Abstract
Streptococcus agalactiae is the leading cause of bacterial sepsis and meningitis in neonates and also the causative agent of different serious infections in immunocompromised adults. The wide range of diseases that are caused by S. agalactiae suggests regulatory mechanisms that control the formation of specific virulence factors in these bacteria. The present study describes a gene from S. agalactiae, designated rogB, encoding a protein with significant similarity to members of the RofA-like protein (RALP) family of transcriptional regulators. Disruption of the rogB gene in the genome of S. agalactiae resulted in mutant strain RGB1, which was impaired in its ability to bind to fibrinogen and fibronectin. Mutant RGB1 also exhibited a reduced adherence to human epithelial cells but did not show an altered invasion of eukaryotic cells. By real-time PCR analysis, mutant RGB1 revealed an increased expression of the cpsA gene, encoding a regulator of capsule gene expression. However, strain RGB1 exhibited a reduced expression of the rogB gene and of two adjacent genes, encoding putative virulence factors in S. agalactiae. Furthermore, mutant RGB1 was impaired in the expression of the fbsA gene, coding for a fibrinogen receptor from S. agalactiae. The altered gene expression in mutant RGB1 could be restored by plasmid-mediated expression of rogB, confirming a RogB deficiency as the cause for the observed changes in virulence gene expression in S. agalactiae. Reporter gene studies with a promotorless luciferase gene fused to fbsA allowed a growth-dependent analysis of fbsA expression in S. agalactiae. These reporter gene studies also suggest that RogB exerts a positive effect on fbsA expression in S. agalactiae.
Streptococcus agalactiae, also named group B streptococcus (GBS), is the leading cause of bacterial sepsis and meningitis in neonates in many industrialized countries (1). In addition, it is the cause of substantial pregnancy-related morbidity and has emerged as an increasingly common cause of invasive disease in the elderly and in immunocompromised persons (51, 53). The spectrum of diseases caused by GBS in adults ranges from urinary tract and soft tissue infections to life-threatening sepsis and meningitis (40). Besides being the causative agent of many different types of infections, GBS can persistently colonize the human skin and mucous membranes without inducing clinical symptoms (41).
Like many other pathogens, GBS can attach to epithelial surfaces by binding to different host cell proteins. Binding of GBS to human laminin is mediated by the lipoprotein Lmb, which has been studied on the molecular level (43). Although GBS does not bind soluble fibronectin on its surface (4), adherence of the bacteria to immobilized fibronectin has been convincingly demonstrated (45). In a recent study, Beckmann et al. (3) identified C5a peptidase from GBS to mediate binding of the bacteria to fibronectin. In addition, fibronectin binding of GBS has been shown to mediate the invasion of the bacteria into host cells (7). Binding of GBS to human fibrinogen is brought about by the fibrinogen receptor FbsA, which interacts with fibrinogen by repeptive units and which is widely distributed in different GBS strains (39).
Recently, the genomic sequences of the serotype III GBS strain NEM316 (17) and of the serotype V strain 2603 V/R (47) were published. Analysis of the obtained sequence data revealed the presence of several putative virulence genes, including bacterial surface proteins and virulence regulators. Although a few regulatory systems from GBS have been studied on the molecular level (10, 34, 42), the targets and stimuli of most transcriptional regulators from GBS are still unknown.
Pathogenic bacteria often use global regulatory networks to control the expression of different virulence factors in response to changing environmental cues throughout the infection process. In Streptococcus pyogenes several regulatory proteins are involved in the transcriptional control of virulence factors. Besides two-component signal transduction systems (14, 21-23, 26) and the multiple gene activator Mga (32), the two regulatory proteins RofA and Nra have been shown to exert a significant effect on the expression of various virulence genes in S. pyogenes (2, 15, 16, 19, 25, 28, 31). RofA and Nra exhibit 62% identity to each other and comprise a novel family of transcriptional regulators (16, 19). Analysis of the genome database identified two further RofA homologous proteins in the chromosome of S. pyogenes and one in the genome of Streptococcus pneumoniae (19). These new members of the RofA-like protein family were named, accordingly, RALPs and are suggested to play a role in the regulatory network of virulence in the two pathogens.
The present study describes a new member of the RALP family of transcriptional regulators from GBS, designated RogB. By insertional inactivation of the rogB gene in the chromosome of GBS the importance of RogB for the binding of the bacteria to host cell proteins and the adherence to and invasion into eukaryotic cells was addressed. Using real-time PCR, the expression of known and putative virulence genes was compared between the rogB mutant and its parental strain. Finally, reporter gene studies addressed the role of RogB for the expression of the fbsA gene, encoding a fibrinogen receptor from GBS.
MATERIALS AND METHODS
Bacterial strains, culture conditions, plasmids, and proteins.
GBS strain 6313 is a serotype III clinical isolate obtained from an infected neonate and has been described previously (49). GBS 6313 fbsA-luc harbors a promotorless luciferase gene, transcriptionally fused to the fbsA gene in the chromosome of this strain. The GBS mutants RGB1 and RGB1 fbsA-luc are rogB::pG+host6 derivates of the GBS strains 6313 and 6313 fbsA-luc, respectively, carrying an insertionally inactivated rogB gene. Escherichia coli DH5α (20) served as host for the GBS pTEX5236 cosmid gene library (35) and for the recombinant pG+host6 and pAT28 plasmids.
The rogB gene was isolated from a pTEX5236-based (46) gene bank of GBS 6313 constructed in E. coli (35). Plasmid pCR-TOPO (Invitrogen) served for cloning and sequencing of the 350-bp rogB-specific PCR product, plasmid pUC18 (50) was used for subcloning the rogB encoding region in E. coli, and the vector pG+host6 (Appligene) served for the disruption of rogB in the genome of GBS 6313. Plasmid pAT28 is an E. coli-Streptococcus shuttle vector (48) and served for the construction of the rogB-carrying plasmid pATrogB that was used for complementation analysis of GBS strain RGB1. For the construction of plasmid pATrogB, the rogB gene was amplified from chromosomal DNA of GBS 6313 by PCR with the primers CCGCGGATCCCAACTCCTATTGTGCCG and CGGCACGAGCTCGTCACTCCATGAATCTCTTG. The BamHI and SacI restriction sites used for cloning are underlined. The rogB-containing PCR product and plasmid pAT28 were digested with BamHI and SacI, ligated, and transformed into E. coli DH5α. The plasmids pAT28 and pATrogB were subsequently transformed into the GBS strains 6313, RGB1, 6313 fbsA-luc, and RGB1 fbsA-luc.
GBS was cultivated at 37°C in Todd-Hewitt yeast broth (THY) consisting of Todd-Hewitt broth (Oxoid) supplemented with 1% of yeast extract. GBS strains carrying the pG+host6 plasmid inserted into the chromosomal copy of rogB were selected on THY medium containing erythromycin (5 μg/ml). GBS strains carrying the plasmids pAT28 or pATrogB were grown in the presence of spectinomycin (200 μg/ml). E. coli was grown at 37°C in Luria broth. Recombinant E. coli clones carrying pTEX5236-, pG+host6-, pAT28-, or pUC18-based plasmids were selected in the presence of chloramphenicol (15 μg/ml), ampicillin (50 μg/ml), spectinomycin (100 μg/ml), or ampicillin (100 μg/ml).
