Abstract
hilA encodes an activator of Salmonella enterica serovar Typhimurium virulence genes and is transcriptionally modulated by environmental conditions. We show that H-NS represses hilA under low-osmolarity conditions. H-NS, HU, and Fis also appear to affect the derepression of hilA by HilD. Modulation of hilA by counteracting repressing and derepressing mechanisms may allow Salmonella serovar Typhimurium to regulate its virulence genes in response to different situations in vivo.
Genes on Salmonella pathogenicity island 1 (SPI1) are required for Salmonella enterica serovar Typhimurium to cause gastroenteritis (27). SPI1 encodes components of a type III secretion system that translocates bacterial effector proteins into the cytosol of mammalian cells and modifies host cell signaling pathways (18). A large number of environmental conditions and regulatory factors control the expression of the SPI1 secretion system by regulating hilA expression (18). hilA is located on SPI1 and encodes an OmpR/ToxR transcriptional regulator, which coordinately activates the genes that encode the SPI1 secretion system (18).
It has been proposed that counteracting repressing and derepressing mechanisms regulate hilA expression. The presence of an upstream DNA sequence represses a plasmid-borne hilA promoter (23). HilC and HilD, two SPI1-encoded AraC/XylS family members, have been called derepressors and are thought to counteract the repression of hilA by binding to sites within the upstream repressing sequence (URS) (24). However, recent results appear to contradict this model and support the idea that HilD interacts with the α subunit of RNA polymerase to activate hilA transcription (4). For example, replacement of the URS with 84 bp of unrelated sequence or a precise deletion of the URS in the Salmonella serovar Typhimurium chromosome was shown to reduce, rather than increase, hilA expression (4; unpublished results). Unfortunately, we could not rescue our deletion with the intact URS and wondered if a deleterious effect of the URS deletion selects for secondary genetic changes that both reduce hilA-lacZ expression and prevent replacement of the URS allele (unpublished results). Still, it is intriguing to consider that under certain conditions HilC and HilD may be able to activate hilA expression and that under other conditions they may be able to increase hilA expression by derepressing hilA transcription. Studies showing that HilC and HilD are not required for hilA expression from a plasmid in in vitro transcription assays or in the absence of the URS suggest that HilC and HilD can derepress hilA expression, possibly by antagonizing the repressing effects of proteins bound to the URS (21, 23).
We suspected that small nucleoid-binding proteins, such as H-NS, HU, and Fis, might control hilA expression by affecting its repression and/or derepression. Small nucleoid-binding proteins have been shown to regulate gene expression in response to specific environmental conditions that also control hilA expression (1, 18, 22). H-NS can silence promoters by interacting with distal sequences (5) and has been shown to be counteracted by AraC/XylS factors (8). HU may also antagonize the effects of H-NS on gene expression (6). The small nucleoid-binding protein Hha has been reported to bind the URS and repress hilA expression (4, 11). In contrast, Fis appears to have the opposite regulatory effect, since Wilson et al. have shown that a mutation in fis reduces hilA expression (28).
Effect of H-NS on hilA expression.
