Abstract
Ecteinascidin-743 (ET-743) is a tetrahydroisoquinoline alkaloid isolated from the tunicate Ecteinascidia turbinata currently under phase II clinical trials for its potent anticancer activity. ET-743 binds DNA in the minor groove and forms covalent adducts with some sequence specificity. It selectively inhibits in vitro binding of the CCAAT box factor NF-Y. In this study, we assayed ET-743 function in vivo on the HSP70 promoter. On heat induction, the drug blocks transcription rapidly at pharmacological concentrations and in a CCAAT-dependent manner, whereas the activity of the CCAAT-less simian virus 40 promoter is not affected. The effect is exerted at the mRNA level. The distamycin-like alkylating tallimustine is inactive in these assays. Binding of NF-Y and of the heat-shock factor is normal in ET-743-treated cells. Run-on analysis of several endogenous genes further proves that the drug has rapid, profound, and selective negative effects on transcription. Thus, this marine-derived compound is a promoter-specific, transcription-interfering agent.
Keywords: drug, DNA binding, transcription
Ecteinascidin-743 (ET-743) is a marine tetrahydroisoquinoline alkaloid isolated from the tunicate Ecteinascidia turbinata with a potent cytotoxic activity against a variety of tumor cell lines in vitro and against several rodent tumors and human tumor xenografts in vivo (1). ET-743 is under phase II clinical investigation in Europe and the U.S. as a potential anticancer drug (2, 3). Its primary mode of action is poorly understood. ET-743 binds to the minor groove of DNA with some degree of sequence specificity (4), forming covalent adducts by reacting with the N-2 of guanine to its carbinolamine moiety (refs. 5–7; reviewed in ref. 8). At high concentrations, ET-743 (and the related synthetic drug phthalascidin) targets DNA topoisomerase I in vivo (9, 10).
In theory, all DNA-binding drugs could interfere with crucial cellular functions, such as DNA repair, replication, and transcription (8). Transcription is regulated by promoter and enhancer elements recognized by gene-specific DNA-binding proteins, by general transcription factors, and, at a higher level, by chromatin structures (11). Impairment of the complex interactions between activators and their DNA targets could lead to a change in the pattern of gene expression. Interference at this level by alkylating drugs might help explain their profound biological functions. Minor groove binders, such as the distamycins and CC-1065, have a high affinity for AT-rich DNA sequences and were reported to inhibit DNA binding of sequence-specific factors involved in transcriptional activation (12–19). We initially tested whether ET-743 was able to inhibit the binding of several known transcription factors (19); most of them were not inhibited, even at very high concentrations, up to 300 μM. Only SRF/TCF and NF-Y showed some level of sensitivity to the drug, albeit at suprapharmacologic concentrations. Interestingly, preincubation of ET-743 with NF-Y yielded inhibition of DNA binding at lower concentrations compared with preincubating the drug with DNA, suggesting that the protein rather than the CCAAT box is a target.
Several considerations have since led us to focus our attention on NF-Y. NF-Y activates the CCAAT element, present in 25% of eukaryotic promoters (20), including many regulated during the cell cycle (21). NF-Y is composed of three different subunits, NF-YA, NF-YB, and NF-YC, all of which are necessary for DNA binding. NF-YB and NF-YC are related to core histones, sharing a histone fold motif. NF-YB–NF-YC dimerization is a prerequisite for NF-YA association and subsequent DNA binding. Methylation interference and IC-substitution mutations on the target CCAAT sequence suggest that NF-Y makes important contacts in the minor groove (17). Two unrelated cytotoxic drugs (i.e., genistein and HMN-154) inhibit in vitro binding of NF-Y to the CCAAT box by targeting the NF-YB subunit (22, 23). Whether their antiproliferative effects are related to inhibition of NF-Y activity remains to be elucidated. Finally, ET-743 is produced by a marine tunicate, presumably as an antifeedant or antimicrobial agent for protection in its natural environment. NF-Y proteins are among the most conserved throughout evolution and are found in distantly related phyla: vertebrates, invertebrates, plants, fungi, and parasites. In fungi, for example, deletion of NF-Y genes causes a phenotype of slow growth (24). For the above reasons, we decided to set up an in vivo approach to understand whether NF-Y-mediated transcriptional activation is a possible pharmacological target of ET-743.