Fibrinogen, fibronectin, laminin, collagen I, and collagen IV were purchased from Sigma-Aldrich. Fibrinogen was passed through a gelatin-Sepharose column to remove contaminating fibronectin.
Construction of an fbsA-luc transcriptional fusion in GBS.
A promotorless luciferase gene (luc) was isolated from plasmid pFW11-luc (31) by BamHI/HindIII digestion. The luc box was subsequently ligated into the BamHI/HindIII-digested vector pG+host6, resulting in plasmid pGluc, in which the luciferase gene is flanked by two multiple cloning sites. Subsequently, the 3′ end of fbsA was amplified by PCR from chromosomal GBS DNA by PCR with the primers 5′-CCGCGGATCCGTAGGTCAACTTATAGGG and 5′-CCGCGGATCCATTATACTTAATTTTCATTGCG. The BamHI restriction sites used for cloning are underlined. After digestion of the obtained PCR fragment and of plasmid pGluc with BamHI, the 3′ fbsA fragment was ligated into pGluc and transformed in E. coli DH5α. Insert-carrying clones were subsequently sequenced to identify clones with the correct orientation of the 3′ end of fbsA in pGluc. The resultant plasmid was termed pGlucfbsA3′. The downstream region of fbsA was amplified by PCR from chromosomal GBS DNA with the primers 5′-TGGCACAAGCTTCAATCATTTAGTAACTATATATAATG and 5′-GAGCGGGGTACCGTTTCACTTGTTCTATTGG. The HindIII and KpnI restriction sites used for cloning are underlined. The PCR product and plasmid pGlucfbsA3′ were digested with HindIII and KpnI, ligated, and transformed in E. coli DH5α. The resultant plasmid, pGluc-fbsA, was transformed in GBS 6313 with subsequent erythromycin selection at 30°C. Cells in which pGluc-fbsA had integrated into the chromosome were selected by growth of the transformants at 37°C with erythromycin selection as described previously (27). Four of such integrant strains were serially passaged for 3 days in liquid medium at 30°C without erythromycin selection to facilitate the excision of plasmid pGluc-fbsA, leaving the desired promotorless luciferase box in the chromosome. Dilutions of the serially passaged cultures were plated onto agar plates and single colonies were tested for erythromycin sensitivity to identify pGluc-fbsA excisants. Chromosomal DNA of GBS 6313 and of 24 erythromycin-sensitive GBS excisants was tested by Southern blotting after HindIII digestion by using a digoxigenin-labeled fbsA gene probe obtained with the primers 5′-GTCCTGTATCTGCTATGGATAGTGTTGG and 5′-ACATTTTGATCATCACCTG.
Construction of the GBS rogB mutants RGB1 and RGB1 fbsA-luc.
The thermosensitive plasmid pG+host6 (Appligene) was used for targeted disruption of rogB in GBS strains 6313 and 6313 fbsA-luc to construct mutants RGB1 and RGB1 fbsA-luc, respectively. An internal rogB fragment was amplified by PCR with the primers 5′CGCGGATCCATGATTCAGGCAGGTTACC and 5′TGGCACAAGCTTGGAAGTAAGGTAAGCAAG. The BamHI and HindIII restriction sites used for cloning are underlined. The resulting PCR product and plasmid pG+host6 were digested with BamHI and HindIII, ligated, and transformed into E. coli DH5α. Plasmid pGrogB was subsequently transformed into the GBS stains 6313 and 6313 fbsA-luc by using the method of Ricci et al. (36), and transformants were selected by growth on erythromycin agar at 30°C. Cells in which the plasmid had integrated into the GBS chromosome were identified by growth at 37°C with erythromycin selection as described previously (27). Successful disruption of rogB was confirmed by Southern blotting with AccI-digested chromosomal DNA of the GBS parental strains 6313 and 6313 fbsA-luc and their mutants, RGB1 and RGB1 fbsA-luc, by using a digoxigenin-labeled rogB fragment obtained with the primers 5′-CACTTGGTTGCAATGTTTG and 5′-CTTACTGATAAGCCCGAGG.
Quantification of specific transcripts with LightCycler real-time PCR.
GBS strains were grown in 50 ml of THY broth to exponential growth phase (optical density at 600 nm [OD600] = 0.30), and RNA was isolated by using the RNeasy kit (Qiagen) as described previously (35). Contaminating DNA was degraded by digestion with DNase as described elsewhere (18). To exclude the possibility of DNA contamination during RNA preparation, RNA samples were subjected to PCR amplification without prior reverse transcription. However, no amplificates were obtained. Reverse transcription of RNA was performed with random hexanucleotides and the RevertAid First strand cDNA synthesis kit (MBI Fermentas) according to the instructions of the manufacturer. For expression analysis of the genes cfb, lmb, sodA, cpsA, hylB, lytR, gyrA, rogB, fbsA, sag1478/gbs1408, and sag1477/gbs1407, the primers listed in Table 1 were used. The temperature profile for template amplification was essentially as described elsewhere (18). In brief, the sample was initially denatured for 1 cycle at 95°C for 30 s, following 44 cycles of denaturation at 95°C for 1 s, annealing at 50°C for 15 s, and extension with fluorescence acquisition at 72°C for 30 s. The temperature transition during the amplification was set to 20°C/s. Melting-curve analysis was performed at between 65 and 95°C with stepwise fluorescence acquisition and a temperature transition of 0.1°C/s. Sequence-specific standard curves were generated by using 10-fold serial dilutions (105 to 108 copies) of genomic DNA. The quantitiy of cDNA for the investigated genes was normalized to the quantity of gyrA cDNA in each sample. The gyrA gene was chosen as an internal standard since gyrase genes in streptococci and staphylococci represent ubiquitously expressed housekeeping genes that are frequently used for the normalization of gene expression in quantitative reverse transcription-PCR experiments (6, 18). Each experiment was performed at least four times with two independent RNA preparations.
TABLE 1.
