated and dissolved in nuclease-free water. RNA concentrations and purity were determined by measuring absorbance at 260 and 280 nm.
Quantitative RT PCR.
Relative RNA concentrations were determined by quantitative reverse transcriptase (RT) PCR using a Perkin-Elmer TaqMan PCR system (Applied Biosystems). Using Primer Express 1.5 software (ABI), the primers and probe were designed on the basis of published M. catarrhalis sequences. The primer sequences were as follows. For bipA, the forward primer sequence was 5′-GCCCTGCCAGCACTGTCT-3′, the reverse primer was 5′-AAATGGTGAGTCGTTAACCTGAAAA-3′, and probe sequence was 5′-TTGATGAGCCGACGGTCTCGATGA-3′. For purH, the forward primer sequence was 5′-GGTGAGTTGCCACAGCCG-3′, the reverse primer sequence was 5′-AGTAGACCGCCATTGACTCGTT-3′, and the probe sequence was 5′-CACAGCGGGCAGCTCAATTTGACCTA-3′. Probes were 5′ labeled with 6-carboxyfluorescein and 3′ labeled with the “black hole quencher” BHQ1 (Biosearch Technologies). The primers and probes were synthesized by Integrated DNA Technologies, Inc.
Each 10-μl PCR mixture contained 100 ng of RNA, 1× TaqMan PCR Master Mix, 1× Multiscribe and RNase inhibitor mix (omitted in RT-negative reactions), 300 nM concentrations (each) of the forward and reverse primers, and 250 nM probe. For each sample, both RT-positive and RT-negative reactions were tested in duplicate. The reactions were carried out using a Lightcycler (Roche) with optimized cycle conditions as follows: 48°C for 30 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. Using the −ΔΔCt method as described by the manufacturer (ABI user bulletin no. 2), amounts of purH were determined relative to bipA mRNA levels.
Nucleotide sequence analysis.
Plasmid DNA was sequenced with an ABI Prism 310 genetic analyzer for automated DNA sequencing (Perkin-Elmer Biosystems). The oligonucleotide primer KAN-2 FP1 (Epicentre), which binds to nucleotides (nt) 1127 to 1151 of the EZ::TN <KAN-2> transposon, was used to sequence the plasmid pMHO35ETN2. The nucleotide sequence data were analyzed with GeneTool Lite software (BioTools Inc.) and the Gapped BLAST 2.0 BLASTX search tool through the National Center for Biotechnology Information service using the nonredundant protein database.
Statistics.
All statistical analyses were performed using a Student t test and GraphPad Prism 2.01 software. P values of <0.05 were considered statistically significant.
RESULTS
The M. catarrhalis O35E uspA1 mutant binds to human lung and middle ear cells in vitro.
As previously shown (1, 33, 34), the lack of UspA1 expression was found to considerably decrease adherence of M. catarrhalis O35E to Chang human conjunctival cells (Table 2). However, no significant reduction in the binding of uspA1 isogenic mutant strain O35E.1 to human lung (A549) or middle ear (HMEE) cells was observed (Table 2). These results imply that a molecule other than the previously characterized adhesin UspA1 is involved in the adherence of M. catarrhalis O35E to A549 and HMEE cells.
TABLE 2.
Adherence of M. catarrhalis strains to human cell monolayers in vitro
| M. catarrhalis strain | Description | UspA1 expressiona | Hag expressiona | % Adherence (±SD) to:
|
||
|---|---|---|---|---|---|---|
| A549 cells | Chang cells | HMEE cells | ||||
| O35E | Wild type | + | + | 33.7 ± 1.6 | 22.6 ± 5.7 | 17.6 ± 6.8 |
| O35E.1 | uspA1 mutant | − | + | 28.1 ± 6.1 | 4.2 ± 2.6b | 13.1 ± 7.1 |
| O35E.TN2 | TN mutant | + | − | 1.8 ± 1.1b | 23.6 ± 2.1 | 0.4 ± 0.2b |
| O35E.TN19 | TN mutant | + | − | 2.0 ± 1.1b | NDc | ND |
| O35E.TN34 | TN mutant | + | − | 2.2 ± 0.3b | ND | ND |
| O35E.TN107 | TN mutant | + | − | 2.3 ± 2.2b | ND | ND |
| O35E.TN114 | TN mutant | + | − | 1.9 ± 1.3b | ND | ND |
| O35E.TN115 | TN mutant | + | − | 3.9 ± 2.8b | ND | ND |
| O35E.TN184 | TN mutant | + | − | 2.2 ± 1.5b | ND | ND |
| O35E.TN246 | TN mutant | + | − | 2.7 ± 2.0b | ND | ND |
| O35E.TN532 | TN mutant | + | − | 3.7 ± 2.2b | ND | ND |
As determined by Western blot analysis.
