Abstract
To find genes involved in natural competence in Helicobacter pylori, we used a bioinformatics database search and found two transformation-related open reading frames (ORFs): a comE3 homologue (HP1361 ORF) of Bacillus subtilis and a comL homologue (HP1378 ORF) of Neisseria gonorrhoeae. We failed to obtain an HP1378 ORF knockout mutant, while an HP1361 ORF knockout mutant was obtained by transposon shuttle mutagenesis. The DNA transformation abilities of both natural transformation and electroporation were severely impaired (frequency, <10−9) in the HP1361− mutant. Complementation with a pHel2 vector carrying the HP1361 ORF restored the capabilities of natural competence (to a frequency of 4.21 × 10−7) and electroporation (to 3.62 × 10−7). The HP1361− mutant showed impairment in DNA binding and uptake. The results suggest that HP1361 is a comE3 homologue and is required for DNA binding and uptake during DNA transformation.
Most Helicobacter pylori strains are naturally competent. Gene transfer between H. pylori strains is extremely common and causes a high degree of diversity (21). Besides contributing to the development of genetic diversity, natural competence in H. pylori is also helpful in the adaptation of H. pylori to changing environments and may shed light on the clinically important issues of virulence and the development of antibiotic resistance (4, 13). Resistance to metronidazole, a key component of widely used combination regimens for H. pylori eradication, was recently shown to be associated with natural competence (24). To study genes involved in natural transformation, we searched two complete genome sequences of H. pylori and found two transformation-related open reading frames (ORFs): a comE3 homologue (HP1361 ORF) of Bacillus subtilis and a comL homologue (HP1378 ORF) of Neisseria gonorrhoeae (1, 22). We tried to study these two ORFs by knockout analysis and complementation. However, only the HP1361 mutant could be obtained; we therefore tried to characterize the HP1361 gene and its role in the natural transformation of H. pylori.
The bacterial strains used in this study are listed in Table 1. H. pylori cells grown on Columbia blood agar plates for 48 h were harvested for subsequent experiments. The HP1361 ORF was amplified by PCR from a naturally competent clinical isolate (NTUH-C1) by using a primer pair, comE3f (5′-GGGTGTGTGGGGTGTTTTTAAGCCTTT-3′) and comE3r (5′-ACCTACAATTAAGCTAGGCATTATAAT-3′); the HP1361 ORF was then ligated into a pILL570 vector. This ORF had a full length of 1,251 bp encoding 417 amino acids. The transposon miniTnKm was transposed into the cloned HP1361 ORF fragments in Escherichia coli HB101 by two consecutive conjugation steps as described previously (17). After natural transformation and kanamycin selection, we obtained the HP1361 knockout mutant NTUH-C1-Em. The insertion of the miniTnKm cassette within the HP1361 ORF was confirmed by PCR using primer pair comE3f and comE3r and primer pair KmCf (5′-ATATCYCGRGGATAAACCCAGCGAACCATT-3′) and KmCr (5′-TATACYCGRGCTCGACATACTGTTCTTCCC-3′).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic(s) | Reference or source |
|---|---|---|
| H. pylori strains | 24 | |
| NTUH-C1 to NTUH-C15 | Wild type, naturally competent | 24 |
| NTUH-I1 to NTUH-I8 | Wild type, noncompetent | 24 |
| NTUH-C1-Em | NTUH-C1/HP1361::miniTnkm | This study |
| NTUH-C1pC1 | NTUH-C1 carrying pC1 | This study |
| NTUH-C1-EmpC1 | NTUH-C1/HP1361::miniTnkm carrying pC1 | This study |
| E. coli strains | ||
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15lacX74 recA1 deoR araD139Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| HB101 | hsdR hsdM recA supE44 leuZ4 leuB6 proA2 thi-1Sm | 17 |
| NS2114 | recA Sm100; contains a λ-cre prophage, Rifr F − | 17 |
| DH1 | F−supE44 recA1 endA1 gurA96 thi-1 hsdR17 T (rK− mK+) relA1 | 17 |
| DH5α | endA1 hsdR17 (rk− mk+) supE44 thi-1 recA gyrA (Nalr) relA1Δ(argF-lacZYA)U169 deoR[φ80dlacΔ(lacZ) M15] | Promega |
| Plasmids | ||
| pC1 | pHel2 containing Pure-HP1361 | This study |
| pCRII-TOPO | ColE1 Ampr Kmr; PCR cloning vector | Invitrogen |
| pILL570 | ReppBR322 mob Spr | 17 |
| pTCA | ReppACYC184 Tc TnpA Imm Tn3 | 17 |
| pILL553 | pox38::TnKm | 17 |
| pHel2 | E. coli-H. pylori shuttle, Cat resistance | 12 |
Natural transformation and electroporation were performed as previously described (5). To determine the transformation frequency, a chloramphenicol acetyltransferase (CAT) cassette (a gift from D. E. Taylor) inserted with the H. pylori gene yxjD (23); a 23S rRNA gene from a clarithromycin-resistant strain, with an A-to-G mutation at nucleotide 2143 (15), in a pCR2.1 vector (Invitrogen, Carlsbad, Calif.); and an E. coli-H. pylori shuttle vector, pHel2 (a gift from R. Haas) (12), which carries the CAT gene, were used as DNA donors. The transformation frequency was defined as the number of resistant colonies divided by the total number of viable bacteria.