Materials and Methods
Drugs and Cell Culture.
ET-743, ET-14, and tallimustine were prepared as a 1-mg/ml stock solution dissolved in ethanol and kept at −20°C. Before use, the drugs were diluted freshly in double-distilled sterile water to the desired concentrations. Mouse NIH 3T3 fibroblasts were maintained in DMEM supplemented with 10% (vol/vol) FCS. Cells were cotransfected with 10 μg of HSP70, HSP70Ym, chloramphenicol acetyltransferase (CAT; a kind gift of N. Landsberger, University of Insubria, Varese, Italy), or the simian virus 40-containing NβGal plasmids and 2 μg of a plasmid containing the hygromycin-resistance gene. For extract preparation, cells were washed in PBS, resuspended in 100 mM Tris⋅HCl (pH 7.8), sonicated, and controlled for protein concentrations by the Bradford reagent (Sigma). CAT assays were carried out with equivalent amounts of extracts by the quantitative method described in ref. 25. β-Galactosidase activity was measured in 60 mM Na2HPO4, 40 mM NaH2PO4, 1 mM MgCl2, 50 mM β-ME, and 0.66 mg/ml O-nitrophenyl β-d-galactopyranoside. The reaction was stopped by adding 1 M Na2CO3, and the A420 was measured. Results from four to six independent experiments were plotted; standard deviations were <20%.
Electrophoretic Mobility-Shift Assay.
Nuclear extracts were prepared according to the method described in ref. 26. Electrophoretic mobility-shift assays were performed as described in refs. 18 and 19. Nuclear extracts (4 μg) were incubated for 15 min at room temperature in the presence of 300 ng of poly(dI-dC) with the Y box oligonucleotide or with an oligonucleotide containing the Xenopus HSP70 proximal heat-shock element (HSE; 5′-ACGAAATGGAAGCCTCGGGAAACTTCGGGTCGG).
Run-On Assays.
Run-on transcription on isolated NIH 3T3 nuclei was performed by treating cells with 30 nM of ET-743 for 2 and 4 h. Isolation of nuclei from cells, in vitro labeling of nascent mRNA with [α-32P]UTP, and isolation of mRNA were carried out according to the methods described in ref. 27. Plasmids (5 μg) containing the indicated cDNAs were spotted onto nitrocellulose, which was then hybridized with labeled RNAs and washed as described (27). Autoradiographs of the filters were quantified by densitometry with a Molecular Dynamics Laser Densitometer and imagequant software.
Results
Transcriptional Interference Effect of ET-743 on the Transfected HSP70 Promoter.
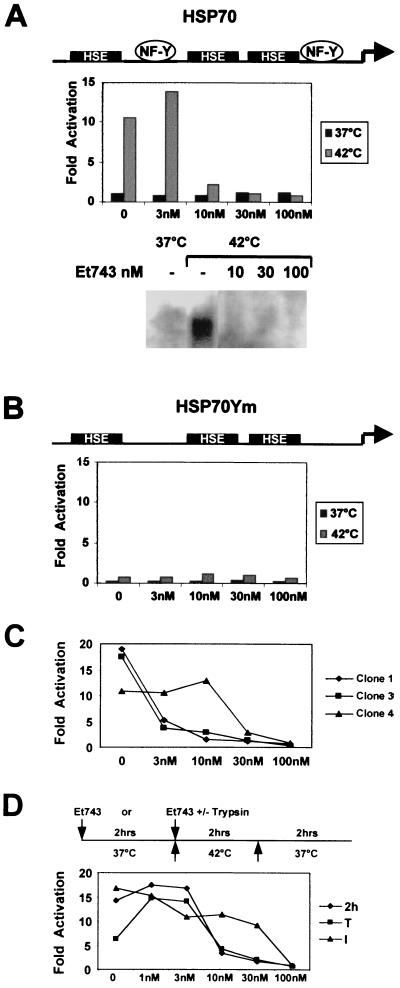
The structure of ET-743 is shown in Fig. 1. We chose the Xenopus HSP70 promoter for the following reasons: (i) it has few important elements, consisting of three HSEs binding to the heat-shock factor (HSF; refs. 28 and 29) and two CCAAT boxes activated by NF-Y (30, 31); (ii) it is a very efficient and rapidly inducible promoter, enabling evaluation of a direct role of the drug in activation; and (iii) information about the role of NF-Y in chromatin configuration and interactions with coactivators is available (30, 31). We cotransfected mouse NIH 3T3 fibroblasts with constructs containing a reporter CAT gene driven either by a wild-type HSP70 promoter or by a promoter mutated in the two CCAAT boxes (see Fig. 2 A and B), together with a vector containing the hygromycin-resistance gene. A pool of more than 100 clones was selected as well as several individual clones. The 3T3-HSP70 and 3T3-HSP70Ym cells were grown and induced at 42°C for 2 h to verify the function and inducibility on heat treatment