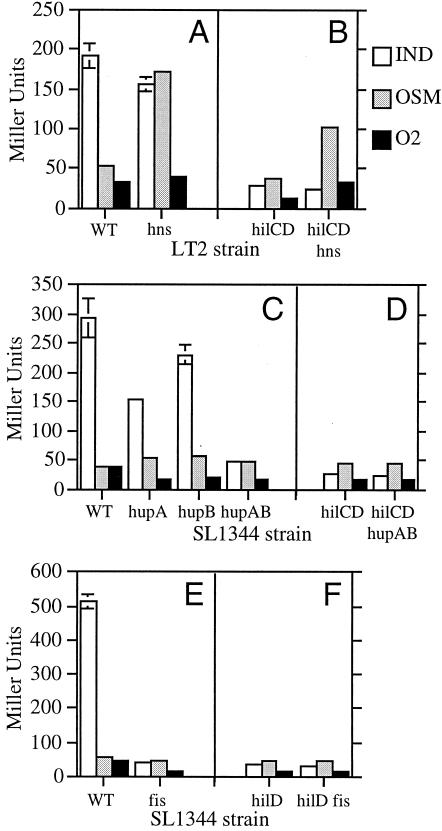
To test whether H-NS regulates hilA expression in Salmonella serovar Typhimurium, we examined the effect of the hns-1::kan mutation on the expression of the chromosomal hilA080::Tn5lacZY (Tetr) transcriptional fusion (2, 12). Although we routinely analyze hilA expression in Salmonella serovar Typhimurium SL1344, we found that hns mutations cause this strain to become mucoid and grow poorly. Therefore, we analyzed hilA-lacZ expression in Salmonella serovar Typhimurium LT2 (SGSC1412), which appears to better tolerate hns mutations. β-Galactosidase assays were performed on bacterial cultures grown under inducing or repressing conditions for hilA expression, as previously described (20, 23). Although hilA regulation in LT2 might not be identical to that in SL1344, we found that, as in SL1344, aerobic conditions and low-osmolarity conditions repress hilA-lacZ expression in LT2 (Fig. 1A). Interestingly, the hns-1::kan mutation increases hilA-lacZ expression under the low-osmolarity conditions but not under the aerobic growth conditions (Fig. 1A). Due to the counteracting nature of the repressing URS and derepressing HilC and HilD proteins that directly control hilA expression, the hns-1::kan mutation may increase hilA expression under the low-osmolarity conditions (i) by decreasing its repression and/or (ii) by increasing its derepression by HilC or HilD.
FIG. 1.
Effects of small nucleoid-binding proteins on hilA-lacZ expression. Salmonella serovar Typhimurium LT2 and SL1344 strains containing the hilA080::Tn5lacZY chromosomal fusion were grown under inducing (IND; white bars), repressing low-osmolarity (OSM; gray bars), or repressing aerobic (O2; black bars) conditions. β-Galactosidase activities are reported in Miller units and are the averages (± standard deviations) of values obtained from the results of at least two independent cultures during one representative experiment. Similar results were obtained in repeated experiments. (A) Fusion strain (wild type [WT]) and its hns-1::kan (hns) derivative; (B) hilC1::cam hilD1::kan (hilCD) and hilC1::cam hilD1::kan hns-1::kan (hilCD hns) derivatives; (C) fusion strain (WT) and its hupA::kan (hupA), hupB::kan (hupB), and hupA::kan hupB::kan (hupAB) derivatives; (D) hilC1::cam hilD1::kan (hilCD) and hilC1::cam hilD1::kan hupA::kan hupB::kan (hilCD hupAB) derivatives; (E) fusion strain (WT) and its fis3::cam (fis) derivative; (F) hilD1::kan (hilD) and hilD1::kan fis3::cam (hilD fis) derivatives.
To test whether hilC and hilD are required for the hns mutation to increase hilA expression in LT2, we introduced hilC1::cam and hilD1::kan mutations into the LT2 hns-1::kan strain. The hilC1::cam mutation was constructed by allele replacement with pLS112, a pLD55 derivative that contains a chloramphenicol resistance gene cassette inserted into the BclI site of hilC (20). The hilD1::kan mutation was constructed previously (24). The hns-1::kan hilC1::cam hilD1::kan hilA080::Tn5lacZY mutant was constructed by transducing LT2 hns-1::kan with a hilC1::cam hilD1::kan hilA080::Tn5lacZY P22 lysate (7, 23). Because hilD1::kan is located between hilC1::cam and hilA080::Tn5lacZY, chloramphenicol- and tetracycline-resistant transductants of LT2 hns-1::kan also contain the hilD1::kan mutation. The presence of the hilD1::kan mutation was confirmed by PCR.
Our analysis of hilA-lacZ expression in the hilC hilD mutant background shows that, even in the absence of hilC and hilD, the hns-1::kan mutation increases hilA expression under low-osmolarity conditions (Fig. 1B). These results are consistent with the idea that the hns mutation decreases the repression of hilA under these conditions. These results also provide further support for the idea that HilC and HilD function as derepressors, rather than activators, of hilA transcription. The hns mutation may also influence the derepression of hilA by HilC or HilD, since its ability to increase hilA-lacZ expression is partially attenuated in the hilCD mutant background (Fig. 1B).