PCR products amplified by the primer pair comE3f and comE3r were used to determine the sequences of the HP1361 ORF in clinical isolates. Inverse PCR was used for the determination of the miniTnKm insertion site. Chromosomal DNA from NTUH-C1-Em was extracted, subsequently completely digested with restriction endonuclease HindIII, and self-ligated by T4 DNA ligase. Primers KmCf and KmCr were used for inverse PCR; sequences were determined by using primer Km-seq3 (5′-TGGTAACTGTCAGACCAAGTTTACTC-3′). Automatic sequencing was performed with an ABI PRISM 377 genetic analyzer (Applied Biosystems) as described previously (5).
Total RNAs of H. pylori cells were extracted as described previously (3). Ten micrograms of total RNA from each H. pylori isolate was transferred onto a nylon membrane (Roche Diagnostics GmbH, Mannheim, Germany) by using a slot blot system (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom). The membranes were prehybridized, hybridized with RNA probes, and detected with a digoxigenin (DIG) luminescence detection kit (Roche Diagnostics). Antisense 23S rRNA and antisense HP1361 RNA probes were transcribed in vitro by using cloned genes in a pCRII-TOPO vector and were labeled with DIG by a DIG RNA labeling kit (SP6/T7) (Roche Diagnostics). Densitometry was analyzed with an NIH Image version 1.62 software program by using 23S rRNA as the internal standard (3).
For complementation of HP1361 ORF, a promoter of the urease operon (HP0073 to HP0072) (Pure) and a DNA fragment containing the HP1361 ORF, its upstream 233 bp, and its downstream 108 bp were amplified by using PCR primer pair Puref-NotI (5′-GCGGCCGCTCAGTTGGTAGAGCAC-3′) and Purer-XhoI (5′-CTCGAGCTTATTCTCCTATTCTTA-3′) and primer pair HP1362f-XhoI (5′-GAACTCGAGGCTGAAATCATTGTGGCT-3′) and HP1360r-XbaI (5′-ATGTCTAGAAAATATCGTATGCTCC-3′), respectively. These two DNA fragments were ligated into a pCRII-TOPO vector sequentially by using the unique restriction enzyme sites NotI, XhoI, and XhoI and XbaI, respectively. The Pure-HP1361 fragment was amplified by using a primer pair, Puref-NotI and HP1360r-XbaI, cloned into a pCRII-TOPO vector by another TA cloning procedure, excised from the plasmid, and finally ligated into the pHel2 vector by using EcoRV sites. This transcomplementation plasmid was named pC1. pC1 was introduced into NTUH-C1 by natural transformation and resulted in NTUH-C1pC1. Chromosomal DNA from NTUH-C1-Em was transformed into NTUH-C1pC1, and selection of chromosomal HP1361 knockout strains was performed to obtain the complementation strain NTUH-C1-EmpC1.
DNA binding and uptake assays were performed as described previously (6) with minor modifications. A 294-bp DNA fragment was labeled with [α-32P]dCTP by PCR using primer pair Puref-NotI and Purer-XhoI. The purified PCR product had a specific activity of 1.4 × 107 cpm/μg. Twenty-five microliters of the bacterial suspension (corresponding to 108 CFU) was mixed with 5 ng of a 32P-labeled DNA fragment and incubated on plates for 24 h. The cells were then scraped and resuspended in 200 μl of phosphate-buffered saline (PBS). For uptake assays, the samples were treated with 100 μg of DNase I for 1 h at room temperature in order to degrade exogenous DNA. The cells were washed with PBS three times and resuspended in 25 μl of PBS. Each sample was added to 3 ml of scintillation liquid, and cell-associated radioactivity was measured on a scintillation counter. A total of six experiments were performed for each uptake and binding assay.