of the integrated CAT genes. The 3T3-HSP70 cells, but not 3T3-HSP70Ym cells, showed a 10- to 15-fold induction of CAT activity (see below). We therefore tested ET-743 in this system. First, we incubated increasing concentrations of ET-743 with 3T3-HSP70 or 3T3-HSP70Ym for 2 h at 37°C, followed by a heat shock of 2 h at 42°C and subsequent recovery at 37°C. Cells were then harvested, and CAT activity was measured. A clear dose-dependent negative effect of ET-743 was evident with 3T3-HSP70 already at 10 nM concentration (Fig. 2A); Northern blot analysis indicated that the effect is indeed at the level of mRNA (Fig. 2A). With 3T3-HSP70Ym cells, only a small level of induction was observed, consistent with previous results (30, 31); essentially, this level was unaffected by ET-743 treatment (Fig. 2B). We then performed this type of experiment with three individual clones of 3T3-HSP70. Results were similar to those of the pooled cells, but the ET-743 concentrations required to inhibit activity were somewhat more variable among individual clones (Fig. 2C). Clones 1 and 3 were inhibited significantly at 3 nM, whereas clone 4 required 30 nM. On individual clones of 3T3-HSP70Ym, no effect of the drug was observed (not shown). These results suggest that the site or sites of integration play some role in ET-743 sensitivity. Induction of the HSP70 promoter is known to be very rapid and is already detectable after 2 min (28, 29). To verify the rapidity of the ET-743 inhibitory response, we modified the experimental conditions, eliminating the 2-h preincubation period and adding the drug immediately before placing plates at 42°C either to cells attached to the plastic (Fig. 2D, see I) or to trypsinized cells (Fig. 2D, see T). The former protocol required a 100-nM dose, whereas the latter showed little difference in the inhibition profile compared with the 2-h pretreatment (Fig. 2D, 2h). Thus, the effect of the drug is clearly dose-dependent, exerted at the level of mRNA production, and visible within minutes from addition.
Figure 1.
Structures of Et-743 and tallimustine.
Figure 2.
Inhibition of Xenopus HSP70 promoter activity by ET-743. (A Top) The scheme depicts the Xenopus HSP70 promoter used to obtain stable cell lines (28). (A Middle) Dose response of ET-743 on the 3T3-HSP70 pool; black bars are CAT activities in noninduced cells; gray bars represent CAT activities after heat induction. ET-743 was added to exponentially growing cells 2 h before the 2-h shock at 42°C; cells were then placed for 2 h at 37°C to recover. Values are calculated as fold of activation over basal, non-heat-shocked conditions. (A Bottom) Northern blot analysis of CAT mRNA of 3T3-HSP70 cells extracted from untreated cells kept at 37°C (lane 1) and at 1 h after heat induction either without (lane 2) or with pretreatments for 2 h with the indicated concentrations of ET-743 (lanes 3–5). (B) Same conditions as described for A, except that the 3T3-HSP70Ym pool was used. (C) Dose response of ET-743 from experiments similar to the ones described for A and B, except that three individual clones of HSP70 were used. (D) Comparison between the inhibitory curves of ET-743 on the 3T3-HSP70 pool. Cells were pretreated for 2 h as described for A (2h), pretreated for 1 min and then placed at 42°C (I), or trypsinized before adding ET-743 and then immediately (within 1 min) placed at 42°C (T).
Specificity of the ET-743 Effect.