The difference in the P value compared to the value for wild-type strain O35E was found to be statistically significant using a Student t test.
ND, not determined.
Isolation of M. catarrhalis mutants with reduced adherence to A549 human lung cells.
To identify the adhesin(s) mediating the attachment of M. catarrhalis O35E to A549 cells, we used an EZ::TN <KAN-2> Transposome system (Epicentre) to mutagenize strain O35E. The Transposome consists of an inactive complex between a transposon and its transposase. Upon electroporation of the Transposome into bacteria, the presence of magnesium ions activates this complex which in turn leads to the random insertion of the transposon into the genome of the organism under study. The transposon used in this system contains a KAN resistance gene, which is an effective antimicrobial marker for M. catarrhalis (2, 33, 35, 40). Southern blot analysis of randomly chosen mutants performed with a transposon-specific probe clearly demonstrates that the EZ::TN <KAN-2> transposon inserted randomly into the genome of M. catarrhalis (data not shown).
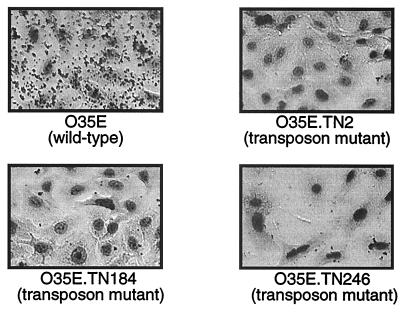
The ability of individual transposon mutants to attach to A549 cells was tested using a microcolony formation assay. Out of 1,000 Kanr colonies screened, nine mutants were impaired in attachment to A549 cells. Figure 1 illustrates the results of this screening for three mutants. Quantitative assays confirmed these observations and revealed an 8- to 19-fold reduction in adherence (Table 2). Note that these transposon mutants were obtained from independent mutagenesis experiments.
FIG. 1.
Binding of M. catarrhalis strains to A549 cells in a visual microcolony formation assay.
The M. catarrhalis adherence-negative mutants do not express the Hag protein.
Southern hybridization analysis of the mutants revealed that they contained a single transposon insertion (data not shown). Furthermore, the data suggested that all nine adherence-negative isolates contained a transposon in the same locus. Southern blot experiments revealed that a transposon-specific probe bound to PvuII DNA 10-kb fragments and to NcoI fragments migrating with identical 8-kb molecular masses (data not shown). Thus, these results imply that all these mutants showed decreased binding to A549 cells because they all contained transposons (in or upstream of the gene) that encode the same adhesin.
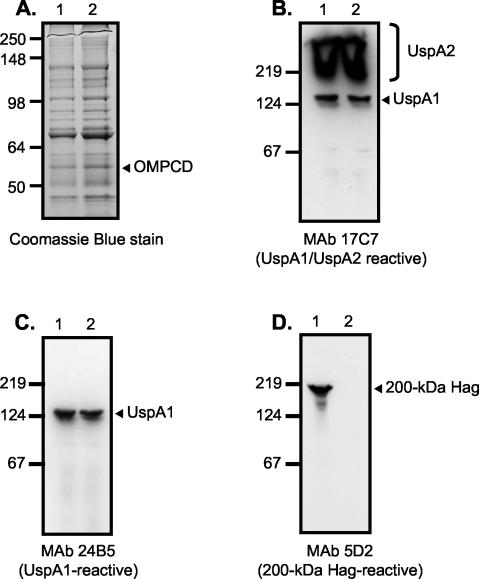
Since this adherence factor is likely a surface-exposed molecule and therefore associated with the outer membrane of M. catarrhalis, outer-membrane vesicles were isolated from the adherence mutants to determine whether they were missing a particular protein. The proteins present in these preparations were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then stained with Coomassie blue. This procedure identifies only abundant proteins. The protein profiles of the mutants were indistinguishable from that of the parent strain O35E (Fig. 2A). The mutants appear to express wild-type levels of the OMPCD protein (Fig. 2A). This molecule is a heat-modifiable antigen which migrates aberrantly in SDS-PAGE (24, 42) and has been shown to bind to human middle ear mucin (49) and thus might have functioned as an adhesin for the A549 cells.
