Abstract
Tomato bushy stunt virus (TBSV), a tombusvirus with a nonsegmented, plus-stranded RNA genome, codes for two essential replicase proteins. The sequence of one of the replicase proteins, namely p33, overlaps with the N-terminal domain of p92, which contains the signature motifs of RNA-dependent RNA polymerases (RdRps) in its nonoverlapping C-terminal portion. In this work, we demonstrate that both replicase proteins bind to RNA in vitro based on gel mobility shift and surface plasmon resonance measurements. We also show evidence that the binding of p33 to single-stranded RNA (ssRNA) is stronger than binding to double-stranded RNA (dsRNA), ssDNA, or dsDNA in vitro. Competition experiments with ssRNA revealed that p33 binds to a TBSV-derived sequence with higher affinity than to other nonviral ssRNA sequences. Additional studies revealed that p33 could bind to RNA in a cooperative manner. Using deletion derivatives of the Escherichia coli-expressed recombinant proteins in gel mobility shift and Northwestern assays, we demonstrate that p33 and the overlapping domain of p92, based on its sequence identity with p33, contain an arginine- and proline-rich RNA-binding motif (termed RPR, which has the sequence RPRRRP). This motif is highly conserved among tombusviruses and related carmoviruses, and it is similar to the arginine-rich motif present in the Tat trans-activator protein of human immunodeficiency virus type 1. We also find that the nonoverlapping C-terminal domain of p92 contains additional RNA-binding regions. Interestingly, the location of one of the RNA-binding domains in p92 is similar to the RNA-binding domain of the NS5B RdRp protein of hepatitis C virus.
Viral-coded replicase proteins are essential for replication of tombusviruses, similar to other positive-strand RNA viruses (20, 32, 45). Tomato bushy stunt virus (TBSV), the prototypical tombusvirus, codes for p33 and p92 replicase proteins (reviewed in reference 43), which are essential for TBSV replication (32, 45). Based on the presence of the signature motifs for RNA-dependent RNA polymerase (RdRp) within the C-terminal portion of p92, the predicted function of the p92 is to synthesize viral RNA progenies (29, 45). The function(s) of p33 in TBSV replication is currently unknown.
An interesting feature of tombusviruses is that the larger replicase protein is expressed from the genomic RNA via a ribosomal readthrough mechanism of the p33 termination codon (43, 45). Due to the expression strategy, the N-terminal portion of p92 overlaps with p33. Both p33 and p92 have been proposed to be part of the TBSV replication complexes (45). Accordingly, Western blot analysis of the partially purified TBSV RdRp preparation revealed the presence of both p33 and p92 in the transcriptionally active RdRp fractions (J. Pogany and P. D. Nagy, unpublished data).
Both replicase proteins are localized to membranous structures in infected cells—the putative sites of tombusvirus replication (8, 45). The membrane localization domains in the replicase proteins are present within the N-terminal overlapping domain (41, 42). Based on their essential roles in tombusvirus RNA replication, it is possible that both tombusvirus replicase proteins can bind to the viral RNA in infected cells. For example, the viral genomic RNA must be recruited to the viral replicase complex after translation (1, 6, 7). In addition, the viral replicase complex, which likely contains both viral- and host-coded proteins, must synthesize a complementary (minus-strand) RNA on the genomic (+) RNA. This is followed by robust plus-strand RNA synthesis utilizing the minus-strand intermediates (19). Tombusviruses also synthesize subgenomic RNAs for the expression of their 3′-proximal genes (43). In addition, tombusviruses are frequently associated with defective interfering (DI) RNAs that are generated from the genomic RNA by multiple deletions (reviewed in reference 52). Synthesis (i.e., replication) of all these RNAs requires both replicase proteins in vivo (8, 32, 45). Accordingly, a recently developed in vitro assay based on partially purified RdRp from tombusvirus-infected cells was used to demonstrate that the TBSV RdRp could synthesize cRNA on both plus- and minus-stranded TBSV templates (28). Further studies confirmed that the tombusvirus RdRp (obtained for either TBSV or the closely related cucumber necrosis virus) could bind to TBSV RNA in vitro and recognize cis-acting sequences, such as the genomic and complementary promoters (33, 34) and a replication enhancer (35), which leads to RNA transcription in vitro.