HP1361 knockout mutant NTUH-C1-Em was obtained by transposon mutagenesis. DNA sequencing of NTUH-C1-Em showed that the miniTnKm cassette was inserted between nucleotides 1095 and 1096 of the HP1361 gene. The natural transformation capability of NTUH-C1-Em was profoundly decreased (no colony on selective medium, i.e., a frequency of <10−9) (Table 2). Because the knockout of the HP1361 ORF abolishes the transferring DNA by either natural transformation or electroporation, the complementation should be done by adding a transcomplementation plasmid prior to knocking out the wild-type HP1361 gene. The transformation frequency in NTUH-C1pC1, a wild-type NTUH-C1 strain carrying plasmid pC1, decreased from 10−4 to 2.42 × 10−7 compared with that of NTUH-C1 (Table 2). The complementation strain NTUH-C1-EmpC1 restored the natural transformation frequency to 4.21 × 10−7; although lower than that for NTUH-C1, it was almost the same as that for NTUH-C1pC1 (Table 2). The transformation frequencies of electroporation for NTUH-C1, NTUH-C1pC1, and NTUH-C1-EmpC1 were similar to the frequencies of natural transformation (Table 2). These results suggested that the HP1361 ORF was a comE3 homologue and was essential to DNA transformation in H. pylori.
TABLE 2.
Transformation frequencies of NTUH-C1 and its derivatives
| Method | Donor DNAe | Transformation frequency
for indicated recipient of the H. pylori
straina
|
|||
|---|---|---|---|---|---|
| NTUH-C1 | NTUH-C1-Emd | NTUH-C1pC1 | NTUH-C1-EmpC1 | ||
| Natural transformation | CM resistance cassetteb | 10−4 | <10−9 | ND | ND |
| CLA resistance cassettec | 10−4 | <10−9 | 2.42 × 10−7 | 4.21 × 10−7 | |
| pHel2d | 10−4 | <10−9 | ND | ND | |
| Electroporation | CM resistance cassette | 10−4 | <10−9 | ND | ND |
| CLA resistance cassette | 10−4 | <10−9 | 3.40 × 10−7 | 3.62 × 10−7 | |
| pHel2 | 10−4 | <10−9 | ND | ND | |
Where the frequency was <10−9, no colony grew on selective medium. ND, not done.
Chloramphenicol acetyltransferase expression cassette inserted with the H. pylori gene yxjD (HP0691).
23S rRNA gene from a clarithromycin-resistant strain (with an A-to-G mutation at nucleotide 2143) in a pCR2.1 vector.
E. coli-H. pylori shuttle vector pHel2.
CM, chloramphemicol; CLA, clarithromycin.
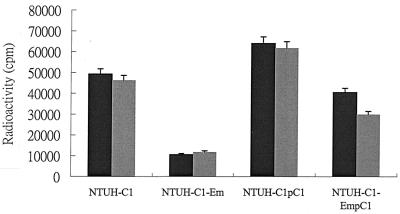
We could not obtain any transformant by natural transformation or by electroporation in HP1361 knockout mutant NTUH-C1-Em, even when plasmid pHel2 was used as the DNA donor (Table 2). This result indicated that knockout of the HP1361 ORF may interfere with DNA binding or uptake. The results of DNA binding and uptake assays showed that the HP1361 ORF is important for DNA uptake but is also involved in extracellular DNA binding. When the HP1361 ORF was knocked out, DNA uptake ability dropped to 21.3% and binding ability dropped to 25.4% compared to those of the wild-type strain (Fig. 1). . NTUH-C1-EmpC1 restored partial binding and uptake abilities to 82.1% (uptake) and 64.6% (binding).
FIG. 1.
Results of DNA binding and uptake assays. The values are the means ± the standard deviations of cell-associated radioactivity values from six experiments. The black columns show the results of the DNA binding assay, and the gray columns show the results of the DNA uptake assay.