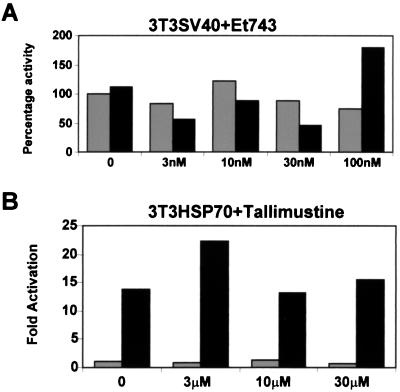
To verify the specificity of the drug for the HSP70 promoter, we selected a pool of NIH 3T3 cells in which the CCAAT-less simian virus 40 promoter-enhancer driving a β-galactosidase reporter gene was stably introduced. After standard treatments, we observed that the simian virus 40 transcription unit was not affected by the presence or absence of ET-743, either at 37°C or after a 42°C shock, determining that the negative effect of the drug could not be unmasked under these conditions (Fig. 3A). Next, we used the cytotoxic agent tallimustine on 3T3-HSP70 cells; tallimustine shares some features in common with ET-743, including the capacity to bind in the minor groove, to alkylate DNA (32), and to inhibit NF-Y binding (17). Results of these experiments are shown in Fig. 3B; neither basal nor heat-induced transcription of the 3T3-HSP70 pool is influenced significantly, even at a high cytotoxic drug concentration (30 μM). An ET-743 analog, ET-14, also showed no inhibition of heat-mediated transcriptional activation (not shown). Taken together, these data strongly indicate that the ET-743 effect does not occur by virtue of widespread Pol II inhibition. This effect is different from that seen with another minor groove binder, which is inactive in transcriptional interference.
Figure 3.
Specificity of the ET-743-HSP70 effect. (A) A pool of 3T3-simian virus 40 cells were treated with the indicated amounts of ET-743 for 2 h and then either kept at 37°C or heat treated at 42°C. Extracts were prepared after 2 h and assayed for β-galactosidase activity (23). Results are plotted as percentage of untreated cells. Gray bars, 37°C; black bars, 42°C heat shock. (B) Dose response of tallimustine on the 3T3-HSP70 pool; the indicated amounts of the drug were added 2 h before the heat treatment. Gray bars, 37°C; black bars, activities after heat shock.
Function of DNA-Binding Activators of HSP70 Transcription in ET-743-Treated Cells.
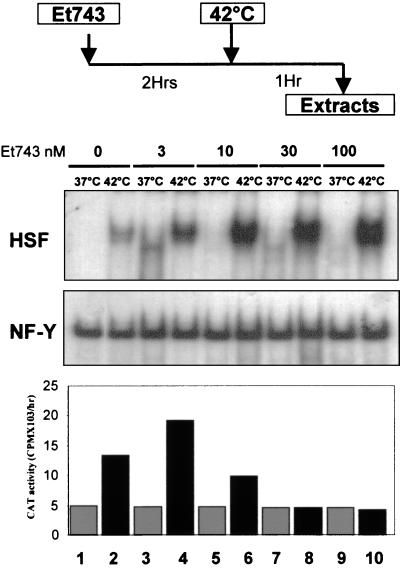
A possible explanation for the lack of HSP70 induction is that the drug negatively affects NF-Y-or HSF-binding capacity (28–31). In particular, HSF is known to exist in non-DNA-binding monomeric forms, and only after increasing the temperature, are trimerization and efficient DNA binding achieved. The previous results thus could be explained by assuming that ET-743 inhibits HSF trimerization and/or HSE binding. After a 2-h incubation with increasing ET-743 concentrations, NIH 3T3 cell lines were shocked for 1 h, after which nuclear extracts were prepared. As expected, HSF binding was present only in heat-shocked extracts (Fig. 4, lanes 1 and 2), increasing 5-fold in ET-743-treated cells in a dose-dependent manner and reaching a maximum at 100 nM (Fig. 4, compare lanes 2 with 4, 6, 8, and 10). NF-Y-binding activity was unchanged, whether the cells were heat-shocked (Fig. 4, lanes 2, 4, 6, 8, and 10) or kept at 37°C (Fig. 4, lanes 1, 3, 5, 7, and 9). The corresponding promoter activity showed the usual inhibition of heat-shock induction, even after only a 1-h incubation at 42°C and in the absence of a 2-h recovery period (Fig. 4 Bottom). These results rule out negative effects on HSF trimerization and suggest that lack of DNA binding by the HSP70 activators NF-Y and HSF might be not to be a contributing reason for ET-743 promoter inhibition.
