Abstract
Aims
To investigate the relationship between proguanil metabolic ratio (MR, proguanil/cycloguanil) and CYP2C19 genotype in a Caucasian population.
Methods
Ninety-nine Caucasians (age range: 18–55 years, 54 female, 45 male) were genotyped for CYP2C19 and phenotyped for proguanil oxidation by collecting urine for 8 h after taking 100 mg proguanil hydrochloride. Proguanil and cycloguanil concentrations were measured by h.p.l.c. PCR was employed for CYP2C19 genotyping.
Results
The three (3%) individuals who were homozygous for CYP2C19*2 (*2/*2) had the highest proguanil MRs (range: 8.0–134.6). Seventy-three (74%) individuals were homozygous for the wild-type allele (*1/*1) and 23 (23%) were heterozygous (*1/*2). The *1/*1 individuals had lower MRs (median = 1.4, range: 0.23–5.9, P = 0.003, Mann–Whitney U-test) than the *1/*2 subjects (median = 2.5, range: 0.88–7.3).
Conclusions
A CYP2C19 gene-dose effect for proguanil oxidation to cycloguanil was observed, confirming a role for CYP2C19 in cycloguanil formation in vivo. However, there was substantial overlap of proguanil MRs in subjects of different CYP2C19 genotypes, due possibly to variability in the activity of other enzymes contributing to the formation of cycloguanil.
Keywords: CYP2C19, gene-dose effect, genotype, proguanil, metabolic ratio
Introduction
Cytochrome P450 2C19 (CYP2C19) is the major enzyme catalysing the 4′-hydroxylation of S-mephenytoin [1, 2] and the polymorphic expression of this enzyme is responsible for the genetic polymorphism in the metabolism of S-mephenytoin [3]. CYP2C19 is important for the oxidation and elimination of a number of other drugs, including omeprazole, diazepam and proguanil [4].
The gene that codes for CYP2C19 has been mapped to chromosome 10 [5].CYP2C19*1 refers to the wild-type allele [6] and four mutations of this gene, which produce inactive P450 enzymes, have been identified [7–10]. The CYP2C19*2 (m1 mutation) accounts for 75 and 85% of Oriental and Caucasian mutant alleles, respectively, and is a single base pair G→A mutation in exon 5 of CYP2C19 [7]. The CYP2C19*3 allele (m2 mutation) accounts for 10–25% of Oriental mutant alleles and is rare in Caucasians [8, 11, 12]. This mutation is a single base pair G→A mutation in exon 4 which results in a premature stop codon. An individual who inherits two mutant CYP2C19 alleles has a reduced capacity to metabolize CYP2C19 substrates and is termed a poor metabolizer (PM). Individuals who are homozygous or heterozygous for CYP2C19*1 and who are efficient metabolizers of CYP2C19 substrates are termed extensive metabolizers (EMs). Results of recent population studies, which have utilized both CYP2C19 phenotyping and genotyping techniques, suggest that heterozygous EMs have a capacity to oxidize the CYP2C19 substrates omeprazole and mephenytoin that is intermediate between PMs and homozygous EMs—a phenomenon which is indicative of a CYP2C19 gene-dose effect [11, 13].
Proguanil is oxidized to a major and pharmacologically active triazine metabolite, cycloguanil, which inhibits plasmodial dihydrofolate reductase, and a minor metabolite, 4-chlorophenylbiguanide. The formation of both metabolites is diminished in poor metabolizers of S-mephenytoin, indicating that the pathways are catalysed by CYP2C19 [14, 15]. The urinary recovery of cycloguanil relative to proguanil has therefore been used as a phenotypic probe to assess the activity of CYP2C19 in different ethnic groups [15–17]. In vitro studies using human liver microsomes have confirmed that CYP2C19 catalyses this pathway of metabolism but also indicate that cytochrome P450 3A (CYP3A) plays a role [18, 19] which recent in vivo data suggest is only minor [19]. The large inter and intraindividual variability in CYP3A activity in vivo [20] and hence variability in its contribution to proguanil oxidation, may mask interindividual differences in CYP2C19 activity. Furthermore, as the proguanil MR and 4′-hydroxymephenytoin index are not correlated in EMs of mephenytoin [21, 22] the relationship between cycloguanil and the formation of 4′-hydroxymephenytoin in vivo and CYP2C19 genotype requires further elucidation.