FIG. 2.
SDS-PAGE and Western blot analysis of M. catarrhalis parent strain O35E (lane 1 in all panels) and representative adherence-negative isolate O35E.TN2 (lane 2 in all panels). Proteins present in outer-membrane vesicles were resolved by SDS-PAGE and stained with Coomassie blue (A). Whole-cell lysates were prepared and analyzed by Western blotting with the UspA1- and UspA2-reactive MAb17C7 (B), the UspA1-specific MAb 24B5 (C), and the 200-kDa Hag-specific MAb 5D2 (D). Note that whole-cell lysates were heated at 100°C for 15 min prior to Western blot analysis. Under these conditions, UspA1 migrates as a 125-kDa antigen and UspA2 migrates as an aggregate of very high molecular weight that is expressed in large amounts (bracket on the right side of panel B) (12). The mutants appear to express wild-type levels of the OMPCD protein (arrow in panel A). Positions of molecular mass markers (in kilodaltons) are shown to the left of each panel.
It has been shown that expression of UspA2 does not confer adherence to Chang cells (33). Nevertheless, UspA2 binds to the extracellular matrix protein vitronectin (36) and shares sequence similarities with the Yersinia adhesin YadA (2, 12, 33). It is therefore possible that UspA2 expression confers binding to other cell lines such as A549. However, Western blot analysis with the UspA2-reactive MAb 17C7 demonstrated that the representative O35E.TN2 strain expressed wild-type levels of UspA2 (Fig. 2B). Furthermore, immunoblot analysis with MAb 24B5 indicated that O35E.TN2 expressed wild-type levels of the UspA1 adhesin (Fig. 2C). Thus, the reduced binding of mutants to A549 cells cannot be attributed to a defect in UspA1 or UspA2 expression. All nine adherence-negative isolates expressed wild-type levels of UspA1 and UspA2 (data not shown).
Similar Western blot experiments utilizing Hag-specific MAb 5D2 revealed that all nine adherence mutants lacked the Hag protein (Fig. 2D). Therefore, our data suggest that the adherence-negative isolates contained a transposon in the hag gene or a gene that regulates its expression. To address this, a plasmid library of DNA fragments isolated from the representative M. catarrhalis mutant O35E.TN2 was introduced into E. coli and then selected for Kanr transformants. This selection yielded the plasmid pMHO35ETN2, which harbors the transposon as well as flanking O35E.TN2 genomic DNA. This construct was used to sequence outside of the transposon and into M. catarrhalis genomic DNA. DNA sequence analysis revealed that the transposon was inserted between nt 5419 and 5420 of the M. catarrhalis O35E hag gene (accession number AY77637), which would disrupt translation at codon 1717 of the 1,964-amino-acid open reading frame (ORF).
To verify that all nine adherence mutants contained a transposon in the hag gene, these isolates were analyzed by PCR. The hag-specific oligonucleotide primers F1 and R1 yielded a 6.4-kb amplicon in the parent strain O35E and a 7.6-kb amplicon in the transposon mutants (data not shown). These results confirm that the 1,221-bp EZ::TN <KAN-2> transposon was inserted in the hag gene of all nine independently isolated mutants. We also cloned the transposon insertion of strains O35E.TN114 and O35E.TN532 as described above. Sequence analysis of the plasmids pMHO35ETN114 and pMHO35ETN532 indicated that the transposon was inserted between nt 4746 and 4747 and nt 5269 and 5270 of O35E-hag, respectively. These results show that the hag mutants were independent insertions.