Oligonucleotide primers for quantitative reverse transcription-PCR by using a LightCycler
| Gene | Sequence (5′-3′)
|
|
|---|---|---|
| Forward primer | Reverse primer | |
| gyrA | GACGTTCAGGTATTCAC | TCAAACTGAGGTACGACG |
| cfb | TGAGGCTATTACTAGCGTGG | AAGTCGACAGCATCACACG |
| lmb | ATGGAAGTCACACAAGGC | ATAGCAGCAACTGAGCCG |
| sodA | CATCATGATAAGCACCATGC | TGGAGTATCTTGATTGGCAG |
| cpsA | GGTGATAGTCAAGCTATGG | TCTATCGTTATCGCCTCC |
| hylA | CCTATTATCCAACGTACCG | GAACCTGTAACTGATAACGG |
| scpB | AACAGTAGCAGATGACGC | AGCTAGTGCAGCATTACC |
| lytR | GATGATGAACCAGTTGCACG | TGCCATTGTTGTAGTGAGCC |
| rogB | GCAGTTGCACAAGATAGTC | TTTGAGAGAGAGTTTCTG |
| fbsA | GTAGGTCAACTTATAGGG | ATACTTAATTTTCATTGCG |
| sag1408 | TTCGGCACAATAGGAGTTG | CTTAACTTGCCAAGTCTGG |
| sag1407 | TGGTGACTTATGGACG | TGTACCAATACCACCTG |
General DNA techniques.
Chromosomal GBS DNA was isolated according to the method of Pospiech and Neumann (33). Conventional techniques for DNA manipulation, such as restriction enzyme digests, PCR, ligation, transformation by electroporation, and Southern blotting, were performed as described by Sambrook et al. (38).
Determination of luciferase activity.
For assessment of luciferase activity from fbsA-luc transcriptional fusions in GBS, the bacteria were grown aerobically in THY broth at 37°C with shaking. For determining the luminescence of the bacterial culture, 1-ml samples were taken at different time points to determine the OD600 of the culture and to measure the luciferase activity. Luciferase activity was measured essentially as described by Podbielski et al. (31). Briefly, 150 μl of bacterial cell suspension was transferred in a sample tube of the Flash'n'Glow luminometer (Berthold). The reservoirs of the luminometer were filled with 2.5× assay buffer (62.5 mM glycyl-glycin [pH 7.8], 25 mM MgCl2) and 330 μM d-luciferin. Then, 200 μl of 2.5× assay buffer and 200 μl of luciferin solution were automatically added to the bacterial suspension in the sample tube, and the luminescence was immediately measured for 15 s at 22°C. The relative light units (RLU) at the different time points were obtained by subtraction of the luminescence at the beginning of the experiment from the luminescence at later time points.
Binding of FITC-labeled GBS to immobilized human matrix proteins.
Terasaki microtiter plates were coated with fibronectin, fibrinogen, laminin, or collagens I or IV, and the binding of fluorescein isothiocyanate (FITC)-labeled GBS to the immobilized proteins was measured essentially as described by Podbielski et al. (31). In brief, 10 μl of a 100-μg/ml stock solution of human fibronectin, fibrinogen, laminin, or collagen I or IV was added to each well, followed by incubation overnight at room temperature in a moist chamber. Subsequently, the microtiter plates were washed with phosphate-buffered saline (PBS), and residual buffer was carefully removed. FITC-labeling of GBS was performed with cultures in the exponential (OD600 = 0.5) and in the stationary (OD600 = 1.5) growth phases. A total of 12 ml of bacterial culture was pelleted by centrifugation, washed with 12 ml of PBS, and resuspended in 2 ml of FITC-solution (1 mg of FITC/ml in 50 mM sodium carbonate buffer [pH 9.2]). After a 20-min incubation in the dark, the cells were pelleted by centrifugation, washed twice with PBS, and sonicated for 20 s to disrupt bacterial chains. The bacterial suspension was adjusted to an OD600 of 1.0 with PBS to ensure an equal number of bacteria per volume for the different strains. Subsequently, the suspension was vortexed vigurously and kept in the dark until use. Then, 10 μl of FITC-labeled streptococci were added to each Terasaki well coated with different human proteins. After a 60-min incubation at 37°C, unbound bacteria were removed by five washes with PBS, and bound bacteria were fixed with 0.5% glutaraldehyde for 5 min. The plates were finally washed twice with PBS, and the fluorescence of each well was determined in an automated Cyto-Fluor II fluorescence reader (PerSeptive Biosystems) at excitation and detection wavelengths of 485 and 530 nm, respectively. The efficiency of FITC labeling of the bacteria was determined by incubating 500 μl of the FITC-labeled bacteria for 60 min at 37°C, followed by three washes of the bacteria with PBS, resuspension of the cells in 500 μl of PBS, and measurement of the fluorescence of 10-μl aliquots of the suspension in uncoated Terasaki mitrotiter plates. The amount of bound bacteria per well was calculated as the percentage of total labeled bacteria added to each well. Each assay was measured in triplicate and repeated at least four times.
Binding of soluble 125I-labeled human proteins to GBS.
The human proteins fibrinogen, fibronectin, laminin, and collagens I and IV were radiolabeled with 125I by using the chloramine T method (24). Binding of the labeled proteins to GBS was performed essentially as described by Chhatwal et al. (9). Briefly, cultures in the exponential (OD600 = 0.5) or stationary (OD600 = 1.5) growth phase were pelleted by centrifugation, washed twice with PBS supplemented with 0.02% Tween 20 (PBST), and adjusted photometrically to a transmission of 10% at 600 nm to ensure an equal number of bacteria per volume for the different strains. A total of 0.2 ml of the bacterial suspension was added to 20 μl of 125I-labeled protein (50,000 cpm). After an incubation for 1 h at room temperature, the streptococci were sedimented by centrifugation and washed with 1 ml of PBST. The radioactivity of the pellet was finally measured in a gamma counter (Packard Instruments). The amount of bacterium-bound protein was calculated as the percentage of total radiolabeled protein added to the bacteria. Each experiment was repeated at least three times in triplicate.
Epithelial cell adherence and internalization assay.
Adherence of GBS to epithelial cells and invasion into epithelial cells was assayed essentially as described elsewhere (5, 37). Since GBS reveals growth in tissue culture medium, thereby influencing the number of bacteria that can adhere to and invade the host cells, the number of bacteria after growth for 2 h in tissue culture medium was set as the input inoculum as described elsewhere (13). To determine the input inoculum of each strain, 1 ml of RPMI tissue culture medium was inoculated with 5 × 106 bacteria of the different strains, and the total number of bacteria after growth for 2 h was quantitated by plating serial dilutions onto THY agar plates. Since the different strains revealed identical growth in RPMI medium (ν = 1.75 h−1), the total number of bacteria after growth for 2 h was 5.5 ± 0.2 × 107. This value was taken as input inoculum for the different GBS strains and used to determine the multiplicity of infection in the adherence assays.
For adherence and invasion assays, A549 cells were transferred to 24-well tissue culture plates at 4 × 105 cells per well and cultivated overnight in RPMI tissue culture medium (Gibco-BRL) supplemented with 10% of fetal calf serum. The medium was subsequently replaced with 1 ml of fresh RPMI medium.