Recently, a small study of 10 subjects of different ethnic groups reported that CYP2C19 genotype does relate to the formation of cycloguanil in vivo [23]. The present study was performed to investigate the relationship between CYP2C19 genotype and proguanil metabolism in a population of healthy Caucasian subjects in vivo.
Methods
Subjects
Ninety-nine unrelated, healthy volunteers participated in this study which was approved by the Royal North Shore Hospital Human Research Ethics Committee (HREC Protocol no. 9502–027 m (CTN)). All subjects reported that all four grandparents were Caucasians. Fifty-four females and 45 males were studied. The median age of subjects was 27 years (range: 18–55) and body mass index was 22.5 (range: 17.1–31.5). Subjects were drug-free for 7 days prior to the study (except oral contraceptives OC, n = 15, hormone replacement therapy HRT, n = 3 and cigarette smoking, n = 7) and were also alcohol-free for 24 h prior to and during the study.
Phenotyping
After voiding their bladder all subjects took 100 mg proguanil hydrochloride (Paludrine®, kindly donated by ICI Pharmaceuticals, Macclesfield, Cheshire, UK; 345 μmol) orally with 200 ml of water and then collected their urine for the following 8 h (mean 8.0±0.5 h). A 30 mg (81 μmol) dextromethorphan hydrobromide (Parke Davis, Sydney, New South Wales, Australia) dose was coadministered as a probe for cytochrome P450 2D6 (CYP2D6). Previous studies have demonstrated that coadministration of dextromethorphan with proguanil does not influence the formation of cycloguanil from proguanil [21, 24]. The 8 h urine volume was measured and 10 ml aliquots of urine were stored at −20° C pending analysis. No severe adverse effects were reported by the volunteers during the phenotyping procedure.
Proguanil and dextromethorphan chromatographic analysis
Concentrations of proguanil and cycloguanil in urine were measured using reversed-phase high performance liquid chromatography (h.p.l.c.) [25]. The between day reproducibilities of the proguanil (CV < 7% 10.4, 13.6 μg ml−1, n = 6) and cycloguanil assays (CV < 14% 0.8, 5.2 μg ml−1, n = 6) were good. The limits of quantification for proguanil and cycloguanil were 0.1 μg ml−1 and 0.2 μg ml−1, respectively. Cycloguanil was not detected in the urine of an individual who was a poor metabolizer of both proguanil and dextromethorphan and the concentration was assumed to be 0.2 μg ml−1. To assess CYP2D6 phenotype urinary dextromethorphan and dextrorphan concentrations were also measured using this h.p.l.c. technique [25]. The between day reproducibilities of the dextromethorphan and dextrorphan assays were (CV < 17% 0.12, 0.54 μg ml−1, n = 6) and (CV < 4% 0.36, 7.8 μg ml−1, n = 6), respectively. The limits of quantification for dextromethorphan and dextrorphan were 0.2 μg ml−1 and 0.1 μg ml−1, respectively.
CYP2C19 genotyping
All subjects were tested for the m1 mutation and 73 subjects were tested for the m2 mutation. For CYP2C19 genotyping, a blood sample (10 ml) was collected from each subject (K3 EDTA Vacutainers, Greiner Labortechnik, Germany) and stored at 4° C. DNA was released from 2 μl of blood that was diluted 1:1 with sterile 0.9% sodium chloride (Delta West Pty Ltd, Bentley, Western Australia, Australia) using 20 μl of GeneReleaserTM (BioVentures Incorporated, Murfreesboro, TN) (20 μl) in accordance with the manufacturers instructions. Polymerase chain reaction (PCR) techniques were performed as previously described by de Morais et al. [7, 8, 26] (Omn-E Thermocycler, Hybaid Ltd, Teddington, Middlesex, UK). The m1 and m2 PCR products were digested as previously described by de Morais et al. [7, 8] and the restricted PCR products (14 μl) were analysed on a 4% agarose gel (Molecular Biology grade, Promega Corporation, Madison, WI).