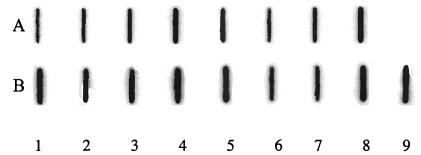
There are two H. pylori 26695 HP1361 homologues which are reported to associate with natural transformation: ComEC of B. subtilis (GenBank accession number CAB14499.1) and Rec2 of Haemophilus influenzae (GenBank accession number AAC21739.1). A comparison of these amino acid sequences showed that HP1361 of NTUH-C1 shared 22.90% similarity to the ComEC N terminus (34 of 495 amino acids) and 21.97% similarity to the Rec2 N terminus (51 of 534 amino acids) (Fig. 2). . We randomly chose 23 clinical isolates for PCR analysis; 15 were naturally competent and 8 were noncompetent. All of these strains were positive by PCR using primer pair comE3f and comE3r. Comparison of amino acid sequences from the DNA sequencing data of four naturally competent (NTUH-C1, NTUH-C2, NTUH-C3, and NTUH-C4) and four noncompetent (NTUH-I1, NTUH-I2, NTUH-I3, and NTUH-I4) clinical isolates revealed 95 to 98% similarity to H. pylori 26695 HP1361 and 94 to 98% similarity within these clinical isolates. There is no significant difference between these two groups (data not shown). RNA slot blot analysis revealed that HP1361 mRNA was expressed in all 23 strains; however, there was no difference in RNA expression between 15 competent and 8 noncompetent strains (Fig. 3). .
FIG. 2.
Multiple alignments of amino acid sequences sharing homology with NTUH-C1 HP1361. The sequences are from H. pylori 26695 HP1361 (26695HP1361; GenBank accession number AAD08401.1), H. pylori J99 jhp1279 (J99jhp1279; accession number AAD06851.1), B. subtilis 168 ComEC (B. sub comEC; accession number CAB14499.1), and H. influenzae KW20 Rec2 (H. inf rec2;accession number AAC21739.1).
FIG. 3.
RNA slot blot results for HP1361 from competent and noncompetent strains with labeled antisense HP1361 RNA (A) or labeled antisense 23SrRNA (B) as the probe. RNA expression levels were measured by densitometry with rRNA as the internal control. Lanes 1 to 4, RNA from naturally competent H. pylori strains NTUH-C1, NTUH-C2, NTUH-C3, and NTUH-C4; lanes 5 to 8, RNA from noncompetent H. pylori strains NTUH-I1, NTUH-I2, NTUH-I3, and NTUH-I4; lane 9, RNA from a comE3 deletion mutant. comE3 and its flanking regions were cloned into a plasmid. After inverse PCR and transformation, the mutant was obtained by replacing comE3 with a CAT gene (5, 23).
H. pylori is one of the most diverse bacterial species. New genotypes of H. pylori are generated by recombination that is fast enough to essentially eliminate the effect of clonal descent on the population structure (20). One of the mechanisms of horizontal gene transfer used by H. pylori is natural transformation. HP0333, comB, comH, a virB4 homologue, and the type IV secretion system of H. pylori were reported to be associated with natural transformation (2, 5, 14, 18, 19). The process of natural transformation has been well studied in model organisms such as B. subtilis, Streptococcus pneumoniae, H. influenzae, and N. gonorrhoeae (7, 8, 9, 10). First, extracellular DNA binds the surfaces of bacteria; next, the DNA is usually restricted into fragments of a suitable length and then transported into the cytoplasm through a channel or translocator protein. In the model for natural transformation in H. pylori put forward by Smeets and Kusters (20), ComB proteins may form a pore-like complex for DNA translocation, but how the proteins perform the DNA processing before transport through the cell wall is still unclear.
In B. subtilis, ComE3 is involved in natural transformation and is specifically required for DNA uptake (11). Although there are no experimental data to demonstrate associated ATPase activities, ComE3 is predicted to be a DNA translocator (11). In this study, we demonstrated that the comE3 homologue, the HP1361 ORF, is also essential for natural transformation in H. pylori. Knockout of this gene resulted in the abolishment of the ability of natural transformation (frequency, <10−9). Significantly, DNA binding and uptake assays revealed that the HP1361 ORF played an important role in these two processes. This finding may aid in understanding the early steps of Smeets and Kusters's transformation model (20). Using the GES hydrophobicity scale data from The Institute for Genomic Research (http://www.tigr.org), HP1361 was predicted to be a transmembrane protein. This was in agreement with the prediction that HP1361 was a membrane- or periplasm-associated protein. Like ComE3 of B. subtilis, it may function as a DNA translocator (11).