For adherence assays, the A549 cells were infected with 5 × 106 bacteria, and the infected cells were incubated in RPMI tissue culture medium for 2 h at 37°C, resulting in an input inoculum of (5.5 ± 0.2) × 107 bacteria and a multiplicity of infection of 138:1 (see above). The epithelial cells were subsequently detached from the well by the addition of trypsin-EDTA and lysed by the addition of 300 μl of distilled water. The number of cell-adherent bacteria was determined by plating appropriate dilutions of the lysate onto THY agar plates. Due to the lysis of the eukaryotic cells in this approach, the calculation of cell-adherent bacteria also included bacteria that had invaded the host cells. Therefore, the number of invasive bacteria was subtracted from the obtained numbers of cell-adherent bacteria to calculate the actual number of adherent bacteria. To determine the number of adherent bacteria per eukaryotic cell, the number of cell-adherent bacteria was divided by the number of A549 cells per assay.
For invasion assays, the epithelial cells were infected with 5 × 106 streptococci, incubated for 2 h at 37°C, and washed three times with PBS. Subsequently, the infected cells were incubated for 2 h in tissue culture medium supplemented with penicilling G (10 U) and streptomycin (0.01 mg) to kill extracellular bacteria. After three washes with PBS, the epithelial cells were detached by the addition of trypsin-EDTA and lysed in 300 μl of distilled water. The amount of invasive bacteria was quantitated by plating serial dilutions of the lysate onto THY agar plates. The invasion index (13) was calculated as follows: (number of invasive/number of adherent bacteria) × 100%. Each experiment was repeated at least three times in triplicate.
Nucleotide sequence accession number.
The nucleotide sequence of the rogB encoding region from S. agalactiae was submitted to the EMBL nucleotide sequence database and was assigned accession no. AJ279088.
RESULTS
Isolation and characterization of the rogB gene from GBS.
In an approach to identify virulence-associated transcriptional regulators in GBS before the genome sequence of GBS had been published, RofA-like protein (RALP) sequences from different origins, including two newly identified RALPs in the unfinished genomic sequence of Streptococcus equi (http://www.sanger.ac.uk/) were aligned, resulting in the identification of two regions with high similarity, i.e., the region from amino acids 146 to 154 and from amino acids 256 to 260 in the RofA sequence from S. pyogenes (Fig. 1). Based on the two conserved peptide sequences, the degenerate primers rog1 (GAGTATCGWATWCGWTWYYT) and rog2 (ATAWATWARAAAWADRTARTC) were synthesized and used for PCR with chromosomal DNA from GBS, resulting in the amplification of a fragment of 350 bp. The PCR product was cloned in the pCR-TOPO vector, and the insert from one clone was subsequently sequenced. Analysis of the obtained sequence revealed an incomplete open reading frame (ORF); the deduced amino acid sequence of this ORF exhibited significant similarity to RALP sequences (not shown). This result indicated that GBS possesses a RALP that was termed regulator of group B streptococcus (RogB). To isolate the entire rogB gene from GBS, the 350-bp PCR product was used as a digoxigenin-labeled probe to screen a GBS cosmid library in E. coli. Of 723 cosmid clones, 6 hybridized to the 350-bp rogB probe. One of the cosmids was partially digested, and fragments ranging in size between 2 to 4 kb were ligated in the E. coli plasmid pUC18. The digoxigenin-labeled rogB probe was subsequently used to identify clones carrying the rogB gene in pUC18, and the 2.4-kb insert of one subclone was finally sequenced. Analysis of the obtained sequence revealed one complete ORF and one incomplete ORF (Fig. 2). Within the complete ORF the entire sequence of the rogB-specific 350-bp PCR product could be identified. This ORF was thus designated rogB. Inspection of the rogB gene identified two potential start codons: one at bp 373 (ATG) and the other at bp 398 (TTG). The translational start of the rogB gene is postulated to be located at bp 398 because of the presence of a reasonable ribosome-binding site (GAGAGAGA) in front of this start codon. Using bp 398 as the translational start, the rogB gene has a size of 1,427 bp and encodes a protein of 509 amino acids, with a predicted molecular mass of 60,261 Da. In front of the rogB gene, several A- and T-containing stretches could be identified that are typical for noncoding regions in GBS. The incomplete ORF extends from the end of the sequenced fragment at bp 2398 to bp 1980 and codes for a polypeptide of 137 amino acids. Computer-assisted database analysis of its deduced polypeptide revealed significant similarity to galactosyltransferases from other organisms. The respective ORF was therefore designated galT. Between the two ORFs, at positions 1926 to 1987, a potential rho-independent terminator structure (ΔG = −42.2 kcal) could be identified, indicating transcriptional termination of the two ORFs at this site.
FIG. 1.
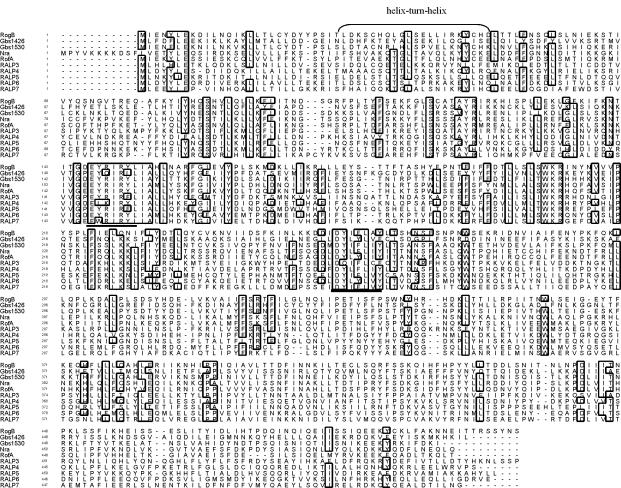
Alignment of the amino acid sequence of RogB from GBS with RofA, Nra, RALP3, and RALP4 from S. pyogenes, RALP5 from S. pneumoniae, RALP6 and RALP7 from S. equi, and the RALP encoding ORFs Gbs1426 and Gbs1530 in the genome of GBS NEM316. Identical amino acid residues between at least 7 of the 10 aligned proteins are boxed. The putative helix-turn-helix motif is indicated by a bracket. Underlined are two highly conserved regions within the aligned proteins that were used for the design of degenerate primers to isolate the rogB gene from GBS 6313.
FIG. 2.
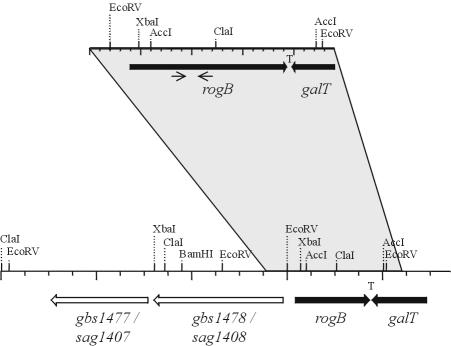
Restriction map of the rogB-encoding region in GBS. The upper restriction map displays the RogB-encoding region in GBS 6313, while the lower restriction map shows the respective region in the genomes of GBS NEM316 and GBS 2603 V/R, respectively. The shaded area marks identical regions in the genome of the different strains. Black arrows indicate the positions of the rogB (starting at bp 398) and the galT gene, and a “T” represents the proposed transcriptional terminator between the two genes. The light arrows below the rogB gene indicate the location of the primers used for amplification of the partial rogB sequence from GBS 6313. Open arrows represent the genes gbs1477/sag1407 and gbs1478/sag1408 in the genomes of GBS NEM316 and GBS 2603 V/R, respectively.