Data analysis
Urinary recoveries of cycloguanil and proguanil were calculated as the product of the molar concentration of analyte in urine and the volume of urine collected and expressed as a percentage of the proguanil dose (% CG and % PG). The rate of urinary excretion was calculated to account for variation in the urine collection interval (% CG/h and % PG/h). The proguanil MR was calculated as the ratio of the molar recovery of proguanil to cycloguanil in the urine collected (PG/CG). The Hardy–Weinberg equation was used to determine the expected frequency of mutant and wild-type (CYP2C19*1) alleles in the population [27]. Dextromethorphan MR was calculated as the ratio of the urinary molar recovery of dextromethorphan to dextrorphan. Based on the frequency distribution of metabolic ratios in this population, an antimode of 1.0 was used to distinguish CYP2D6 poor and extensive metabolizers. Differences in the calculated parameters between individuals with different CYP2C19 genotypes, between poor and extensive metabolizers of dextromethorphan, and between individuals of different hormonal status were assessed using the nonparametric Mann–Whitney U-test. Where significant differences were observed the 95% confidence interval for the difference (95% CI of diff) between the population means is also reported. Spearman-Rank correlation coefficients were calculated to investigate the relationship between dextromethorphan MR and proguanil MR in individuals who were not taking an OC or HRT and who were extensive metabolizers of dextromethorphan and either homozygous or heterozygous for CYP2C19*1.
Results
The frequencies of the CYP2C19 genotypes observed in the 99 Caucasian subjects are listed in Table 1. Three (3%) individuals were homozygous for the CYP2C19*2 allele (*2/*2), 23 (23%) were heterozygous (*1/*2) and 73 (74%) were homozygous for the wild-type allele (*1/*1). None of the 73 subjects tested possessed a CYP2C19*3 allele. The observed frequencies of the *1/*1 and *1/*2 genotypes are in agreement with those predicted according to the Hardy–Weinberg rule from the frequency of the *2/*2 genotype, indicating that the population studied is in equilibrium with the Hardy–Weinberg rule for population genetics.
Table 1.
The frequencies [95% confidence interval] of CYP2C19 genotypes observed in the Caucasian population and the frequencies expected based on the Hardy–Weinberg Law.

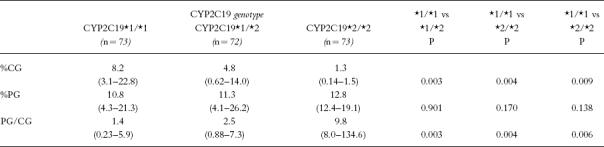
The frequency distribution histograms of the proguanil MRs in subjects with different CYP2C19 genotypes are illustrated in Figure 1. The urinary recoveries of cycloguanil and proguanil and the proguanil MRs of individuals with each CYP2C19 genotype are listed in Table 2. The urinary recovery of proguanil was similar in subjects of all 3 CYP2C19 genotypes. The urinary recovery of cycloguanil in the *2/*2 individuals was lower (P = 0.009; 95% CI of diff = −8.83–−0.27) than that in the *1/*2 and *1/*1 (P = 0.004; 95% CI of diff = −12.09–−2.87) individuals. The urinary recovery of cycloguanil in the group of *1/*2 individuals was lower (P = 0.003; 95% CI of diff = −4.77–−1.09) than that with the *1/*1 genotype. The genotype differences in urinary cycloguanil recovery were maintained when adjusted for the urine collection interval (%CG/h, P = 0.002; data not shown). The proguanil MRs of the *2/*2 individuals were higher than those of individuals with the *1/*2 (P = 0.006; 95% CI of diff = 21.38–74.64) and *1/*1 genotypes (P = 0.004; 95% CI of diff = 35.01–63.16). The proguanil MRs of the *1/*2 individuals were higher (P = 0.003; 95% CI of diff = 0.44–1.71) than those of *1/*1 subjects.
Figure 1.