Phenotypic characterization of the hag mutants.
To rule out the possibility that the transposon mutants did not attach well to human cells due to a growth defect, the growth of strain O35E.TN2 was compared to that of the parent O35E in broth and tissue culture media. Interestingly, the mutant appeared to grow slightly better than the parent strain (data not shown). The lack of Hag expression by M. catarrhalis O35E has been shown previously to result in a loss of clumping properties but to have no adverse effect on adherence to cells of the Chang, HEp-2, NCIH292, and 16HBE14o− tissue culture cell lines (47). Consistent with these data, the autoagglutination properties of strain O35E.TN2 were severely impaired (data not shown) and the transposon mutant attached at nearly wild-type levels to the aforementioned human cells (Table 2 [Chang cell results] and data not shown [for the other cell lines]). In contrast, Hag expression was found to be necessary for M. catarrhalis O35E to adhere to HMEE cells at wild-type levels (Table 2).
The transposon insertion in O35E-hag does not affect expression of the gene located directly downstream.
BLAST searches of the patented M. catarrhalis genome through the National Center for Biotechnology Information service identified the hag ORF (nt 42694 to 48534 of AX0674457) as well as an ORF located directly downstream (nt 48964 to 50593 of AX0674457). This ORF is predicted to encode a protein with 70% identity (82% similarity) to Pseudomonas putida phosphoribosylaminoimidazolecarboxamide formyltransferase PurH (NP_746927.1). To demonstrate that the adherence phenotype of O35E.TN2 was not due to a polar effect on expression of the gene downstream of hag, we used quantitative real-time PCR to measure M. catarrhalis purH expression. As a control, we monitored expression of the unlinked M. catarrhalis gene bipA (nt 77514 to 75670 of AX067466). This ORF is predicted to encode a protein with 70% identity (83% similarity) to the Salmonella enterica serovar Typhimurium GTP-binding elongation factor family protein BipA (NP_462889.1). We found that purH expression was not significantly different between the wild type (relative expression ± standard error of the mean, 1.4 ± 0.07) and the hag mutant (1.2 ± 0.4). The signal in the RT-negative control was less than 5% of the signal observed in the presence of RT. Previous work and these data make it unlikely that the adherence phenotype of the hag mutants was due to changes in transcription of purH. Although other studies have shown that a mutation in hag leads to loss of adherence, this is the first report testing the possibility that these hag mutations affect the transcription of the purH gene.
DISCUSSION
Little is known about the gene products involved in the adherence of M. catarrhalis to human cells. To date, only the UspA1 (33, 36) and UspA2H (33) proteins and the recently described McaP (61) protein have been shown to directly mediate binding to some tissue culture cell lines in vitro. While the vast majority of M. catarrhalis isolates express UspA1 (33, 37), only 20% of the strains contain a uspA2H gene (33, 37). To effectively adhere to host surfaces, most organisms produce multiple adhesins. For example, nontypeable H. influenzae expresses at least six distinct adherence factors, namely, Hia (6), Hap (23, 58), Oap (48), hemagglutinating pili (20, 27), and the high-molecular-weight adhesins HMW1 and HMW2 (5, 13, 57). It is therefore likely that M. catarrhalis expresses other adherence factors in addition to UspA1, UspA2H, and McaP. One potential adhesin, OMPCD, binds to middle ear mucin (49), and the bacterial gene product most similar to OMPCD, the Pseudomonas aeruginosa OprF protein (41), was recently shown to function as an adhesin for A549 cells (4). Other M. catarrhalis adhesin candidates include UspA2, which is similar to the Yersinia adhesin YadA (2, 12, 33), and the Hag protein, which has been shown to bind erythrocytes (16, 17, 47) and human IgD (47). Even though the lack of Hag expression does not adversely affect the adherence of the M. catarrhalis O35E strain to various human cell lines (47), M. catarrhalis strain Bc5 expresses a Hag homologue (designated MID) which was shown to bind human B cells (19, 21).