Analysis of the genome sequences of the meanwhile sequenced GBS strains NEM316 and 2603 R/V identified gene gbs1479 in the genome of strain NEM316 and gene sag1409 in the genome of strain 2603 V/R to encode the RogB protein. In both genomes the rogB gene is followed by an antiparallel-oriented gene, encoding a putative galactosyltransferase. Interestingly, the rogB gene is preceded in both genome sequences by two genes encoding putative cell surface proteins (Fig. 2). In the genome of GBS strain NEM316 these ORFs were designated Gbs1478 and Gbs1477, and in the genome of strain 2603 V/R these ORFs were termed Sag1408 and Sag1407. Although the exact function of these ORFs is unknown, their deduced polypeptides reveal similarity to fibronectin- and collagen-binding proteins from streptococci and staphylococci and, therefore, these ORFs represent putative virulence factors from GBS. For further analysis, we adapted the gene designation of GBS strain NEM316 for these genes, i.e., they were termed gbs1478 and gbs1477.
RogB is similar to other RALPs.
Basic local alignment search tool (BLAST) databank analysis of the deduced RogB protein revealed 50.2, 49.9, 36.2, and 24.8% identities to the RALP transcriptional regulators RofA, Nra, RALP3, and RALP4, respectively, from S. pyogenes; 25.0 and 24.9% identities to RALP6 and RALP7, respectively, from S. equi; and 22.9% identity to RALP5 from S. pneumoniae (Fig. 1). In the genome sequence of GBS NEM316 and GBS 2603 R/V, two additional putative RALP-encoding ORFs were identified. In the genome of GBS NEM316 the deduced amino acid sequence from the genes gbs1426 and gbs1530 exhibited 36.6 and 52.0% identities, respectively, to RogB. The deduced polypeptide from gene sag1356 in GBS 2603 R/V reveals 36.6% identity to RogB. In GBS 2603 R/V, the sag1463 gene is highly identical to gene gbs1530 from NEM316; however, sag1463 from 2603 R/V carries a nonsense mutation at bp 621, resulting in the permature termination of protein synthesis. The similarity of RogB to the above-listed proteins ranged from 42 to 84%. Using the method of Dodd and Egan (12), a putative helix-turn-helix DNA-binding motif could be identified in the N-terminal region of RogB (Fig. 1). The location of the helix-turn-helix motif in RogB corresponds to the respective motifs in the other RALPs (16, 19, 31). The homology data and the putative DNA-binding motif suggest that RogB is a transcriptional regulator in GBS.
Disruption of rogB impairs the binding of GBS to host proteins.
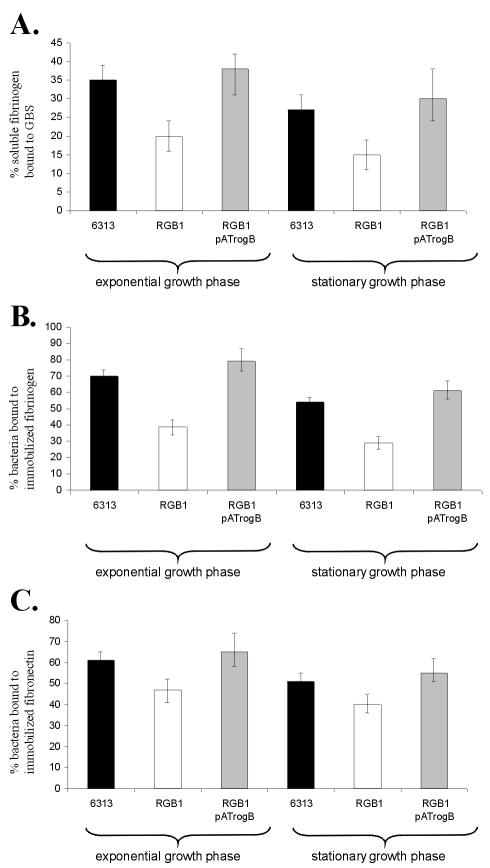
In S. pyogenes, the transcriptional regulators RofA and Nra control the interaction of the bacteria with extracellular matrix (ECM) proteins. To analyze the importance of RogB for the binding of GBS to human proteins, the rogB gene was insertionally inactivated in GBS 6313, resulting in GBS mutant RGB1. Southern blot analysis confirmed the successful disruption of the rogB gene in the chromosome of RGB1 (data not shown). Since the rogB gene is followed by a strong rho-independent terminator and the galT gene, which is oriented in opposite direction to rogB, disruption of rogB in mutant RGB1 does not exert a polar effect on the expression of downstream genes. No difference in the growth rate and final optical cell density between GBS RGB1 and its parental strain was observed (data not shown). To address the influence of RogB on the formation of putative adhesins in GBS, the ability of the GBS strains 6313 and RGB1 to bind to soluble or immobilized ECM proteins was quantitated. By using cells from the exponential (OD600 = 0.5) and the stationary (OD600 = 1.5) growth phases, the importance of the growth phase on the adhesive properties of GBS was analyzed. Both GBS strains revealed no binding of soluble or immobilized collagen I, collagen IV, or laminin and no binding of soluble fibronectin (not shown). However, GBS 6313 and GBS RGB1 accumulated soluble fibrinogen on their surface and bound to immobilized fibrinogen and fibronectin, respectively (Fig. 3). As shown in Fig. 3, both strains revealed a growth-phase dependency in their binding to fibrinogen and fibronectin, i.e., in exponentially growing cells the interaction with fibrinogen or fibronectin was increased by ca. 30 and 20%, respectively. However, compared to the parental strain, the GBS mutant RGB1 exhibited a 45% reduced binding to soluble and immobilized fibrinogen (Fig. 3A and B). Similarly, the binding of GBS mutant RGB1 to immobilized fibronectin was reduced by ca. 25% (Fig. 3C). The introduction of the vector pAT28 into the GBS strains 6313 or RGB1 had no influence on the different binding of the two strains to fibrinogen and fibronectin (data not shown). However, pAT28-mediated expression of rogB in strain RGB1 pATrogB restored its growth-phase-dependent binding to fibrinogen and fibronectin to the wild-type level (Fig. 3). These data indicate that binding of GBS to human fibrinogen and fibronectin is regulated by RogB and the growth phase of the bacteria.
FIG. 3.
Binding of soluble fibrinogen to GBS and attachment of GBS to immobilized fibrinogen and fibronectin, respectively. The assays were performed with GBS cells from the exponential and from the stationary growth phase. The ordinate represents the mean binding of total 125I-labeled soluble fibrinogen to the GBS strains 6313, RGB1, and RGB1 pATrogB, respectively (A), or the mean binding of the total bacteria of the GBS strains 6313, RGB1, or RGB1 pATrogB to immobilized fibrinogen (B) or fibronectin (C). Each assay was performed at least four times in triplicate.