To gain insights into the functions of the tombusvirus replicase proteins, we tested their abilities to bind RNA in vitro, and we report the results in this paper. We demonstrated that both p33 and p92 of TBSV could bind to TBSV-derived RNA sequences in vitro. Gel mobility shift assays performed with a series of truncated recombinant p33 purified from Escherichia coli revealed that an arginine- and proline-rich motif (termed RPR motif), which is conserved among tombusviruses, is critical for efficient RNA binding. The corresponding region in the overlapping domain of p92 may also bind to RNA, although this has not been confirmed. The lack of confirmation is because we also found that p92 had additional RNA-binding domains within its nonoverlapping C-terminal region; therefore, mutations within the RPR motif in p92 did not abolish RNA binding. Overall, these experiments supply direct evidence that both TBSV replicase proteins bind to viral RNA, and this feature might be important for the functions of these proteins during tombusvirus infection.
MATERIALS AND METHODS
Construction of expression plasmids.
The full-length TBSV cDNA clone (T-100; generous gift of Andy White [16a]) was used to amplify the p33 open reading frame (ORF) using primers 3 and 4 (Table 1). The PCR product was kinased, digested with XmnI, and cloned into a protein expression vector pMAL-c2X (NEB) to generate expression construct p33 (Fig. 1 and Table 2). Construct p33 contains the in-frame fusion of the maltose-binding protein (MBP) and the p33 ORF. To express the p92 ORF, we used a mutant TBSV clone (pHS-175; supplied by H. Scholthof [45]), which had the amber (UAG) termination codon replaced by a tyrosine codon at the end of the p33 ORF. The mutated p92 gene was amplified via PCR using primers 3 and 5 (Table 1) and template pHS-175. The obtained PCR product was cloned into pMAL-c2x to generate construct p92 (Fig. 1 and Table 2) as described for construct p33 above. A similar strategy was used to generate construct p92C (Fig. 1) by PCR using T-100 template (16a) and primers 6 and 11 (Table 1), except that we applied XbaI for digestion of the PCR product. All other deletion constructs of p33 and p92 (see Fig. 7 and 8) were generated by the method described for p33, with the exception that the obtained PCR products were digested with EcoRI and XbaI for cloning (Table 2). The primers used to generate the expression plasmids are listed in Table 1.