Using a commercially available system (Epicentre) in an effort to identify M. catarrhalis gene products involved in adherence, we developed the ability to randomly mutagenize this bacterium. This system works well with M. catarrhalis strain O35E, routinely yielding 500 to 1,000 transposon mutants per 20 ng of Transposome. To our knowledge, this is the first report of a transposon mutagenesis system for M. catarrhalis and should provide a useful tool for studying this organism. Next, transposon mutants were screened to identify those that no longer bound A549 human lung cells in microcolony formation assays. A549 cells were utilized because our data suggested that the already characterized adhesin UspA1 does not mediate adherence to these human lung cells. Furthermore, strain O35E was previously shown to contain a uspA2 gene and therefore does not express the other characterized adhesin, UspA2H (33). We reasoned that at least some M. catarrhalis transposon mutants that no longer bound to A549 cells would likely contain a transposon in or upstream of a gene encoding a novel adherence factor.
Even though a relatively small number of transposon mutants (i.e., 1,000) were screened, we were able to identify nine isolates that exhibited significantly decreased binding to A549 cells and did not exhibit an obvious growth defect. This frequency (0.09%) is higher than would be expected unless the disrupted gene were larger than others or mutants had a slight growth advantage. These nine mutants expressed wild-type levels of UspA1, which is consistent with our finding that the lack of expression of this protein did not adversely affect adherence to A549 cells in our assays. Furthermore, our results indicate that decreased adherence is not due to a defect in expression of OMPCD, UspA2, or purH. Notably, none of the mutants expressed Hag (Fig. 2D). Because expression of this 200-kDa antigen has been previously shown to be phase variable (K. Sasaki, L. Myers, S. M. Loosmore, and M. H. Klein, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., p. 89, 1999), we used nucleotide sequence analysis and PCR to verify that the transposon insertion was indeed in the hag gene. These data therefore demonstrate that the lack of Hag expression considerably decreases the adherence of M. catarrhalis O35E to A549 cells as well as to primary cultures of HMEE, which in turn suggests that Hag is an adhesin for these cells. Interestingly, the hag ORF is large (5,895 nt) and the mutant O35E.TN2 grows slightly better than the parent O35E. This might partly explain the relatively high frequency of hag mutants that were isolated (0.09%).
Our results are consistent with those of Pearson et al., who showed that a lack of Hag expression results in loss of autoagglutination capabilities but does not affect adherence to cells of the Chang, HEp-2, 16HBE14o−, or NCI-H292 cell lines (47). These investigators did not measure the adherence of their isogenic hag mutant to A549 or HMEE cells. Our representative hag mutant, O35E.TN2, no longer autoagglutinates and binds at wild-type levels to Chang as well as to HEp-2, 16HBE14o−, and NCI-H292 cells. Pearson et al. also demonstrated that Hag forms projections that cover the entire surface of M. catarrhalis cells (47). It is therefore possible that the absence of this major outer membrane protein could affect the proper surface display of the actual adhesin for A549 and HMEE cells. Similar membrane destabilization effects have been reported previously (32, 62). However, the finding that the Hag homologue protein MID, which is expressed by M. catarrhalis strain Bc5, directly binds B cells in vitro suggests that Hag functions as an adhesin for certain human cell types (19, 21). While this article was being revised, Forsgren and colleagues published a report in which they demonstrated that the M. catarrhalis MID protein (a Hag homologue) is an adhesin for A549 cells (18). The region of this protein containing the adherence epitope (MID764-913) has a high level of sequence similarity (73% identity and 81% similarity; data not shown) to residues 715 to 863 of O35E-Hag. Our results constitute independent proof of the role of Hag (MID) as an adhesin. Our findings also add to those of Forsgren by showing that this 200-kDa molecule also confers adherence to human middle ear cells.
Due to the high incidence and recurrence of M. catarrhalis infections, a vaccine for this organism is desirable. Interference with adherence is an approach with a high likelihood of substantially reducing the risks of these infections, because adherence is a crucial step that occurs early in bacterial pathogenesis (7, 26, 56). Furthermore, adhesins are attractive vaccine candidates because they are surface located and thus are readily accessible to antibodies. For these reasons, it is important to identify the repertoire of adhesins expressed by M. catarrhalis to identify the best vaccine candidate(s). Our results indicate that Hag is involved in adherence to human lung and middle ear cells in vitro, which is relevant to pathogenesis by this upper-respiratory-tract pathogen. Furthermore, recent studies have shown that Hag (and its homologue MID) are well conserved and surface exposed (38, 47). These observations warrant further investigation of this 200-kDa protein as a potential vaccine antigen.