RogB is required for efficient eukaryotic cell adherence.
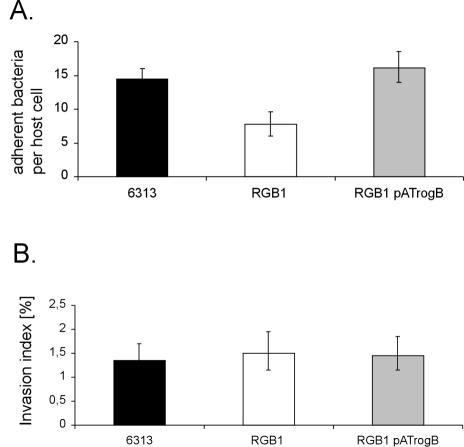
A decreased interaction of a microorganism with ECM proteins often impairs its capability to adhere to and to invade host cells. Therefore, the GBS strains 6313, RGB1, and RGB1 pATrogB were tested for their adherence to and invasion of the human epithelial cell line A549. Adherent and invasive bacteria were quantitated by plate viability counts. As depicted in Fig. 4A, the GBS wild-type strain 6313 revealed an adherence of 14.8 ± 1.1 bacteria per A549 cell. In contrast, the adherence of mutant RGB1 was reduced by 46% compared to the wild-type strain. However, plasmid-mediated expression of rogB in GBS strain RGB1 pATrogB restored its adherence to A549 cells to the wild-type level (Fig. 4A). These data suggest that the rogB gene is involved in the binding of GBS to epithelial cells.
FIG. 4.
Eukaryotic cell adherence of the GBS strains 6313, RGB1, and RGB1 pATrogB and determination of the invasion indices of these strains. (A) The adherence of the different strains to A549 cells is given as number of adherent bacteria per eukaryotic cell. (B) The invasion index was obtained by relating the number of invasive bacteria to the number of adherent bacteria. Each experiment was performed at least three times in triplicate.
To compare the invasion of the different GBS strains into A549 cells, the number of invasive bacteria of each strain was related to the number of adherent bacteria, resulting in the calculation of the invasion index (13). As depicted in Fig. 4B, GBS strains 6313, RGB1, and RGB1 pATrogB revealed similar invasion indices, indicating that rogB is not involved in the invasion of epithelial cells by GBS.
Expression profiling of virulence genes in GBS 6313 and GBS RGB1.
The previous results suggested a significant effect of RogB on virulence mechanisms in GBS. Since in S. pyogenes members of the RALP family control a variety of virulence genes at the transcriptional level, the expression profile of important virulence genes was analyzed by real-time PCR in GBS RGB1 and its parental strain 6313. Expression profiling was performed in a LightCycler with the cfb, lmb, sodA, cpsA, hylB, scpB, lytR, gyrA, rogB, and fbsA genes encoding CAMP factor, the laminin-binding protein Lmb, superoxide dismutase, an activator of capsule gene expression, hyaluronate lyase, C5a peptidase, an autolysin response regulator, gyrase subunit A, RogB regulator protein and fibrinogen receptor FbsA, respectively. Also compared in the two GBS strains was the expression profile of the genes gbs1478 and gbs1477, preceding rogB, and encoding putative virulence factors from GBS. Equal amounts of total RNA from exponential-phase (OD600 = 0.3) cultures of GBS 6313 and GBS RGB1 were reverse transcribed and used to quantitate the transcript levels of the above-mentioned genes by real-time PCR. The obtained data were normalized to the expression of the gyrA gene in the two strains. A twofold difference in transcription was interpreted as a significant difference in expression between the two strains. Relative to GBS 6313, transcription of the genes cfb, lmb, sodA, hylB, scpB, and lytR was unaltered in the rogB mutant RGB1. However, expression of the cpsA gene was (4.78 ± 0.55)-fold increased in mutant RGB1 compared to its parental strain. In contrast, the transcription levels of the genes rogB, fbsA, gbs1478, and gbs1477 were (3.29 ± 0.51)-, (3.13 ± 0.34)-, (3.35 ± 0.47)-, and (2.74 ± 0.49)-fold higher in GBS 6313 than in GBS RGB1. This result suggests that RogB exerts a negative effect on the transcription of cpsA in GBS and that it activates the expression of the adjacent genes gbs1478 and gbs1477, and that of fbsA, encoding a fibrinogen receptor from GBS. In addition, the rogB gene appears to be autoregulated by the RogB protein. However, disruption of rogB may also decrease the stability of the truncated rogB transcript.
Plasmid-mediated expression of rogB in strain RGB1 pATrogB resulted in (0.76 ± 0.44)-, (0.18 ± 0.36)-, and (3.23 ± 0.59)-fold-increased expression of the genes rogB, fbsA, and gbs1477, respectively, compared to the GBS wild-type strain 6313. The expression of the genes gbs1478 and cpsA was reduced (0.62 ± 21) and (0.13 ± 0.67)-fold, respectively, in strain RGB1 pATrogB compared to the wild-type strain. This finding shows that in strain RGB1 pATrogB the expression of the genes rogB, fbsA, gbs1478, and cpsA is restored to about the wild-type level. However, plasmid-mediated expression of rogB appears to result in an elevated expression of the gene gbs1477.
Expression of fbsA is controlled by RogB and the growth phase of GBS.
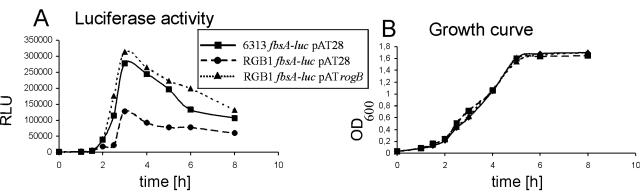
Among the genes that were shown to be under transcriptional control of RogB, the fbsA gene represents the best-studied virulence factor from GBS. Therefore, the influence of RogB on fbsA expression was characterized in more detail by transcriptionally fusing the fbsA gene in the chromosome of GBS with a promotorless luciferase gene. After two recombination events, a luciferase cassette without vector sequences was placed behind the fbsA gene in the chromosome of GBS 6313. In the resultant GBS strain, 6313 fbsA-luc, expression of fbsA can be directly quantitated by measuring its luciferase activity. Correct insertion of the reporter gene into the genome of GBS 6313 was confirmed by Southern blot hybridization (data not shown). To analyze the importance of rogB in controlling the expression of fbsA in GBS, the rogB gene was insertionally inactivated in the chromosome of GBS 6313 fbsA-luc, and the mutant strain was termed RGB1 fbsA-luc. Southern blot analysis confirmed the successful disruption of the rogB gene in the chromosome of mutant RGB1 fbsA-luc (results not shown). For complementation studies, the GBS strains 6313 fbsA-luc and RGB1 fbsA-luc were transformed with the vector pAT28, and strain RGB1 fbsA-luc was transformed with the rogB-carrying plasmid pATrogB. The synthesis of luciferase was subsequently measured in the resultant GBS strains during growth of the bacteria in complex media, and the results were plotted as RLU and OD600, respectively, against time. As shown in Fig. 5B, the three strains exhibited identical growth behavior in complex medium. In all three strains, transcription of fbsA increased significantly during the early exponential growth phase, peaked in the middle of exponential growth, and decreased moderately at the transition from the exponential to the stationary growth phase (Fig. 5A). However, expression of fbsA in mutant RGB1 fbsA-luc pAT28 was on average 50% lower than in strain 6313 fbsA-luc pAT28. Plasmid-mediated expression of rogB in strain RGB1 fbsA-luc pATrogB increased the fbsA transcription to values comparable to those of the wild-type strain 6313 fbsA-luc pAT28. These findings confirm our results obtained by real-time PCR and show that the presence of RogB stimulates the transcription of the fbsA gene in GBS.