TABLE 1.
List of primers used for PCR to generate expression constructs
| Primer | Nucleotide positionsa | Sequence |
|---|---|---|
| 3 | 169-186 | GAGACCATCAAGAGAATG |
| 4 | 1056-1039 | CT(A/T)TTTGACACCCAGGGA |
| 5 | 2622-2605 | TC(A/C)AGCTACGGCGGAGTCGG |
| 6 | 1057-1075 | GGAGGCCTAGTACGTCTAC |
| 10 | 1056-1039 | GCAGTCTAGACTATTTGACACCCAGGGA |
| 11 | 2622-2605 | GCAGTCTAGATCAAGCTACGGCGGAGTC |
| 48 | 616-632 | GAGGAATTCTACGCTACCCTACCTAG |
| 49 | 1477-1493 | GAGGAATTCGTCTCCGAGAGGGATAG |
| 50 | 2016-2002 | GAGTCTAGATCAAATCCCCAGATGACG |
| 75 | 706-723 | GAGGAATTCTGTCTGGTGGTTGAGCCG |
| 76 | 796-813 | GAGGAATTCACTGGGCGCCCTCGTCGA |
| 77 | 952-969 | GAGGAATTCGATGTCATATTGCCTTTG |
| 78 | 871-888 | GAGTCTAGACTATCTATTCTCTGGACTGTT |
| 79 | 1381-1399 | GAGGAATTCGCCCACCGACTTGGGTATG |
| 92 | 973-990 | GAGTCTAGACTAGACAAAACAGCATCCAAT |
| 93 | 1306-1323 | GAGGAATTCACTCCACAACCTACCAAA |
| 139 | 1624-1641 | GAGTCTAGACTAAACAGCTTTCATCAGCTT |
| 183 | 667-678 | GAGGAATTCGCACGAGCACACATGGAG |
| 184 | 727-738 | GAGTCTAGACTATTTACCCTTAAGTTCCCT |
| 383 | 820-837 | GAGGAATTCTATGCGGCAAAGATCGCA |
| 385 | 1537-1554 | GAGTCTAGACTAAGGTGCTGGGTCACCCTT |
| 664 | 2059-2076 | GAGTCTAGACTAATTCCTGCGTTCGACAAT |
| 665 | 2209-2226 | GAGTCTAGACTAGTTTCGAACCATCTTCCA |
| 777 | 1357-1371 | GCGGGCTAGCGCTGCAGCGGTTTGTGAGAAGG |
| 778 | 1319-1335 | GCGGGCTAGCAGCCGCAGCTCCAAAGGCTCCTTTG |
| 779 | 1450-1465 | GCGGGCTAGCGCTGCAGCCGTGGAGAGTCTGC |
| 780 | 1412-1428 | GCGGGCTAGCAGCCGCAGCCGCACCGCTGTAGTATG |
| 781 | 1498-1513 | GCGGGCTAGCGCTGCCTTGACTACCTTCGTAA |
| 782 | 1466-1482 | GCGGGCTAGCCGCGGAGACAGGTGTGATAT |
| 783 | 1558-1573 | GCGGGCTAGCGCTGCGGTGATTCAGCCTCGAA |
| 784 | 1520-1536 | GCGGGCTAGCAGCCGCAGCAGACGTCGATATCTTCT |
| 785 | 1582-1597 | GCGGGCTAGCGCTGCGTACAATGTGGAACTTG |
| 786 | 1550-1566 | GCGGGCTAGCGGCCTGAATCACCCGAGGTG |
| 787 | 1615-1630 | GCGGGCTAGCATGGAATCCAAGCTGA |
Position on the TBSV genomic RNA.
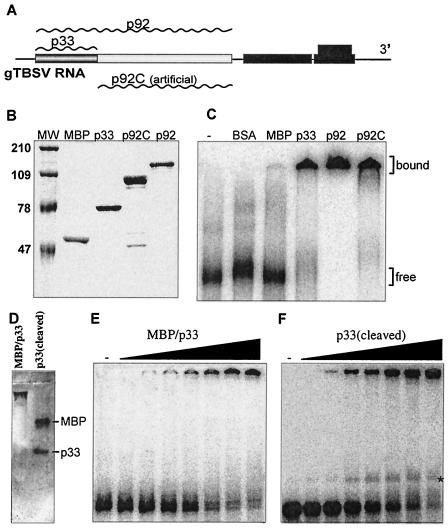
FIG. 1.