Acknowledgments
This study was supported in part by institutional startup funds from the Medical College of Ohio, a grant from the Thrasher Research Fund (award number 02816-6), and a grant from NIH/NIAID (1 RO1 AI051477-01 A1) to E.R.L.
We thank Eric Hansen at the University of Texas Southwestern Medical Center in Dallas for providing M. catarrhalis strains and antibodies. We also thank Thomas DeMaria at Ohio State University for providing cultures of human middle ear cells. We also thank Eric Hansen, Robert Blumenthal, and Mark Wooten for their helpful comments on the manuscript.
Editor: D. L. Burns
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Thoracic Society. 1995. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 152:S77-S121. [PubMed] [Google Scholar]
- 4.Azghani, A. O., S. Idell, M. Bains, and R. E. Hancock. 2002. Pseudomonas aeruginosa outer membrane protein F is an adhesin in bacterial binding to lung epithelial cells in culture. Microb. Pathog. 33:109-114. [DOI] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 7.Beachey, E. H. 1981. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J. Infect. Dis. 143:325-345. [DOI] [PubMed] [Google Scholar]
- 8.Berner, R., R. F. Schumacher, M. Brandis, and J. Forster. 1996. Colonization and infection with Moraxella catarrhalis in childhood. Eur. J. Clin. Microbiol. Infect. Dis. 15:506-509. [DOI] [PubMed] [Google Scholar]
- 9.Cappelletty, D. 1998. Microbiology of bacterial respiratory infections. Pediatr. Infect. Dis. J. 17:S55-61. [DOI] [PubMed] [Google Scholar]
- 10.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3:293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, J. J. 1999. Moraxella (Branhamella) catarrhalis: clinical, microbiological and immunological features in lower respiratory tract infections. APMIS Suppl. 88:1-36. [PubMed] [Google Scholar]
- 12.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawid, S., S. Grass, and J. W. St. Geme III. 2001. Mapping of binding domains of nontypeable Haemophilus influenzae HMW1 and HMW2 adhesins. Infect. Immun. 69:307-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Beccaro, M. A., P. M. Mendelman, A. F. Inglis, M. A. Richardson, N. O. Duncan, C. R. Clausen, and T. L. Stull. 1992. Bacteriology of acute otitis media: a new perspective. J. Pediatr. 120:81-84. [DOI] [PubMed] [Google Scholar]
- 15.Faden, H. 2001. The microbiologic and immunologic basis for recurrent otitis media in children. Eur. J. Pediatr. 160:407-413. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald, M., R. Mulcahy, S. Murphy, C. Keane, D. Coakley, and T. Scott. 1997. A 200 kDa protein is associated with haemagglutinating isolates of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 18:209-216. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald, M., R. Mulcahy, S. Murphy, C. Keane, D. Coakley, and T. Scott. 1999. Transmission electron microscopy studies of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 23:57-66. [DOI] [PubMed] [Google Scholar]
- 18.Forsgren, A., M. Brant, M. Karamehmedovic, and K. Riesbeck. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect. Immun. 71:3302-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsgren, A., M. Brant, A. Mollenkvist, A. Muyombwe, H. Janson, N. Woin, and K. Riesbeck. 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J. Immunol. 167:2112-2120. [DOI] [PubMed] [Google Scholar]
- 20.Gilsdorf, J. R., K. W. McCrea, and C. F. Marrs. 1997. Role of pili in Haemophilus influenzae adherence and colonization. Infect. Immun. 65:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gjorloff Wingren, A., R. Hadzic, A. Forsgren, and K. Riesbeck. 2002. The novel IgD binding protein from Moraxella catarrhalis induces human B lymphocyte activation and Ig secretion in the presence of Th2 cytokines. J. Immunol. 168:5582-5588. [DOI] [PubMed] [Google Scholar]