FIG. 5.
Growth curve and expression profile of an fbsA-luciferase transcriptional fusion in the GBS strains 6313 fbsA-luc pAT28, RGB1 fbsA-luc pAT28, and RGB1 fbsA-luc pATrogB. The bacteria were grown aerobically in THY liquid medium, and at different time points samples were withdrawn for the determination of the luciferase activity (A) and the OD600 (B) of the culture. Luciferase activity is expressed in RLU.
DISCUSSION
GBS adapts to and survives at different locations within the human host by sensing the changing surroundings and regulating the expression of virulence genes in response to environmental signals. In the present study, a putative regulatory protein, RogB, was identified and characterized that reveals significant similarity to members of the RALP family of transcriptional regulators (19), among which RofA and Nra from S. pyogenes have been extensively studied at the molecular level (2, 15, 16, 25, 28). Insertional inactivation of rogB in the chromosome of GBS resulted in a reduced binding of the bacteria to soluble fibrinogen and to immobilized fibrinogen and fibronectin, respectively. This indicates that RogB stimulates the attachment of GBS to ECM and plasma proteins. Also, the regulator RofA from S. pyogenes has been shown to enhance the binding of the bacteria to human fibronectin and fibrinogen (15, 16, 25). However, the Nra protein acts predominantly as a repressor of virulence gene transcription and downregulates binding of S. pyogenes to type I collagen and fibronectin (31).
The interaction of bacteria with ECM proteins is frequently a prerequisite for the successful colonization of the human host. Interestingly, GBS mutant RGB1 was significantly impaired in its attachment to the human epithelial cell line A549. Also, the disruption of rofA in S. pyogenes M6 decreased the ability of the bacteria to attach to epithelial cells (2), whereas the inactivation of nra in S. pyogenes M49 resulted in an increased binding of the mutant to host cells (28). In S. pyogenes, binding of the bacteria to fibronectin has been shown to mediate bacterial adherence to and invasion into eukaryotic cells (29, 30, 44). Since RofA stimulates and Nra downregulates fibronectin binding in S. pyogenes, the different adherence properties of the rofA and nra mutants were attributed to alterations in their ability to interact with human fibronectin (2, 28). Recently, fibronectin binding of GBS was shown not to play a role in the adherence of the bacteria to epithelial cells (7). This indicates that the decreased adherence of GBS mutant RGB1 to epithelial cells was not caused by its reduced binding to human fibronectin. Although binding of GBS to fibrinogen has been shown to protect the bacteria against opsonophagocytosis (8, 39), the interaction of GBS with fibrinogen may also play a role in the adherence to epithelial cells. It can therefore be speculated that the impaired binding of GBS RGB1 to human fibrinogen resulted in the reduced adherence to human cells. Alternatively, RogB may stimulate in GBS the synthesis of further adhesins, which have not yet been identified.
Real-time PCR analysis revealed a RogB-dependent effect on the expression of known and putative virulence genes in GBS. The presence of rogB was shown to stimulate the expression of the genes gbs1478 and gbs1477, which are located upstream of rogB in antiparallel orientation, and to exert a positive effect on transcription or RNA stability of the rogB gene. In S. pyogenes the RALP-like transcriptional regulators RofA and Nra have been shown to be autoregulated and to control the expression of their upstream located genes cpa and prtF, respectively (2, 15, 16, 19, 31). The gene products of the genes prtF and cpa, encoding the fibronectin-binding protein F and the collagen-binding protein Cpa, represent important virulence factors in S. pyogenes. In analogy to the similar genetic organization and regulation of the nra/rofA region in S. pyogenes and that of rogB in GBS, the genes gbs1478 and gbs1477 represent interesting putative GBS virulence genes, whose role for the virulence of the bacteria is currently under investigation.
C5a peptidase is an important virulence factor on the surface of GBS. This protease specifically cleaves the chemotactic complement component C5a, thereby interfering with the recruitment of granulocytes to the site of infection (11, 52). Recently, C5a peptidase from GBS was shown to mediate binding of the bacteria to immobilized fibronectin (3) and to promote internalization of the bacteria into host cells (7). Although disruption of rogB resulted in a reduced binding of mutant RGB1 to human fibronectin, real-time PCR revealed no effect of RogB on the expression of the C5a peptidase encoding gene scpB in GBS, and there was no difference between the internalization of mutant RGB1 and its parental strain in eukaryotic cells. These findings suggest that RogB controls in GBS 6313 the synthesis of a fibronectin-binding protein that is distinct from C5a peptidase. The presence of several fibronectin-binding proteins has already been suggested by Beckmann et al. (3).
Recently, it was demonstrated that the FbsA protein represents the major fibrinogen receptor in GBS 6313 (39). The reduced binding of mutant RGB1 to human fibrinogen suggests that it is impaired in the synthesis of the FbsA protein. We were able to demonstrate, by real-time PCR and by reporter gene studies, that the disruption of rogB indeed decreased the expression of fbsA in mutant RGB1 by ca. 50%. These findings indicate that RogB has a prominent effect on the expression of fbsA in GBS.
Spellerberg et al. (42) described for the GBS strain O90R a putative quorum-sensing system that consists of the genes rgfBDAC. Disruption of rgfC in GBS O90R caused an altered fibrinogen binding of the mutant depending on the bacterial cell density. Since the fibrinogen binding of GBS O90R is exclusively mediated by FbsA (39; unpublished results), it can be speculated that in GBS O90R the fbsA gene is under the transcriptional control of the rgfBDAC quorum-sensing system. Interestingly, neither GBS 6313 (B. Spellerberg, unpublished data) nor GBS NEM316 (17) carry a functional rgfBDAC gene locus. This suggests that fbsA expression is controlled by several regulatory circuits depending on the genetic background of the GBS strain.
In summary, we have identified and characterized a novel regulatory gene from GBS, termed rogB, which is involved in the expresssion control of known and putative virulence genes in these bacteria. Like other members of the RALP family of transcriptional regulators, RogB appears to regulate the interaction of GBS with its human host. Understanding the mode of action of RogB in GBS will contribute significantly to unravel the virulence mechanisms employed by GBS at different sites in the human body.