Effect of HU and Fis on hilA expression.
To see if HU regulates hilA expression in Salmonella serovar Typhimurium, we examined the effects of single hupA and hupB mutations on chromosomal hilA-lacZ expression in SL1344 (Fig. 1C) (16). Because hupA and hupB encode the α and β subunits of HU, which can form active homodimers, we also examined hilA-lacZ expression in a double hupA hupB mutant strain (Fig. 1C). The Salmonella serovar Typhimurium hupA::kan hupB::kan double mutant was constructed by transducing a hupB::kan purD::Tn10 mutant with a hupA::kan P22 lysate and selecting for purine prototrophs. Because the purD gene is linked to hupA, purD+ hupA::kan hupB::kan transductants were obtained and verified by genetic linkage. As shown in Fig. 1C, chromosomal hilA-lacZ expression is dramatically reduced in the hupA::kan hupB::kan mutant grown under conditions that normally induce hilA expression. Thus, in contrast to H-NS, HU plays a positive role in hilA expression. Fahlen et al. reported that a mutation in hupB increases hilA expression (10), which is contrary to our results. However, we used a hupB::kan mutation that disrupts the hupB open reading frame, while the hupB1::Tn5 mutation used by Fahlen et al. is located 188 bp upstream of the hupB translation start site (10, 16). We speculate that the hupB1::Tn5 mutation actually increases hupB expression and HU levels, which may stimulate hilA expression.
Wilson et al. have shown that a fis::kan mutation reduces hilA expression two- to threefold (28). We found that the fis3::cam mutation reduces chromosomal hilA-lacZ expression 13-fold under our conditions, which normally induce hilA (Fig. 1E). These results suggest that, like HU, Fis is required for full induction of hilA expression in Salmonella serovar Typhimurium. Previous studies have shown that HilD, not HilC, is primarily responsible for derepression of hilA expression when Salmonella serovar Typhimurium is grown under our inducing conditions (19, 23). Thus, the hup and fis mutations may decrease hilA expression (i) by decreasing its derepression by HilD and/or (ii) by increasing its repression. In the first case, we expect that mutations in hupAB or fis would not further reduce hilA-lacZ expression in strains lacking HilD. Consistent with the idea that HU and Fis affect the derepression of hilA, our results show that hilA-lacZ expression is not significantly reduced by the hupAB or fis mutations in a hilCD or hilD mutant strain background (Fig. 1DF). HU or Fis may bind to the hilA promoter and enhance its ability to be derepressed by HilD. Alternatively, HU or Fis may influence hilA indirectly by increasing the levels and/or activity of HilD.
Effect of H-NS, HU, and Fis on hilD mRNA levels.
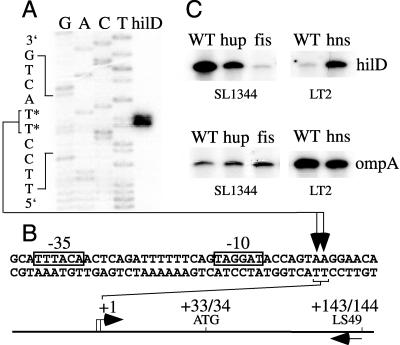
To examine whether H-NS, HU, or Fis affects hilD mRNA levels, we first characterized the hilD transcript by identifying the hilD transcription start site. A primer extension analysis was performed on RNA from wild-type Salmonella serovar Typhimurium grown under inducing high-osmolarity, low-oxygen conditions. An overnight culture of SL1344 was subcultured at a dilution of 1:1,000 in 100 ml of high-salt (1% NaCl) Luria-Bertani medium in a 125-ml Erlenmeyer flask and grown with shaking at 150 rpm to a final optical density at 600 nm of 0.7 to 0.8. Ten micrograms of total RNA, isolated from these cells by using the RNAqueous kit (Ambion Inc.), was annealed to 100 fmol of LS49 (5′-GTCTGACTTTTAATTTGCTGC-3′), which was previously end labeled with [γ-32P]ATP and T4 polynucleotide kinase (NEN Life Science Products, Promega). The primer extension reaction was carried out by using the AMV reverse transcriptase system (Promega) and run on a 6% acrylamide-8 M urea gel; a dideoxy DNA sequencing reaction was carried out by using the fmol DNA sequencing system (Promega) and the end-labeled LS49 primer. Similar to the results of studies by Olekhnovich and Kadner (21), our results indicate that one or two hilD transcription start sites are located 34 or 35 bp upstream of the hilD translation start site and just downstream (6 or 7 bp) of a predicted σ70 binding site (TTTACA-16 bp-TAGGAT) (Fig. 2A and B).