- 22.Helminen, M. E., I. Maciver, J. L. Latimer, J. Klesney-Tait, L. D. Cope, M. Paris, G. H. McCracken, Jr., and E. J. Hansen. 1994. A large, antigenically conserved protein on the surface of Moraxella catarrhalis is a target for protective antibodies. J. Infect. Dis. 170:867-872. [DOI] [PubMed] [Google Scholar]
- 23.Hendrixson, D. R., and J. W. St. Geme III. 1998. The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell 2:841-850. [DOI] [PubMed] [Google Scholar]
- 24.Hsiao, C. B., S. Sethi, and T. F. Murphy. 1995. Outer membrane protein CD of Branhamella catarrhalis: sequence conservation in strains recovered from the human respiratory tract. Microb. Pathog. 19:215-225. [DOI] [PubMed] [Google Scholar]
- 25.Hu, W.-G., J. Chen, J. C. McMichael, and X.-X. Gu. 2001. Functional characteristics of a protective monoclonal antibody against serotype A and C lipooligosaccharides from Moraxella catarrhalis. Infect. Immun. 69:1358-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hultgren, S. J., S. Abraham, M. Caparon, P. Falk, J. W. St. Geme III, and S. Normark. 1993. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell 73:887-901. [DOI] [PubMed] [Google Scholar]
- 27.Kar, S., S. C. To, and C. C. Brinton, Jr. 1990. Cloning and expression in Escherichia coli of LKP pilus genes from a nontypeable Haemophilus influenzae strain. Infect. Immun. 58:903-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 29.Klein, J. O. 2000. The burden of otitis media. Vaccine 19(Suppl. 1):S2-S8. [DOI] [PubMed] [Google Scholar]
- 30.Klein, J. O. 1994. Otitis media. Clin. Infect. Dis. 19:823-833. [DOI] [PubMed] [Google Scholar]
- 31.Klein, J. O., D. W. Teele, and S. I. Pelton. 1992. New concepts in otitis media: results of investigations of the Greater Boston Otitis Media Study Group. Adv. Pediatr. 39:127-156. [PubMed] [Google Scholar]
- 32.Klugman, K. P. 1996. The clinical relevance of in-vitro resistance to penicillin, ampicillin, amoxycillin and alternative agents, for the treatment of community-acquired pneumonia caused by Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis. J. Antimicrob. Chemother. 38(Suppl. A):133-140. [DOI] [PubMed] [Google Scholar]
- 33.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luke, N. R., T. A. Russo, N. Luther, and A. A. Campagnari. 1999. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect. Immun. 67:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMichael, J. C., M. J. Fiske, R. A. Fredenburg, D. N. Chakravarti, K. R. VanDerMeid, V. Barniak, J. Caplan, E. Bortell, S. Baker, R. Arumugham, and D. Chen. 1998. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect. Immun. 66:4374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20:1754-1760. [DOI] [PubMed] [Google Scholar]
- 38.Mollenkvist, A., T. Nordstrom, C. Hallden, J. J. Christensen, A. Forsgren, and K. Riesbeck. 2003. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J. Bacteriol. 185:2285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, T. F., A. L. Brauer, N. Yuskiw, and T. J. Hiltke. 2000. Antigenic structure of outer membrane protein E of Moraxella catarrhalis and construction and characterization of mutants. Infect. Immun. 68:6250-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, T. F., C. Kirkham, and A. J. Lesse. 1993. The major heat-modifiable outer membrane protein CD is highly conserved among strains of Branhamella catarrhalis. Mol. Microbiol. 10:87-97. [DOI] [PubMed] [Google Scholar]
- 42.Murphy, T. F., and M. R. Loeb. 1989. Isolation of the outer membrane of Branhamella catarrhalis. Microb. Pathog. 6:159-174. [DOI] [PubMed] [Google Scholar]
- 43.National Heart, Blood, and Lung Institute. 1998. NHLBI morbidity and mortality chartbook. [Online.] National Heart, Blood, and Lung Institute, Bethesda, Md. http://www.nhlbi.nih.gov/resources/docs/cht-book.htm.