RNA binding by the recombinant p33 and p92 replicase proteins of TBSV in vitro. (A) Schematic representation of the expression strategy of the replicase genes in TBSV. The plus-strand genomic RNA is used in the infected cells to produce the replicase proteins p33 and p92 (wavy lines above the boxes represent the individual replicase proteins expressed from the TBSV RNA). p92 is translated via ribosomal readthrough of the stop codon at the end of the p33 ORF. An artificial deletion derivative of p92, termed p92C, was also generated to produce the unique, nonoverlapping portion of p92 protein, which contains the signature motifs of RNA-dependent RNA polymerases. The other genes shown in black boxes are expressed from subgenomic RNAs (43). (B) SDS-PAGE analysis of purified recombinant TBSV replicase proteins from E. coli. The p33, p92 (the stop codon at the end of p33 was modified to a tyrosine codon to ensure the production of full-length p92; reference 45), and the truncated p92C genes were cloned into pMAL-c2X and expressed as C-terminal fusion proteins with the MBP. The fusion proteins were purified using amylose resin affinity chromatography and analyzed in an SDS-10% polyacrylamide gel. Lane MW shows molecular weight markers (in thousands) on the left, while the other lanes contain samples purified from E. coli: lane 2, MBP/lacZ fusion protein; lane 3, MBP/p33; lane 4, MBP/p92C; lane 5, MBP/p92. (C) A gel mobility shift assay showing interactions between the recombinant TBSV replicase proteins and TBSV RNA. The 82-nt 32P-labeled minus-stranded region III RNA was separately incubated with bovine serum albumin and one of the recombinant proteins (1 μM) as shown, in a binding buffer at 25°C for 30 min and then analyzed in 4% nondenaturing polyacrylamide gel. The unbound, free RNA probe and the shifted (bound) RNA-protein complexes are marked on the right. (D) Mobility of the recombinant MBP/p33 and the p33 (after cleavage with factor Xa) in the absence of RNA probe in a 1% agarose gel. The electrophoresis was performed under the same conditions as in panels E and F. The proteins were stained with Coomassie brilliant blue. (E and F) Comparison of the RNA-binding abilities of two recombinant p33 preparations, which were either fused with MBP or cleaved off the MBP. The gel mobility shift assays were performed as in panel C, except that increasing amounts of MBP/p33 (0.03, 0.06, 0.13, 0.27, 0.65, 1.3, and 2.6 μM protein per lane) or recombinant p33 (cleaved) (0.03, 0.06, 0.13, 0.25, 0.50, 1.0, and 2.0 μM total protein per lane) were applied. The samples were analyzed using 1% agarose gel electrophoresis run at 100 V in a cold room. Note that the faint band located between the fully shifted (top) and free (bottom) RNA bands (marked with *) in panel F was not consistently detectable when we repeated these experiments.
TABLE 2.
List of primers and templates used for PCR to generate expression constructs
| Construct | Primersa | Template |
|---|---|---|
| p92 | 3/5 | pHS175 |
| p33 | 3/4 | T-100 |
| C1 | 48/10 | p33 |
| C2 | 183/10 | p33 |
| C3 | 75/10 | p33 |
| C4 | 76/10 | p33 |
| C5 | 383/10 | p33 |
| C6 | 77/10 | p33 |
| C7 | 183/92 | p33 |
| C8 | 48/78 | p33 |
| C9 | 48/184 | p33 |
| C10 | 75/78 | p33 |
| C11 | 76/78 | p33 |
| C12 | 383/78 | p33 |
| p92C | 6/11 | p92C |
| R3 | 6/50 | p92C |
| R5 | 93/385 | p92C |
| R6 | 93/139 | p92C |
| R7 | 79/139 | p92C |
| R8 | 79/385 | p92C |
| R9 | 79/50 | p92C |
| R10 | 49/50 | p92C |
| R11 | 49/11 | p92C |
| R12 | 661/664 | p92C |
| R13 | 661/665 | p92C |
| R14 | 661/11 | p92C |
| R15 | 93/778, 777/139 | R6 |
| R16 | 93/780, 779/139 | R6 |
| R17 | 93/782, 781/139 | R6 |
| R18 | 93/784, 783/139 | R6 |
| R19 | 93/786, 785/139 | R6 |
| R20 | 93/784, 787/139 | R6 |
| R21 | 93/782, 785/139 | R6 |
| R22 | 93/782, 787/139 | R6 |
Two separate PCRs, performed with two primer pairs, were used to generate several constructs, as described in Materials and Methods.