FIG. 2.
Effect of small nucleoid-binding proteins on hilD mRNA levels. (A) The hilD transcription start site was determined by running primer extension products (hilD) from wild-type Salmonella serovar Typhimurium strain SL1344 RNA alongside dideoxy DNA sequencing reactions (GACT). Nucleotides marked with an asterisk represent two possible hilD transcription start sites on the template strand. (B) The positions of potential −10 and −35 σ70 binding sites, the hilD translation start site (ATG), and the primer used in the primer extension reactions (LS49) are shown below the autoradiograph of the gel in relation to the hilD transcription start sites (+1). (C) hilD and ompA primer extension products were generated by using total RNA from wild-type (WT), hupA::kan hupB::kan (hup), fis3::cam (fis), and hns-1::kan (hns) derivatives of SL1344 or LT2, grown under inducing or low-osmolarity conditions, respectively. The autoradiographs shown are from one representative experiment.
Using this primer extension assay, we compared the hilD mRNA levels in our isogenic wild-type and mutant strains. We purified RNA from wild-type SL1344, the hupA::kan hupB::kan mutant, and the fis3::cam mutant grown under inducing conditions. In contrast, we grew the wild-type LT2 and the hns-1::kan mutant under low-osmolarity conditions, since our previous results showed that the hns-1::kan mutation specifically affects hilA expression in low-osmolarity media (Fig. 1A). Our results indicate that the level of hilD mRNA is reduced in the hupA hupB and fis mutants and increased in the hns mutant (Fig. 2C). Control primer extension assays were also conducted using primer ompA1 (5′-CGAAACCAGCCAGTGCCACTG-3′). ompA encodes a major outer membrane protein, which is important for structural integrity of the outer membrane. In Escherichia coli, ompA is highly expressed and its mRNA levels are only modestly affected by dramatic changes in growth conditions and growth rate (14, 15). As shown in Fig. 2C, each set of wild-type and mutant strains appears to contain similar levels of ompA mRNA. By quantitating the primer extension products obtained in two independent experiments with a phosphorimager, we normalized the hilD product to that of the ompA control and then calculated the level of hilD mRNA in each mutant strain relative to that in its isogenic wild-type strain. This analysis indicates that the hupA::kan hupB::kan mutations and fis3::cam mutation reduce hilD mRNA levels to 29 and 3% of that in wild-type SL1344, respectively. The dramatic reduction in hilD mRNA levels seen in the fis mutant suggests that Fis primarily influences the derepression of hilA by altering the production of HilD. The less dramatic change in hilD mRNA seen in the hupAB mutant indicates that HU may affect the derepression of hilA via more than one mechanism, such as by affecting regulatory pathways that modulate HilD posttranscriptionally (3). Our analysis indicated that the hns-1::kan mutation increases hilD mRNA levels sixfold over that in wild-type LT2. This result suggests that, in addition to decreasing the repression of hilA under low-osmolarity conditions (Fig. 1B), the hns mutation increases the derepression of hilA by HilD. Further studies are needed to understand how H-NS, HU, and Fis alter hilD mRNA levels and affect the derepression of hilA.
Repression of hilA expression.