Transcomplementation was done by adding a pHel2 plasmid containing Pure-HP1361 and then by adding knockout wild-type HP1361. The reasons for using the urease promoter were (i) that no definite promoter upstream of the HP1361 ORF was identified and (ii) that Pure is constitutively expressed in H. pylori. Transformation frequency was only partially restored in the complementation strain NTUH-C1-EmpC1. Nevertheless, in NTUH-C1pC1, the wild-type strain NTUH-C1 added to pC1, the transformation frequency also decreased to approximately that of NTUH-C1-EmpC1. Under the control of the relatively stronger promoter Pure, HP1361 protein could be overexpressed. DNA binding and uptake were increased in the complementation strains; however, transformation frequencies were lower than those for the wild type. This finding suggested that overexpression of HP1361 could interfere with other proteins in subsequent natural processes of transformation.
We failed to express the HP1361 protein in several E. coli expression systems. This protein may be as toxic to E. coli as the ComE protein of B. subtilis (16). There is no significant difference in amino acid sequence or RNA expression between competent and noncompetent strains. Therefore, HP1361 was not the cause of differences in the natural-competence phenotype.
In conclusion, HP1361 is a ComE3 homologue and is required for DNA binding and uptake during DNA transformation of H. pylori.
Acknowledgments
This study was supported by a grant (NSC91-2314-B-002-389) from the National Science Council, Taipei, Taiwan.
Editor: V. J. DiRita
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang, S., C.-Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J.-T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, D. E., R. H. Gilman, J. Lelwala-Guruge, K. Srivastava, Y. Valdez, J. Watanabe, J. Miyagi, N. S. Akopyants, A. Ramirez-Ramos, T. H. Yoshiwara, S. Recavarren, and R. Leon-Barua. 1997. Helicobacter pylori populations in Peruvian patients. Clin. Infect. Dis. 25:996-1002. [DOI] [PubMed] [Google Scholar]
- 5.Chang, K. C., Y. C. Yeh, T. L. Lin, and J. T. Wang. 2001. Identification of genes associated with natural competence in Helicobacter pylori by transposon shuttle random mutagenesis. Biochem. Biophys. Res. Commun. 288:961-968. [DOI] [PubMed] [Google Scholar]
- 6.Chen, I., and E. C. Gotschlich. 2001. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J. Bacteriol. 183:3160-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau, D., and R. Provvedi. 2000. Internalizing DNA. Res. Microbiol. 151:475-480. [DOI] [PubMed] [Google Scholar]
- 9.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 10.Goodgal, S. H. 1982. DNA uptake in Haemophilus transformation. Annu. Rev. Genet. 16:169-192. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, J., G. Inamine, Y. Kozlov, and D. Dubnau. 1993. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol. Microbiol. 10:99-111. [DOI] [PubMed] [Google Scholar]
- 12.Heuermann, D., and R. Hass. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, P. S., A. Goodwin, J. Johnsen, K. Magee, and S. J. O. Veldhuyzen van Zanten. 1996. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J. Bacteriol. 178:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh, P. F., J. C. Yang, J. T. Lin, and J. T. Wang. 1998. Molecular mechanisms of clarithromycin resistance in Helicobacter pylori. J. Formos. Med. Assoc. 97:445-452. [PubMed] [Google Scholar]
- 16.Inamine, G. S., and D. Dubnau. 1995. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J. Bacteriol. 177:3045-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labigne, A. 1997. Random mutagenesis of H. pylori genome, p. 153-163. In C. L. Clayton and H. L. T. Mobley (ed.), Helicobacter pylori protocols. Humana Press, Toyowa, N.J.
- 18.Smeets, L. C., J. J. E. Bijlsma, S. Y. Boomkens, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smeets, L. C., J. J. Bijlsma, E. J. Kuipers, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 27:99-102. [DOI] [PubMed] [Google Scholar]
- 20.Smeets, L. C., and J. G. Kusters. 2002. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends Microbiol. 10:159-162. [DOI] [PubMed] [Google Scholar]
- 21.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 23.Wang, Y., K. P. Poos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]
- 24.Yeh, Y.-C., K.-C. Chang, J.-C. Yang, C.-T. Fang, and J.-T. Wang. 2002. Association of metronidazole resistance and natural competence in Helicobacter pylori. Antimicrob. Agents Chemother. 46:1564-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]