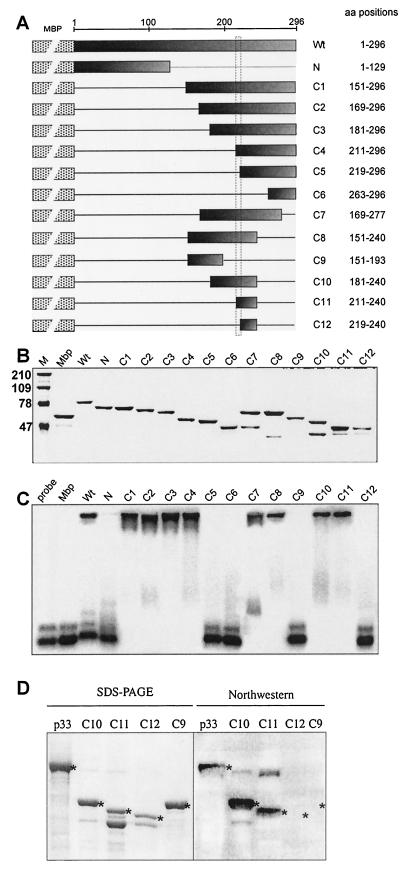
FIG.7.
Mapping the RNA-binding domain in the recombinant p33. (A) A schematic representation of the deletion derivatives of p33. The names of the constructs and the positions of the amino acids present in the truncated proteins are shown on the right. These truncated p33 proteins were expressed in E. coli as fusions to MBP (indicated schematically by a dotted box). The shaded boxes indicate the portions of p33 protein that were present in given expression constructs. The horizontal lines represent the deletions. (B) SDS-PAGE analysis of the purified recombinant proteins in a 10% polyacrylamide gel stained with Coomassie brilliant blue. The lane MW refers to molecular mass markers (in kilodaltons). (C) RNA binding activities of the truncated p33 proteins. The labeled RNA probe and the gel mobility shift assay were as described in the legend to Fig. 1B. Equimolar concentrations (2 μM) of proteins were used for the gel shift assay. (D) Northwestern analysis of selected truncated p33 proteins. The purified recombinant proteins (∼2 μg) were run in SDS-10% PAGE as shown in the left panel, transferred to a PVDF membrane, and then probed with a 32P-labeled probe [region III(-); Fig. 1B]. The positions in the Northwestern blot, which represent a particular recombinant protein, are marked with asterisks.
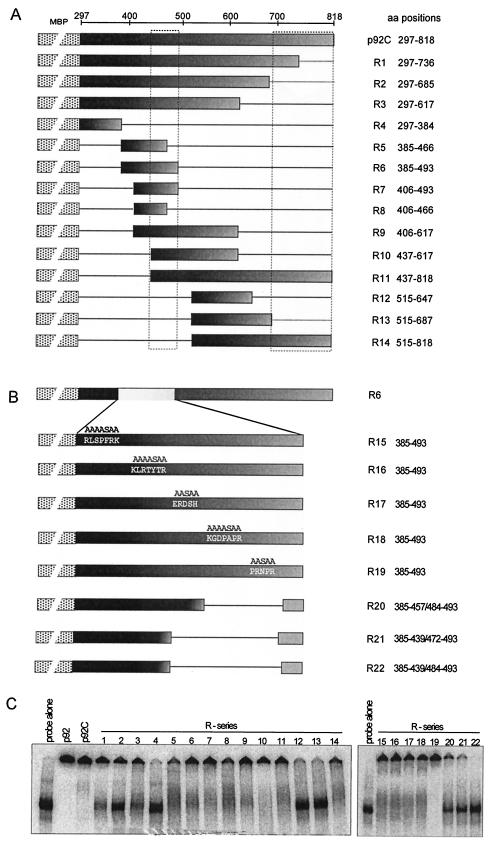
FIG. 8.