We previously proposed that a repressor protein binds to the URS and prevents the expression of hilA under repressing growth conditions (23). Our present results suggest that hilA expression may actually be repressed by different factors under different conditions. While H-NS may directly or indirectly repress the hilA promoter under low-osmolarity conditions, our finding that hilA remains repressed in a hilCD hns mutant grown under inducing or aerobic conditions suggests that other factors may repress hilA expression under these conditions (Fig. 1B). Additional repressors of hilA may include HilE, Pag, and Hha, as strains lacking these proteins exhibit increased hilA expression under inducing conditions (4, 10, 11). Hha may also repress hilA under low-osmolarity conditions (11). To determine whether HilE, Pag, or Hha affects repression, rather than derepression, of hilA, the effect that hilE, pag, and hha mutations have on hilA expression should be examined under repressing environmental conditions in strains lacking HilC and HilD. Since mutations in hilE, pag, and hha do not abolish the repression of hilA expression in Salmonella serovar Typhimurium grown aerobically, other factors may repress hilA expression under aerobic conditions (10, 11).
There are several mechanisms by which H-NS might repress hilA under low-osmolarity conditions. H-NS might bind to the URS and directly occlude transcription initiation from the hilA promoter, as has been proposed for H-NS silencing of the bgl promoter in E. coli (5). Alternatively, H-NS, which bends DNA and affects global DNA supercoiling, may alter the DNA topology at the hilA promoter to repress hilA transcription (16, 17, 25, 26). Interestingly, expression of the SPI1 gene invA is sensitive to chemical inhibitors of DNA gyrase as well as topoisomerase mutations that change DNA supercoiling (13). Because HilA activates invA expression, alterations in DNA supercoiling may affect invA by influencing hilA expression (9).
Acknowledgments
We thank Patrick Higgins, Fred Heffron, Robert Osuna, and Barry Wanner for kindly providing bacterial strains. We also thank Tom Burr for help with the phosphorimager and Robin Lucas for helpful discussions.
This work was supported by NIH grant AI33444.
Editor: V. J. DiRita
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caramel, A., and K. Schnetz. 1998. Lac and λ repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J. Mol. Biol. 284:875-883. [DOI] [PubMed] [Google Scholar]
- 6.Dame, R. T., and N. Goosen. 2002. HU: promoting or counteracting DNA compaction? FEBS Lett. 529:151-156. [DOI] [PubMed] [Google Scholar]
- 7.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichelberg, K., and J. E. Galán. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 11.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falconi, M., V. McGovern, C. Gualerzi, D. Hillyard, and N. P. Higgins. 1991. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 3:615-625. [PubMed] [Google Scholar]
- 13.Galán, J. E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58:1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgellis, D., S. Arvidson, and A. von Gabain. 1992. Decay of ompA mRNA and processing of 9S RNA are immediately affected by shifts in growth rate, but in opposite manners. J. Bacteriol. 174:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgellis, D., T. Barlow, S. Arvidson, and A. von Gabain. 1993. Retarded RNA turnover in Escherichia coli: a means of maintaining gene expression during anaerobiosis. Mol. Microbiol. 9:375-381. [DOI] [PubMed] [Google Scholar]
- 16.Hillyard, D. R., M. Edlund, K. T. Hughes, M. Marsh, and N. P. Higgins. 1990. Subunit-specific phenotypes of Salmonella typhimurium HU mutants. J. Bacteriol. 172:5402-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinton, J. C. D., D. S. Santos, A. Seirafi, C. S. J. Hulton, G. D. Pavitt, and C. F. Higgins. 1992. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol. Microbiol. 6:2327-2337. [DOI] [PubMed] [Google Scholar]
- 18.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 19.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettijohn, D. E. 1996. The nucleoid, p. 158-166. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 23.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 24.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the S. typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 25.Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16:1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tupper, A. E., T. A. Owen-Hughes, D. W. Ussery, D. S. Santos, D. J. P. Ferguson, J. M. Sidebotham, J. C. D. Hinton, and C. F. Higgins. 1994. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 13:258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 28.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]