Figure 4.
DNA-binding activities of HSP70 activators in ET-743-treated cells. Extracts were prepared according to the protocol outlined (Top; ref. 26). Nuclear extracts were assayed in the electrophoretic mobility-shift assays with HSE and Y box probes (Middle), and the corresponding CAT activities in the cytoplasmic extracts are shown (Bottom). Even lanes are extracts from heat-shocked cells; uneven lanes are extracts from cells kept at 37°C.
Effects of ET-743 on Endogenous Genes.
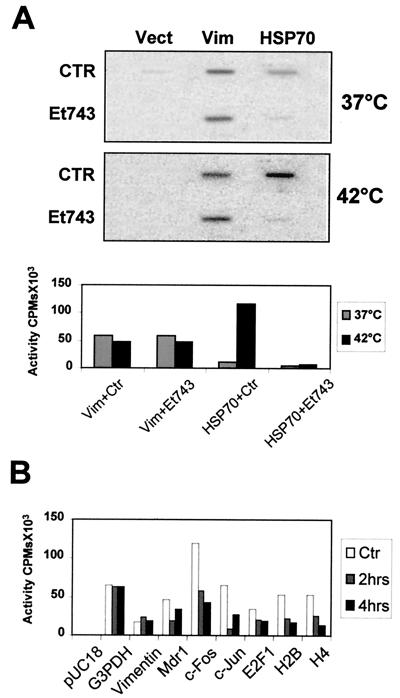
To ascertain whether ET-743 has direct transcriptional effects at pharmacological concentrations on endogenous genes, we performed run-on analysis on isolated NIH 3T3 nuclei. First, we analyzed the transcription rates of mouse HSP70 under basal and induced conditions. A 2-h treatment at 30 nM concentration with ET-743 clearly inhibited gene activation and had no effect on vimentin (Fig. 5A). Next, we decided to extend these observations to other endogenous genes. Fig. 5B shows that short 2- and 4-h treatments induce differential effects on the transcriptional activity of various promoters. Glyceraldehyde-3-phosphate dehydrogenase and vimentin are essentially unchanged; MDR1 and E2F1 are somewhat inhibited, and the rates of c-jun, of histone H2B, of histone H4, all of which contain functionally important CCAAT-boxes, and of c-fos, a CCAAT-less promoter, are affected severely, by 3- to 7-fold. Overall, these in vivo data fully support the hypothesis that ET-743 is not a general Pol II inhibitor but rather has profound, primary, and specific negative effects on gene expression.
Figure 5.
Run-on analysis of endogenous genes. (A) Nuclear run-ons of untreated and heat-shocked NIH 3T3 nuclei. ET-743 (30 nM) was added 2 h before the heat shock, and RNA was extracted after 1 h (27). Three probes were spotted on the filter: vector control (Vect), vimentin (Vim), and HSP70. Quantitation is shown. (B) Same conditions as described for A, except that the indicated cDNAs were analyzed at 37°C; NIH 3T3 cells were pretreated with 30 nM of ET-743 for 2 and 4 h before RNA run-on analysis. G3PDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
We approached the problem of finding the mechanism or mechanisms of action of the cytotoxic agent ET-743. From our original in vitro observation of selective negative effects on some transcription factors to the their DNA targets, we proceeded to investigate the role of ET-743 in vivo. Our experiments on stable transfections of the HSP70 promoter strongly suggest that ET-743 has a specific negative effect on transcription, which is not shared by another minor groove binder alkylating DNA, and that for this activity, the CCAAT-box is required. Two observations strongly indicate that the effect on transcription is primary: (i) the inhibition is extremely rapid, because a 1-min preincubation is sufficient to observe a significant degree of inhibition, and (ii) the effect is observed at pharmacological concentrations. It should be noted that the doses used in this study are within the range achieved in the plasma of patients undergoing clinical trials. To our knowledge, our study is the first demonstration that a cytotoxic alkylating agent is active at this level. Many anticancer drugs seem to influence gene expression by acting on specific promoters: cisplatin on cyclin A, doxorubicin on Id2A, taxol on IL-8, and genistein on GRP78 and HSP70 (33–36). It should be noted, however, that the conditions used in these studies involved long incubation times (24–36 h) at drug concentrations in the high micromolar range, one to three orders of magnitude higher than concentration levels showing cytotoxic effects. This proviso raises the possibility that transcriptional effects might be secondary to impairment of other well known functions: inhibition of microtubule depolymerization by taxol, of protein tyrosine kinases by genistein, and of DNA synthesis by cisplatin, as well as DNA topoisomerase II poisoning by doxorubicin (reviewed in ref. 37).