Mapping the RNA-binding domains within the unique portion of the p92 protein, termed p92C. (A) Schematic representation of the deletion derivatives of p92C. The names of the constructs and the positions of the amino acids present in the truncated proteins are shown on theright. These truncated p92C proteins were expressed in E. coli as fusions to MBP (indicated schematically by a dotted box). The shaded boxes indicate the portions of the p92C protein that were present in given expression constructs. The horizontal lines represent the deletions. (B) Schematic representation of clustered alanine-serine scanning and deletion mutagenesis of a segment in p92C to map the RNA-binding site. The alanine-serine scanning mutations were targeted at five different groups of basic amino acid clusters as shown. Expression constructs R20 to R22 were made by deleting two or more of the basic amino acid clusters as indicated by the straight lines. (C) RNA-binding activities of the truncated recombinant p92C proteins were analyzed in a standard gel mobility shift assay (refer to the legend of Fig. 1B for details). Equimolar concentrations of proteins (∼2 μM) were used in the binding assay in the presence of a 32P-labeled RNA probe [region III(-); Fig. 1B]. The samples containing particular recombinant proteins are indicated above the lanes, using the same numbering as in panels A and B above.
The expression construct N (Fig. 7) was generated by digestion of construct p33 with EcoNI, followed by religation. Constructs R1, R2, and R4 (Fig. 8) were generated by digesting the p92C clone with HindIII, BstBI-HindIII, and EcoNI-HindIII, respectively, followed by religation.
Constructs R15 to R22 (Fig. 8B) were generated using alanine-serine scanning mutagenesis via two separate PCRs for each construct carried out with the R6 template. One PCR product represented sequences that coded for the N-terminal portion of R6, while the other PCR product coded for the C-terminal portion of the R6 protein. The obtained PCR products (the primer pairs used for PCR are shown in Table 2 for R15 to R22) were digested with NheI, and the appropriate PCR products representing the modified N- and C-terminal portions of R6 were ligated together, and then reamplified by PCR. The PCR products were then digested with EcoRI and XbaI before being cloned into pMAL-c2X.
Purification of p33 and p92 proteins and their derivatives from E. coli.
The expression and purification of the recombinant TBSV proteins were carried out as described earlier for the TCV p88 replicase protein (38). Briefly, individual expression plasmids (see above) were transformed into Epicurion BL21-CodonPlus (DE3)-RIL (Stratagene). The overnight cultures from the transformed bacterial cells were diluted to 1:100 in rich growth medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl) containing 0.2% glucose and 100 μg of ampicillin per ml and grown at 37°C until the optical density reached 0.6 to 0.8. Protein expression was then induced at 14°C with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 8 to 10 h. The induced cells were harvested at 4,000 × g at 4°C for 10 min, resuspended in ice-cold column buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 25 mM NaCl, 10 mM β-mercaptoethanol), sonicated on ice to disrupt the cells, and centrifuged at 9, 000 × g for 30 min at 4°C. The supernatant was added to equilibrated amylose resin column (New England Biosciences), washed thoroughly with 20 volumes of column buffer, and eluted with 10 mM maltose in column buffer. All protein purification steps were carried out in a cold room. The purified recombinant proteins were analyzed in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) for their purity. The cleavage of MBP/p33 protein was carried out using factor Xa protease (New England Biosciences) as recommended by the manufacturer (1 μg of factor Xa was applied for 50 μg of MBP/p33). To measure the amount of purified recombinant proteins, we used the Bio-Rad protein assay, which is based on the Bradford method.
Preparation of RNA templates.