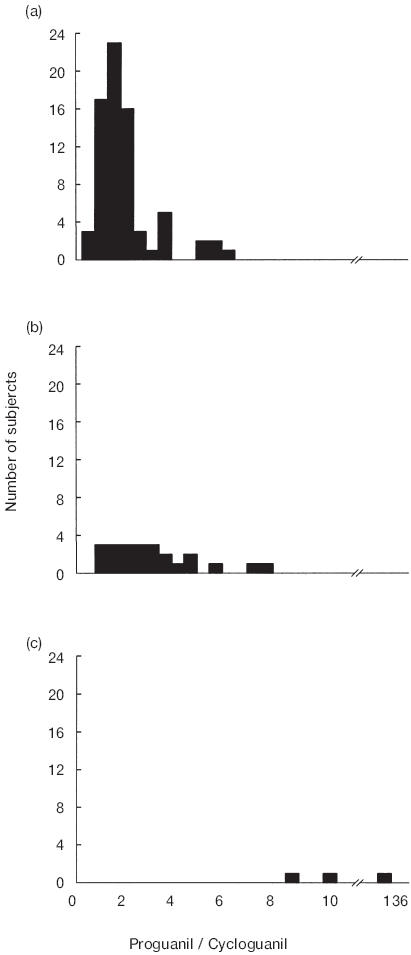
Frequency distribution histograms of proguanil metabolic ratios (proguanil/cycloguanil) for individuals in relation to CYP2C19genotype. (a) CYP2C19*1/*1; (b) CYP2C19*1/*2; (c) CYP2C19*2/*2.
Table 2.
The urinary recoveries of cycloguanil (%CG), proguanil (%PG) and proguanil metabolic ratios (PG/CG) in individuals with different CYP2C19 genotypes (median (range)).
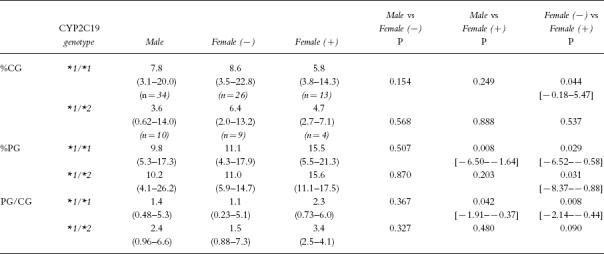
Proguanil MRs in males and females for both the *1/*1 (female n = 39, median = 1.3, range:0.23–6.0) and *1/*2 genotypes (female n = 13, median = 2.5, range:0.88–7.3) were similar (P = 0.782 and P = 0.664, respectively). However, females with the *1/*1 genotype taking OC or HRT had higher proguanil MRs than either *1/*1 females who were not taking hormones (P = 0.008) or males (P = 0.042) (Table 3). For *1/*2 individuals, proguanil MRs were similar in women taking an OC or HRT, women not taking hormones and men.
Table 3.
The urinary recoveries of cycloguanil (%CG) and proguanil (%PG) and the proguanil metabolic ratios (PG/CG) for males, women without exogenous hormones (−) and those taking either an oral contraceptive or hormone replacement therapy (+) (median (range) [95% confidence interval of the difference]).
Seven subjects (7%) were poor metabolizers of dextromethorphan. One poor metabolizer of dextromethorphan, who was also homozygous for CYP2C19*2 and therefore was a poor metabolizer of both proguanil and dextromethorphan, was the only subject in the population studied where cycloguanil could not be detected in urine. The one PM of dextromethorphan with a *1/*2 genotype had a proguanil MR of 3.3. In *1/*1 individuals, there was no difference (P > 0.05) in proguanil MR between the dextromethorphan PMs (n = 5) and EMs (n = 68). When women taking OC or HRT were excluded from the analysis, proguanil MRs were higher in PMs of dextromethorphan (n = 5, median = 2.1, range: 1.1–5.1, P = 0.024; 95% CI of difference = 0.43– 2.06) than in EMs (n = 55, median = 1.3, range: 0.2–5.4). However, no relationship between the dextromethorphan MR and proguanil MR was observed in EMs of dextromethorphan not taking OC or HRT who were either homozygous (rS = 0.056, P > 0.50, n = 55) or heterozygous (rS = 0.168, P > 0.50,n = 18) for CYP2C19*1.