- 44.Neumayer, U., H. K. Schmidt, K. P. Mellwig, and G. Kleikamp. 1999. Moraxella catarrhalis endocarditis: report of a case and literature review. J. Heart Valve Dis. 8:114-117. [PubMed] [Google Scholar]
- 45.Nordstrom, T., A. Forsgren, and K. Riesbeck. 2002. The immunoglobulin D-binding part of the outer membrane protein MID from Moraxella catarrhalis comprises 238 amino acids and a tetrameric structure. J. Biol. Chem. 277:34692-34699. [DOI] [PubMed] [Google Scholar]
- 46.Patrick, C. C., A. Kimura, M. A. Jackson, L. Hermanstorfer, A. Hood, G. H. McCracken, Jr., and E. J. Hansen. 1987. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypeable Haemophilus influenzae. Infect. Immun. 55:2902-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prasadarao, N. V., E. Lysenko, C. A. Wass, K. S. Kim, and J. N. Weiser. 1999. Opacity-associated protein A contributes to the binding of Haemophilus influenzae to Chang epithelial cells. Infect. Immun. 67:4153-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy, M. S., T. F. Murphy, H. S. Faden, and J. M. Bernstein. 1997. Middle ear mucin glycoprotein: purification and interaction with nontypable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116:175-180. [DOI] [PubMed] [Google Scholar]
- 50.Ruuskanen, O., and T. Heikkinen. 1994. Otitis media: etiology and diagnosis. Pediatr. Infect. Dis. J. 13:S23-S26, S50-S54. [PubMed]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 53.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, T. C., D. D. Sledjeski, and M. D. Boyle. 2003. Regulation of protein H expression in M1 serotype isolates of Streptococcus pyogenes. FEMS Microbiol. Lett. 219:9-15. [DOI] [PubMed] [Google Scholar]
- 55.Stefanou, J., A. V. Agelopoulou, N. V. Sipsas, N. Smilakou, and A. Avlami. 2000. Moraxella catarrhalis endocarditis: case report and review of the literature. Scand. J. Infect. Dis. 32:217-218. [DOI] [PubMed] [Google Scholar]
- 56.St. Geme, J. W., III. 1997. Bacterial adhesins: determinants of microbial colonization and pathogenicity. Adv. Pediatr. 44:43-72. [PubMed] [Google Scholar]
- 57.St. Geme, J. W., III. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 62:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.St. Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 59.Stool, S. E., and M. J. Field. 1989. The impact of otitis media. Pediatr. Infect. Dis. J. 8:S11-S14. [PubMed] [Google Scholar]
- 60.Thorsson, B., V. Haraldsdottir, and M. Kristjansson. 1998. Moraxella catarrhalis bacteraemia. A report on 3 cases and a review of the literature. Scand. J. Infect. Dis. 30:105-109. [DOI] [PubMed] [Google Scholar]
- 61.Timpe, J. M., M. M. Holm, S. L. Vanlerberg, V. Basrur, and E. R. Lafontaine. Identification of a Moraxella catarrhalis outer membrane protein exhibiting both adhesin and lipolytic activities. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 62.Tong, H. H., Y. Chen, M. James, J. Van Deusen, D. B. Welling, and T. F. DeMaria. 2001. Expression of cytokine and chemokine genes by human middle ear epithelial cells induced by formalin-killed Haemophilus influenzae or its lipooligosaccharide htrB and rfaD mutants. Infect. Immun. 69:3678-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turner, H. R., M. R. Taylor, and W. R. Lockwood. 1985. Branhamella catarrhalis endocarditis in a patient receiving hemodialysis. South. Med. J. 78:1021-1022. [DOI] [PubMed] [Google Scholar]
- 64.Utsunomiya, T., K. Nakahara, M. Kuramochi, K. Hashiba, Y. Uzuka, and K. Matsumoto. 1984. Branhamella (Neisseria) catarrhalis endocarditis after insertion of a mitral prosthesis: a case report. Nippon Naika Gakkai Zasshi 73:1506-1511. (In Japanese.) [DOI] [PubMed] [Google Scholar]