A recent report by Takebayashi et al. (9) suggests that DNA topoisomerase I is a molecular target of ET-743 in CEM cells at high (10 μM) concentrations. A parallel study by Martinez et al. (10) supports this finding with one caveat: because of the high concentration levels (4 μM in A375 cells), poisoning of topoisomerase I is very unlikely to be the primary effect of ET-743 or of the related synthetic drug phthalascidin, especially because camptothecin-resistant cells retain sensitivity to the two compounds. Our study shows that induction of HSP70 and of some cellular genes is prevented in vivo at concentrations two to three orders of magnitude lower, thus suggesting that poisoning of gene-specific activators is targeted. The two prominent actors in HSP70 are NF-Y and HSF. The drug does not prevent formation of high-affinity HSF trimers (Fig. 4); however, the possibility exists that a DNA-bound HSF could be unable to activate, because uncoupling of the two functions has been reported (38). In an accompanying paper, Scotto and colleagues (39) describe a similar activity for ET-743 evaluated in a different experimental system: cells are the colon carcinoma SW620; the promoter is the NF-Y-dependent human MDR1; the inducers are trichostatin A, UV irradiation, and butyrate; however, the inhibitory concentrations are equally low (10–50 nM). Because the HSP70 and MDR1 promoters have different architectures and because the only obvious common feature is the presence of a CCAAT box on which NF-Y acts (40), it is tempting to speculate that NF-Y is one of the intended targets.
If, indeed, NF-Y is one of the targets, what might be the mechanism or mechanisms? Several lines of reasoning suggest that inhibition of DNA binding is most likely not the reason for the rapid transcriptional inhibition. First, inhibition of NF-Y binding by ET-743 in vitro is seen at very high doses (10 and 30 μM) and actually at higher concentrations when the drug is preincubated with the DNA than when it is incubated with NF-Y (19). Second, nuclear extracts from cells treated in vivo yielded normal NF-Y and actually increased HSF binding, ruling out a generalized negative effect on HSP70 activators. Third, in vivo footprinting analysis of mammalian HSP70 promoters proved that CCAAT boxes are protected—that is, bound by NF-Y- even before heat induction (41, 42) and, indeed, a necessary condition for presetting an active promoter (26). Therefore, we consider it unlikely that NF-Y, stably bound to high-affinity CCAAT boxes, might be displaced rapidly by the low concentrations of ET-743 used in this study. However, two alternative, but not mutually exclusive models could be envisaged to explain the molecular mechanisms of drug action. (i) The apparent necessity for the C ring, which protrudes out from the minor groove in the NMR drug-DNA adducts, suggests that ET-743 might not interfere with DNA binding per se but rather modify the delicate network of neighboring interactions by binding to sites close to transcriptional activators. In this regard, as a consequence of drug binding, DNA is bent toward the minor groove (8). This bending could well change the overall three-dimensional structure of a promoter, and we note that indeed the capacity of NF-Y to induce large directed bends is most likely important for its promoter-organizing activities (43, 44). (ii) In the framework of the many crucial protein–protein interactions known to be essential to activate a given promoter, other levels could be involved that do not depend on the DNA-binding activity of the drug with neighboring activators, coactivators, or the Pol II holoenzyme. For consideration, ET-743 might prevent association of histone acetylases that are normally required for full promoter function. In this respect, we note that, on HSP70, NF-Y recruits and is acetylated by CBP, a coactivator with histone acetylase activity (26), and that, on MDR1, NF-Y recruits P/CAF, another histone acetylase (40).