Discussion
A rank order of proguanil MRs and urinary cycloguanil recovery in relation to CYP2C19 genotype has been observed (Table 2). There is no difference in the urinary recovery of proguanil between individuals who are CYP2C19*1/*1, *1/*2 and *2/*2. The urinary recovery of cycloguanil is higher in *1/*1 individuals than the *1/*2 individuals, who in turn excrete more cycloguanil than *2/*2 individuals. This difference in cycloguanil recovery is reflected in the proguanil MRs, with the *1/*1 individuals having lower proguanil MRs than the *1/*2 individuals, who also had lower MRs than the *2/*2 individuals. This observation suggests a CYP2C19 gene-dose effect for proguanil oxidation to cycloguanil. Consequently the *1/*2 individuals with only one wild-type allele produce less cycloguanil than the *1/*1 individuals because they express less of the active CYP2C19 enzyme. A CYP2C19 gene-dose effect has also been reported for mephenytoin and omeprazole in a number of different ethnic groups [11, 13, 26, 28, 29].
The observation of a CYP2C19 gene-dose effect for proguanil oxidation to cycloguanil in this Caucasian population confirms the recent observations of Coller et al. [23] of a relationship between cycloguanil formation and the CYP2C19 genotype in 10 Caucasian and Asian subjects. In addition, the gene-dose effect provides further evidence for the important contribution of CYP2C19 to the formation of cycloguanil in vivo [19].
Although there was a difference in the proguanil MR between *1/*1 and *1/*2 individuals, substantial overlap was observed and the MR could not be used to distinguish between the 2 genotypes. This phenomenon has been reported previously for mephenytoin and omeprazole [11, 13, 26, 28, 29]. The overlap of proguanil MRs between the *1/*1 and *1/*2 individuals may be attributed to a number of factors. Firstly, cycloguanil formation is also catalysed by CYP3 A [18, 19] and large interindividual variability in the activity of this enzyme may mask a clear distinction between proguanil MRs of the CYP2C19*1/*1 and *1/*2 genotypes. Secondly, there may be additional as yet unidentified mutations of CYP2C19 which could result in an enzyme of altered activity. Thirdly, environmental factors, such as diet or exposure to chemicals, may influence CYP2C19 activity [30, 31]. The present study indicates that OC and HRT influence cycloguanil formation, possibly by decreasing the activity of CYP2C19.
Women taking exogenous hormones, either OC or HRT, had higher proguanil MRs than men and women not taking hormones (Table 3) suggesting that proguanil oxidation to cycloguanil is diminished by OC and HRT, both containing oestrogens. The higher proguanil MRs reflect both the lower recovery of cycloguanil (P < 0.05) and higher excretion of proguanil (P < 0.05) in women taking hormones compared with either the other women or men. OCs are known to inhibit the metabolism of many drugs, including the CYP2C19 substrates propanolol [32], diazepam [33] and imipramine [34] and in vitro studies have demonstrated that ethinyloestradiol and oestradiol inhibit S-mephenytoin 4′-hydroxylation [31]. It has also been reported that peak cycloguanil concentrations are lower and proguanil MRs higher in women when pregnant than 2 months postpartum [35], suggesting that high oestrogen levels, as observed during pregnancy, decrease cycloguanil formation. The present study provides further evidence that the activity of CYP2C19 is diminished by high concentrations of oestrogens.
The frequencies of the CYP2C19 genotypes observed in this population are in agreement with those expected from the Hardy–Weinberg rule (Table 1) and previously reported in Caucasian populations [11, 12]. None of the 73 subjects tested possessed a CYP2C19*3 allele, which is rarely found in Caucasian populations [8, 11].
Two *2/*2 individuals studied had proguanil MRs (MR 8.0,9.8) which were lower than the value of 10, previously suggested by Helsby et al. [36] as appropriate for distinguishing proguanil EMs and PMs in a Caucasian population. The lower proguanil MRs in the present study may be due to the lower dose of proguanil administered (100 mg vs 200 mg) or shorter urine collection interval (8 h vs 12 h). The pharmacokinetics of proguanil are linear [37] and therefore the dose of proguanil administered should not affect the proguanil MR. Our results are in agreement with the observations of others that the frequency distribution of proguanil MRs is skewed and continuous rather than bimodal (Figure 1), making it difficult to assign an antimode [17]. This skewed distribution is probably due to large interindividual variability in the activity of enzymes other than CYP2C19, including CYP3A, which also contribute to the formation of cycloguanil from proguanil.