The DNA template representing the 82-bp region III of DI-72 RNA of TBSV was generated by PCR using DI-72XP as a template (51). The primers used were 253 (5′-TTGGAAATTCTCCTTAGCGAGTAAGACAGACTC-3′) and 23 (5′-GTAATACGACTCACTATAGGGACCCAACAAGAGTAACCTG-3′). The PCR template also included the T7 promoter to facilitate synthesis of RNA probes. Both labeled and unlabeled RNAs for gel mobility shift and competition experiments were prepared in vitro using T7 RNA polymerase, as described previously (26, 27). The labeled RNA probes were obtained using [32P]UTP in the T7 transcription reaction (28), followed by removal of free nucleotides using P-30 micro Bio-Spin columns (Bio-Rad). Template DNA was removed by DNase I, followed by purification of the RNA transcript with phenol-chloroform extraction and 95% ethanol precipitation. The pellet was washed with 70% ethanol to remove residual salts. The RNA transcripts were quantified by UV spectrophotometry (Beckman), followed by either 1% agarose or 5% PAGE (28).
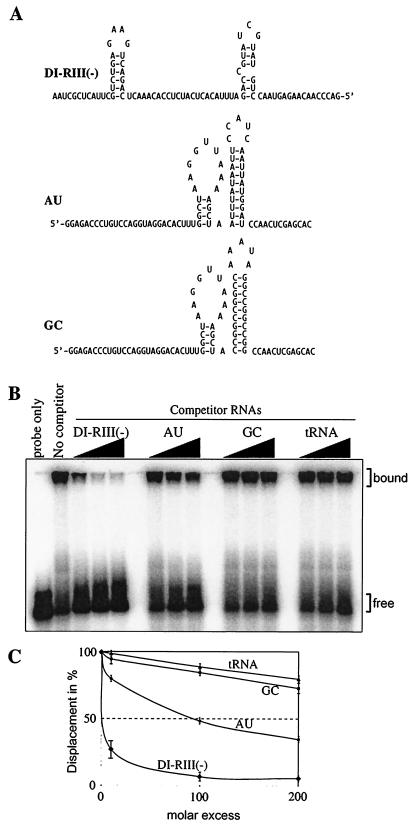
For competition experiments (see Fig. 3), we used unlabeled minus-stranded region III of DI-72 RNA. The dsRNA competitor was generated via annealing of the positive and negative strands of region III RNA. The dsDNA competitor was the PCR-amplified region III DNA fragment, while the ssDNA competitor was an artificially synthesized 51-bp oligo DNA (5′-CCC-AGACCCTCCAGCCAAAGGGTAAATGGGAAAGCCCCCCGTCCGAGGAGG-3′). RNA constructs AU and GC (Fig. 4) were obtained from Chi-Ping Cheng (10).
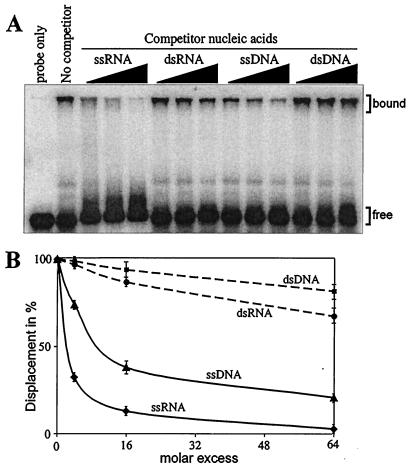
FIG. 3.
Preferential binding of the recombinant p33 to ssRNA. (A) A 1 μM concentration of purified recombinant p33 was incubated with 32P-labeled ssRNA probe, representing the 82-nt minus-stranded region III in the absence or presence of increasing amounts (in 4-, 16-, or 64-fold excess) of unlabeled competitors (as shown at the top of the figure). The experiments were repeated twice. (B) Graphical representation of data obtained in panel A. The extent of competition was quantified as the percentage of displaced labeled RNA probe from the bound complex (indicated on the right in panel A) using a phosphorimager and ImageQuant software (version 1.2).
FIG. 4.