To explain the antiproliferative activity of ET-743, putative targets might be cell-cycle-regulated promoters, many harboring essential NF-Y sites: genes involved in DNA metabolism and/or genes coding for cell-cycle regulators (21). Within the trimeric complex, NF-YA is modulated during the cell cycle, leading to variations in the CCAAT-binding activity (45). A crippled NF-Y would probably hamper proper regulation of these promoters, delaying transcription of crucial genes and affecting the timely progression through the cell cycle. Interestingly, we have recently obtained evidence that ET-743 causes a remarkable delay in S phase progression of the cell cycle, eventually resulting in a G2/M block (E. Erba, G.F., and M.D.'I., unpublished work). Finally, it should be emphasized that NF-Y might just be one of the possible transcription factors involved. Our in vitro DNA-binding inhibition analysis of transcription factors also pointed to SRF, an important activator of the c-fos gene, whose expression is indeed inhibited in the run-on experiments described herein. The assays developed in this study will allow testing of this hypothesis with other promoters and transcriptional activators.
Acknowledgments
We are grateful to K. Scotto for communicating results before publication and for many helpful discussions. S.M. is a recipient of a Fondazione Italiana Ricerca sul Cancro fellowship. We thank N. Landsberger and V. Zimarino for gifts of materials and B. Yen for critically reading the manuscript. This work was supported by grants from Ministero dell'Universitá e della Ricerca Scientifica e Tecnologica to R.M. and from Associazione Italiana per la Ricerca sul Cancro (Italy) to R.M. and M.D.'I.
Abbreviations
- ET-743
ecteinascidin-743
- CAT
chloramphenicol acetyltransferase
- HSE
heat-shock element
- HSF
heat-shock factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rinehart K L, Holt T G, Fregeau N L, Keifer P A, Wilson G R, Perun T J, Jr, Sakai R, Thompson A G, Stroh J G, Shield L S, et al. J Nat Prod. 1990;53:771–792. doi: 10.1021/np50070a001. [DOI] [PubMed] [Google Scholar]
- 2.Guan Y, Sakai R, Rinheart K L, Wang A H. J Biomol Struct Dyn. 1993;10:793–818. [PubMed] [Google Scholar]
- 3.Izbicka E, Lawrence R, Raymond E, Echkardt G, Faircloth G, Jimeno J, Clarke G, Von Hoff D D. Ann Oncol. 1998;9:981–987. doi: 10.1023/A:1008224322396. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn K W. Biochemistry. 1996;35:13303–13309. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- 5.Sakai R, Rinehart K L, Guan Y, Wang A H. Proc Natl Acad Sci USA. 1992;89:11456–11460. doi: 10.1073/pnas.89.23.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore B M, Seaman F C, Hurley L H. J Am Chem Soc. 1997;119:5475–5476. [Google Scholar]
- 7.Seaman F C, Hurley L H. J Am Chem Soc. 1998;120:13028–13041. [Google Scholar]
- 8.Zewail-Foote M, Hurley L H. Anticancer Drug Des. 1999;14:1–9. [PubMed] [Google Scholar]
- 9.Takebayashi Y, Pourquier P, Yoshida A, Kohlhagen G, Pommier Y. Proc Natl Acad Sci USA. 1999;96:7196–7201. doi: 10.1073/pnas.96.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez E J, Owa T, Schreiber S L, Corey E J. Proc Natl Acad Sci USA. 1999;96:3496–3501. doi: 10.1073/pnas.96.7.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:745–779. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 12.Broggini M, Ponti M, Ottolenghi S, D'Incalci M, Mongelli N, Mantovani R. Nucleic Acids Res. 1989;17:1051–1059. doi: 10.1093/nar/17.3.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn A, Affolter M, Muller M, Gehring W J, Leupin W. EMBO J. 1992;11:279–286. doi: 10.1002/j.1460-2075.1992.tb05050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun D, Hurley L H. Gene. 1994;149:165–172. doi: 10.1016/0378-1119(94)90425-1. [DOI] [PubMed] [Google Scholar]
- 15.Chiang S Y, Welch J, Rauscher F J, Beerman T A. Biochemistry. 1994;33:7033–7040. doi: 10.1021/bi00189a003. [DOI] [PubMed] [Google Scholar]