Acknowledgments
We thank S. Sauter for excellent technical assistance.
H. Gutekunst obtained a fellowship from the Landesgraduiertenförderung of Baden-Württemberg. This study was supported as project H1 by the Interdisziplinäre Zentrum für Klinische Forschung, Ulm, Germany.
Editor: V. J. DiRita
REFERENCES
- 1.Baker, C. J., and M. S. Edwards. 1995. Group B streptococcal infections, p. 980-1054. In J. S. Remington and J. O. Klein (ed.), Infectious disease of the fetus and newborn infant. The W. B. Saunders Co., Philadelphia, Pa.
- 2.Beckert, S., B. Kreikemeyer, and A. Podbielski. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect. Immun. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckmann, C., J. D. Waggoner, T. O. Harris, G. S. Tamura, and C. E. Rubens. 2002. Identification of novel adhesins from group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect. Immun. 70:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, K. M., C. J. Baker, and M. S. Edwards. 1987. Interaction of soluble fibronectin with group B streptococci. Infect. Immun. 55:2404-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caparon, M. G., D. S. Stephens, A. Olsen, and J. R. Scott. 1991. Role of M protein in adherence of group A streptococci. Infect. Immun. 59:1811-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, Q., D. Stafslien, S. S. Purushothaman, and P. Cleary. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chhatwal, G. S., I. S. Dutra, and H. Blöbel. 1985. Fibrinogen binding inhibits the fixation of the third component of human complement on surface of groups A, B, C, and G streptococci. Microbiol. Immunol. 29:973-980. [DOI] [PubMed] [Google Scholar]
- 9.Chhatwal, G. S., H. P. Muller, and H. Blöbel. 1983. Characterization of binding of human α2-macroglobulin to group G streptococci. Infect. Immun. 41:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 11.Cleary, P. P., J. Handley, A. N. Suvorov, A. Podbielski, and P. Ferrieri. 1992. Similarity between the group B and A streptococcal C5a peptidase genes. Infect. Immun. 60:4239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-419. [DOI] [PubMed] [Google Scholar]
- 14.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fogg, G. C., and M. G. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogg, G. C., C. M. Gibson, and M. G. Caparon. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of a gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 11:671-684. [DOI] [PubMed] [Google Scholar]
- 17.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 18.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1985. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 21.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath, A., A. Miller, V. J. DiRita, and C. N. Engleberg. 2001. Identification of a major, CsrRS-regulated secreted protein of group A streptococcus. Microb. Pathog. 31:81-89. [DOI] [PubMed] [Google Scholar]
- 23.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46:87-99. [DOI] [PubMed] [Google Scholar]
- 24.Hunter, W. H., and F. C. Greenwood. 1962. Preparation of iodine-131 labeled human growth hormone of high specific activity. Nature 194:495-496. [DOI] [PubMed] [Google Scholar]
- 25.Kreikemeyer, B., S. Beckert, A. Braun-Kiewnick, and A. Podbielski. 2002. Group A streptococcal RofA-type global regulators exhibit a strain-specific genomic presence and regulation pattern. Microbiology 148:1501-1511. [DOI] [PubMed] [Google Scholar]
- 26.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 27.Maguin, E., H. Prevost, S. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molinari, G., M. Rohde, S. R. Talay, G. S. Chhatwal, S. Beckert, and A. Podbielski. 2001. The role played by the group A streptococcal negative regulator Nra on bacterial interactions with epithelial cells. Mol. Microbiol. 40:99-114. [DOI] [PubMed] [Google Scholar]
- 29.Molinari, G., S. R. Talay, P. Valentin-Weigand, M. Rohde, and G. S. Chhatwal. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 65:1357-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada, N., M. Watarai, V. Ozeri, E. Hanski, M. Caparon, and C. Sasakawa. 1997. A matrix form of fibronectin mediates enhanced binding of Streptococcus pyogenes to host tissue. J. Biol. Chem. 272:26978-26984. [DOI] [PubMed] [Google Scholar]
- 31.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 32.Podbielski, A., M. Woischnik, B. Pohl, and K. H. Schmidt. 1996. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med. Microbiol. Immunol. 185:171-181. [DOI] [PubMed] [Google Scholar]
- 33.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 34.Poyart, C., M. C. Lamy, C. Boumaila, F. Fiedler, and P. Trieu-Cuot. 2001. Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J. Bacteriol. 183:6324-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricci, M. L., R. Manganelli, C. Berneri, G. Orefici, and G. Pozzi. 1994. Electrotransformation of Streptococcus agalactiae with plasmid DNA. FEMS Microbiol. Lett. 119:47-52. [DOI] [PubMed] [Google Scholar]
- 37.Rubens, C. E., S. Smith, M. Hulse, E. Y. Chi, and G. van Belle. 1992. Respiratory epithelial cell invasion by group B streptococci. Infect. Immun. 60:5157-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and J. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Schubert, A., K. Zakikhany, M. Schreiner, R. Frank, B. Spellerberg, B. E. Eikmanns, and D. J. Reinscheid. 2002. A fibrinogen receptor from group B streptococcus interacts with fibrinogen by repetitive units with novel ligand binding sites. Mol. Microbiol. 46:557-569. [DOI] [PubMed] [Google Scholar]
- 40.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 42.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, and R. Lütticken. 2002. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect. Immun. 70:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, N. Schnitzler, R. Lütticken, and A. Podbielski. 1999. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talay, S. R., A. Zock, M. Rohde, G. Molinari, M. Oggioni, G. Pozzi, C. A. Guzman, and G. S. Chhatwal. 2000. Co-operative binding of human fibronectin to Sfbl protein triggers streptococcal invasion into respiratory epithelial cells. Cell Microbiol. 2:521-535. [DOI] [PubMed] [Google Scholar]
- 45.Tamura, G. S., and C. E. Rubens. 1995. Group B streptococci adhere to a variant of fibronectin attached to a solid phase. Mol. Microbiol. 15:581-589. [DOI] [PubMed] [Google Scholar]
- 46.Teng, F., B. E. Murray, and G. M. Weinstock. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid 39:182-186. [DOI] [PubMed] [Google Scholar]
- 47.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed]
- 49.Valentin-Weigand, P., P. Benkel, M. Rohde, and G. S. Chhatwal. 1996. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 64:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 51.Waite, D. C., E. J. Alper, and B. J. Mady. 1996. Adult group B streptococcal disease. Ann. Intern. Med. 125:152-153. [DOI] [PubMed] [Google Scholar]
- 52.Wexler, D. E., D. E. Chenoweth, and P. P. Cleary. 1985. Mechanism of action of the group A streptococcal C5a inactivator. Proc. Natl. Acad. Sci. USA 82:8144-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zangwill, K. M., A. Schuchat, and J. D. Wenger. 1992. Group B streptococcal disease in the United States, 1990: report from a multistate active surveillance system. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 41:25-32. [PubMed] [Google Scholar]