One subject in this study was a poor metabolizer of both dextromethorphan and proguanil. Cycloguanil could not be detected in the urine of this individual. Ward et al. [14] also noted that cycloguanil could not be detected in an individual who was a poor metabolizer of both CYP2D6 and CYP2C19. This observation together with the higher proguanil MRs noted in PMs compared with EMs of dextromethorphan (CYP2C19*1/*1 individuals, excluding females taking an OC or HRT) suggests that in vivo CYP2D6 does contribute to the formation of cycloguanil. However, in vitro studies using human liver microsomes have failed to identify a contribution of CYP2D6 to cycloguanil formation [18] and in the present study no relationship between dextromethorphan MR and proguanil MR was observed in *1/*1 and *1/*2 individuals not taking OCs or HRT. Previous studies have shown that coadministration of the probes, dextromethorphan, proguanil and caffeine has no effect on proguanil MR [21, 24] but produces a small increase in dextromethorphan MR, without change in phenotypic assignment. This interaction may be attributable to the inhibition of dextromethorphan metabolism by proguanil (Ki 3.0 μm; unpublished observation). Although the interaction in vivo would be small, at the doses used in the current study, it might have affected the relationship between the actual activity of CYP2D6 in vivo and its assessment as provided by the dextromethorphan MR. Further studies including determination of CYP2D6 genotype are required to clarify the contribution of CYP2D6 to proguanil metabolism in vivo.
In conclusion, a CYP2C19 gene-dose effect for proguanil oxidation to cycloguanil has been observed. In accord with that previously reported for mephenytoin and omeprazole in Caucasian populations. This result confirms both a role for CYP2C19 in cycloguanil formation in vivo and the usefulness of proguanil as a probe for CYP2C19 activity. However, the overlap of proguanil MRs between homozygous wild-type and heterozygous individuals means that the proguanil MR cannot be used to distinguish these two genotypes.
Acknowledgments
Janelle Hoskins was supported by a Westpac Research Fellowship and a Royal North Shore Hospital Centenary Foundation Fellowship (both awarded by Royal North Shore Hospital).
References
- 1.Wrighton SA, Stevens JC, Becker GW, VandenBranden M. Isolation and characterisation of human liver cytochrome P450 2C19: Correlation between 2C19 and S-mephenytoin 4′-hydroxylation. Arch Biochem Biophys. 1994;306:240–245. doi: 10.1006/abbi.1993.1506. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein JA, Faletto MB, Romkes-Sparks M, et al. Evidence that CYP2C19 is the major (S)-mephenytoin 4′-hydroyxlase in humans. Biochemistry. 1994;33:1743–1752. doi: 10.1021/bi00173a017. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson GR, Guengerich FP, Branch RA. Genetic polymorphism of S-mephenytoin hydroxylation. Pharmacol Ther. 1989;43:53–76. doi: 10.1016/0163-7258(89)90047-8. [DOI] [PubMed] [Google Scholar]
- 4.Daniel HI, Edeki TI. Genetic polymorphism of S-mephenytoin 4′-hydroxylation. Psychopharmacol Bull. 1997;32:219–230. [PubMed] [Google Scholar]
- 5.Meehan RR, Gosden JR, Rout D, et al. Human cytochrome P-450 PB-1: A multigene family involved in mephenytoin and steroid oxidations that maps to chromosome 10. Am J Hum Genet. 1989;42:26–37. [PMC free article] [PubMed] [Google Scholar]
- 6.Daly AK, Brockmoller J, Broly F, et al. Nomenclature for human CYP2D6 alleles. Pharmacogenetics. 1996;6:193–201. doi: 10.1097/00008571-199606000-00001. [DOI] [PubMed] [Google Scholar]