- 16.Broggini M, D'Incalci M. Anticancer Drug Des. 1994;9:373–377. [PubMed] [Google Scholar]
- 17.Ronchi A, Bellorini M, Mongelli N, Mantovani R. Nucleic Acids Res. 1995;23:4565–4572. doi: 10.1093/nar/23.22.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellorini M, Moncollin V, D'Incalci M, Mongelli N, Mantovani R. Nucleic Acids Res. 1995;23:1657–1663. doi: 10.1093/nar/23.10.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonfanti M, Caretti G, LaValle E, Sousa Faro J M F, Faircloth G, Mantovani R, D'Incalci M. Anticancer Drug Des. 1999;14:179–186. [PubMed] [Google Scholar]
- 20.Mantovani R. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 21.Zwicker J, Muller R. Trends Genet. 1997;13:3–6. doi: 10.1016/s0168-9525(96)30112-1. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka H, Okshima N, Hidaka H. Mol Pharmacol. 1999;55:356–363. doi: 10.1124/mol.55.2.356. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, Lee A S. J Natl Cancer Inst. 1998;90:381–388. doi: 10.1093/jnci/90.5.381. [DOI] [PubMed] [Google Scholar]
- 24.Brakhage A A, Andrianopoulos A, Kato M, Davis M A, Tsukagoshi N, Hynes M J. Fungal Genet Biol. 1999;27:243–252. doi: 10.1006/fgbi.1999.1136. [DOI] [PubMed] [Google Scholar]
- 25.Seed B, Sheen J Y. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber E, Matthias P, Muller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H-S, Rishi A K, Shao Z-M, Dawson M I, Jong L, Shroot B, Reichert U, Ordonez J, Fontana J A. Cancer Res. 1996;56:5055–5062. [PubMed] [Google Scholar]
- 28.Morimoto R I, Sarge K D, Abravaya K. J Biol Chem. 1992;267:21987–21991. [PubMed] [Google Scholar]
- 29.Wu C. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 30.Landsberger N, Wollfe A P. Mol Cell Biol. 1995;15:6013–6024. doi: 10.1128/mcb.15.11.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wollfe A P. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broggini M, Coley H, Mongelli N, Grandi M, Wyatt M D, Hartley J A, D'Incalci M. Nucleic Acids Res. 1995;23:81–88. doi: 10.1093/nar/23.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spitkovski A, Schulze A, Boye B, Jansen-Durr P. Cell Growth Differ. 1997;8:699–710. [PubMed] [Google Scholar]
- 34.Kurabayashi M, Dutta S, Jeyaseelan R, Kedes L. Mol Cell Biol. 1995;15:6386–6397. doi: 10.1128/mcb.15.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee L-F, Haskill S, Mukaida N, Matsushima K, Ting J P-Y. Mol Cell Biol. 1997;17:5097–5105. doi: 10.1128/mcb.17.9.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moos P J, Fitzpatrick F A. Proc Natl Acad Sci USA. 1998;95:3896–3901. doi: 10.1073/pnas.95.7.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latchman D S. Curr Opin Biotechnol. 1997;8:713–717. doi: 10.1016/s0958-1669(97)80125-5. [DOI] [PubMed] [Google Scholar]
- 38.Jurivich D A, Sistonen L, Kroes R A, Morimoto R I. Science. 1992;255:1243–1246. doi: 10.1126/science.1546322. [DOI] [PubMed] [Google Scholar]
- 39.Jin S, Gorfajn B, Faircloth G, Scotto K W. Proc Natl Acad Sci USA. 2000;97:6775–6779. doi: 10.1073/pnas.97.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin S, Scotto K W. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abravaya K, Phillips B, Morimoto R I. Mol Cell Biol. 1991;11:586–592. doi: 10.1128/mcb.11.1.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorzowski J J, Eckerley C A, Halgren R G, Mangurten A B, Phillips B. J Biol Chem. 1995;270:26940–26949. doi: 10.1074/jbc.270.45.26940. [DOI] [PubMed] [Google Scholar]
- 43.Liberati C, Ronchi A, Lievens P, Ottolenghi S, Mantovani R. J Biol Chem. 1998;273:16880–16889. doi: 10.1074/jbc.273.27.16880. [DOI] [PubMed] [Google Scholar]
- 44.Liberati C, di Silvio A, Ottolenghi S, Mantovani R. J Mol Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- 45.Bolognese F, Wasner M, Lange-zu Dohna C, Gurtner A, Ronchi A, Muller H, Manni I, Mossner J, Piaggio G, Mantovani R, et al. Oncogene. 1999;18:1845–1853. doi: 10.1038/sj.onc.1202494. [DOI] [PubMed] [Google Scholar]