- 7.de Morais SMF, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 8.de Morais SMF, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 9.Xiao ZS, Goldstein JA, Xie HG, et al. Differences in the incidence of the CYP2C19 polymorphism affecting the S-mephenytoin phenotype in Chinese Han and Bai populations and identification of a new rare CYP2C19 mutant allele. J Pharmacol Exp Ther. 1997;281:604–609. [PubMed] [Google Scholar]
- 10.Ferguson RJ, de Morais SMF, Benhamou S, et al. A new genetic defect in human CYP2C19: Mutation in the initiation codon is responsible for poor metabolism of S-mephenytoin. J Pharmacol Exp Ther. 1998;284:356–361. [PubMed] [Google Scholar]
- 11.Chang M, Dahl ML, Tybring G, Götharson E, Bertilsson L. Use of omeprazole as a probe drug for CYP2C19 phenotype in Swedish Caucasians: comparison with S-mephenytoin hydroxylation phenotype and CYP2C19 genotype. Pharmacogenetics. 1995;5:358–363. doi: 10.1097/00008571-199512000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Balian JD, Sukhova N, Harris JW, et al. The hydroxylation of omeprazole correlates with S-mephenytoin metabolism: A population study. Clin Pharmacol Ther. 1995;57:662–669. doi: 10.1016/0009-9236(95)90229-5. [DOI] [PubMed] [Google Scholar]
- 13.Brockmöller J, Rost KL, Gross D, Schenkel A, Roots I. Phenotyping of CYP2C19 with enantiospecific HPLC-quantification of R- and S-mephenytoin and comparison with the intron4/exon5 G→A-splice site mutation. Pharmacogenetics. 1995;5:80–88. doi: 10.1097/00008571-199504000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Ward SA, Helsby NA, Skjelbo E, Brøsen K, Gram LF, Breckenridge AM. The activation of the biguanide antimalarial proguanil co-segregates with the mephenytoin oxidation polymorphism—a panel study. Br J Clin Pharmacol. 1991;31:689–692. doi: 10.1111/j.1365-2125.1991.tb05594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brøsen K, Skjelbo E, Flachs H. Proguanil metabolism is determined by the mephenytoin oxidation polymorphism in Vietnamese living in Denmark. Br J Clin Pharmacol. 1993;36:105–108. doi: 10.1111/j.1365-2125.1993.tb04204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanwimolruk S, Thou MR, Woods DJ. Evidence for the polymorphic oxidation of debrisoquine and proguanil in a Khmer (Cambodian) population. Br J Clin Pharmacol. 1995;40:166–169. doi: 10.1111/j.1365-2125.1995.tb05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanwimolruk S, Pratt EL, Denton JR, Chalcroft SCW, Barron PA, Broughton JR. Evidence for the polymorphic oxidation of debrisoquine and proguanil in a New Zealand Maori population. Pharmacogenetics. 1995;5:193–198. doi: 10.1097/00008571-199508000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Birkett DJ, Rees D, Andersson T, Gonzalez FJ, Miners JO, Veronese ME. In vitro proguanil activation to cycloguanil by human liver microsomes is mediated by CYP3A isoforms as well as by S-mephenytoin hydroxylase. Br J Clin Pharmacol. 1994;37:413–420. doi: 10.1111/j.1365-2125.1994.tb05707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funck-Brentano C, Becquemont L, Leneveu A, Roux A, Jaillon P, Beaune P. Inhibition by omeprazole of proguanil metabolism: Mechanism of the interaction in vitro and prediction of in vivo results from the in vitro experiments. J Pharmacol Exp Ther. 1997;280:730–738. [PubMed] [Google Scholar]
- 20.Kinirons MT, O’Shea D, Downing TE, et al. Absence of correlations among three putative in vivo probes of human cytochrome P4503A activity in young healthy men. Clin Pharmacol Ther. 1993;54:621–629. doi: 10.1038/clpt.1993.199. [DOI] [PubMed] [Google Scholar]
- 21.Funck-Brentano C, Bosco O, Jacqz-Aigrain E, Keundjian A, Jaillon P. Relation between chloroguanide bioactivation to cycloguanil and the genetically determined metabolism of mephenytoin in humans. Clin Pharmacol Ther. 1992;51:507–512. doi: 10.1038/clpt.1992.55. [DOI] [PubMed] [Google Scholar]
- 22.Partovian C, Jacqz-Aigrain E, Keundjian A, Jaillon P, Funck-Brentano C. Comparison of chlorguanide and mephenytoin for the in vivo assessment of genetically determined CYP2C19 activity in humans. Clin Pharmacol Ther. 1995;58:257–263. doi: 10.1016/0009-9236(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 23.Coller JK, Somogyi AA, Bochner F. Association between CYP2C19 genotype and proguanil oxidative polymorphism. Br J Clin Pharmacol. 1997;43:659–660. doi: 10.1046/j.1365-2125.1997.00596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster DJR, Somogyi AA, Gross AS, Bochner F. Dextromethorphan, proguanil and caffeine as phenotyping probes for polymorphism’s in drug metabolism. Proc Aust Soc Clin Exp Pharmacol Toxicol. 1994;1:43. [Google Scholar]
- 25.Hoskins JM, Shenfield GM, Gross AS. Modified high performance liquid chromatographic method to measure both dextromethorphan and proguanil for oxidative phenotyping. J Chromatogr. 1997;696:81–87. doi: 10.1016/s0378-4347(97)00225-9. [DOI] [PubMed] [Google Scholar]
- 26.de Morais SMF, Goldstein JA, Xie HG, et al. Genetic analysis of the S-mephenytoin polymorphism in Chinese population. Clin Pharmacol Ther. 1995;58:404–411. doi: 10.1016/0009-9236(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 27.Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 28.Edeki TI, Goldstein JA, de Morais SMF, et al. Genetic polymorphism of S-mephenytoin 4′-hydroxylation in African-Americans. Pharmacogenetics. 1996;6:357–360. doi: 10.1097/00008571-199608000-00009. [DOI] [PubMed] [Google Scholar]
- 29.Ieiri I, Kubota T, Urae A, et al. Pharmacokinetics of omeprazole (a substrate of CYP2C19): and comparison with two mutant alleles, CYP2C19m1 in exon 5 and CYP2C19m2 in exon 4, in Japanese subjects. Clin Pharmacol Ther. 1996;59:647–653. doi: 10.1016/S0009-9236(96)90004-1. [DOI] [PubMed] [Google Scholar]
- 30.Zhou HH, Anthony LB, Wood AJJ, Wilkinson GR. Induction of polymorphic 4′-hydroxylation of S-mephenytoin by rifampicin. Br J Clin Pharmacol. 1990;30:471–475. doi: 10.1111/j.1365-2125.1990.tb03799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurima M, Inaba T, Kalow W. Mephenytoin hydroxylase activity in human liver: inhibition by steroids. Drug Metab Dispos. 1985;13:746–749. [PubMed] [Google Scholar]
- 32.Walle T, Fagan TC, Walle UK, Topmiller MJ. Stimulatory as well as inhibitory effects of ethinylestradiol on the metabolic clearances of propranolol in young women. Br J Clin Pharmacol. 1996;41:305–309. doi: 10.1046/j.1365-2125.1996.03097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abernethy DR, Greenblatt DJ, Divoll M, Arendt R. Impairment of diazepam clearance with low-dose oral contraceptive steroid therapy. Clin Pharmacol Ther. 1984;35:360–366. [Google Scholar]
- 34.Abernethy DR, Greenblatt DJ, Shader RI. Imipramine disposition in users of oral contraceptive steroids. Clin Pharmacol Ther. 1984;35:792–797. doi: 10.1038/clpt.1984.114. [DOI] [PubMed] [Google Scholar]
- 35.Wangboonskul J, White NJ, Nosten F, ter Kuile F, Moody RR, Taylor RB. Single dose pharmacokinetics of proguanil and its metabolites in pregnancy. Eur J Clin Pharmacol. 1993;44:247–251. doi: 10.1007/BF00271366. [DOI] [PubMed] [Google Scholar]
- 36.Helsby NA, Ward SA, Edwards G, Howells RE, Breckenridge AM. The pharmacokinetics and activation of proguanil in man: consequences of variability in drug metabolism. Br J Clin Pharmacol. 1990;30:593–598. doi: 10.1111/j.1365-2125.1990.tb03818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussein Z, Eaves CJ, Hutchinson DB, Canfield CJ. Population pharmacokinetics of proguanil in patients with acute P. falciparum malaria after combined therapy with atovaquone. Br J Clin Pharmacol. 1996;42:589–597. doi: 10.1111/j.1365-2125.1996.tb00114.x. [DOI] [PubMed] [Google Scholar]