Testing binding preference of the recombinant p33 to RNA. (A) Predicted secondary structures of the competitor RNAs, including the 82-nt minus-stranded region III of TBSV [DI-RIII(-)] and the similarly sized artificial AU and GC templates (10). Note that the double- versus single-stranded regions are comparable in length in these competitor RNAs. (B) Unlabeled competitor RNAs at increasing amounts (in 10-, 100-, or 200-fold excess) were added to the mixture containing the labeled probe (82-nt minus-stranded region III; Fig. 1B) and 1 μM purified recombinant p33 and the bound complexes were analyzed in gel mobility shift assay. The tRNA was from yeast. (C) Graphical representation of data obtained in panel B. The quantification of the experiment was done as described in the legend to Fig. 3. The experiments were repeated twice.
Gel mobility shift assay.
Approximately 1 μM protein was incubated with 5 ng of radioactively labeled minus-stranded region III RNA probe (see above) in a binding buffer (50 mM Tris-HCl [pH 8.2], 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 200 ng of yeast tRNA [Sigma], and 2 U of RNase inhibitor [Ambion]) at 25°C for 30 min (38). After the binding reaction, the samples were analyzed by either 4% nondenaturing PAGE performed at 200 V or 1% agarose gel electrophoresis run at 100 V in Tris-borate-EDTA buffer in a cold room (38). The gels were dried, exposed and analyzed in a phosphorimager, and quantified using ImageQuant version 1.2 (Amersham). For competition experiments, unlabeled competitors (applied in molar excess as indicated in Fig. 3 and 4) were added simultaneously with the labeled RNA probe to the binding reaction. The experimental binding curves were statistically fit to data using a Microsoft Excel spreadsheet.
Northwestern assay.
Approximately equal amounts (∼2 μg) of recombinant proteins were run in an SDS-10% PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes (47). The membranes were renatured at room temperature in a renaturation buffer (10 mM Tris-Hcl [pH 7.5], 1 mM EDTA, 50 mM NaCl, 0.1% Triton X-100, and 1× Denhardt's reagent [44]) with three changes of buffer for 20 min each. The membranes were probed with 32P-labeled RNA (minus-stranded region III; see above) for 1 h, washed three times with the renaturation buffer, air dried, and analyzed using a phosphorimager.
Biosensor analysis.
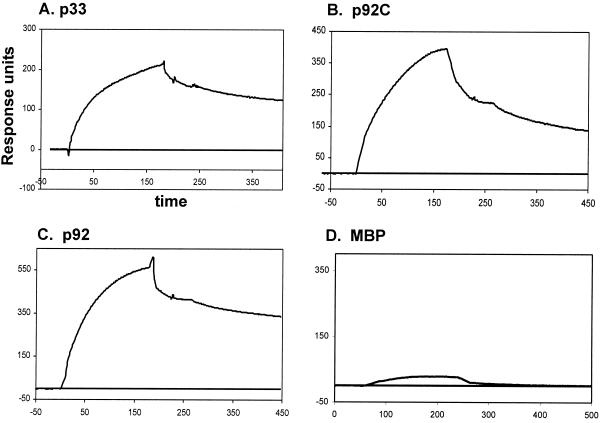
The surface plasmon resonance (SPR) experiments were carried out using BIACORE X (Biacore, Inc., Piscataway, N.J.) at 25°C as recommended by the manufacturer. Briefly, the running buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.05% surfactant P-20) was filtered and degassed every time before use. An SA sensor chip (Biacore) was used to immobilize a 20-mer RNA from the 3′ end of gTBSV RNA with a biotin label at the 5′ end (5′-UGUAACGUCUUUACGUCGGG-3′; Dharmacon, Inc.). The surface of the chip was first preconditioned with three 1-min pulses of 50 mM NaOH in 1 M NaCl. Flow cell 1 (Fc1) was used to immobilize 440 resonance units (RU) of RNA, while flow cell 2 (Fc2) was kept as a control surface to account for nonspecific binding and bulk refractive index changes upon injection of protein samples as suggested by the manufacturer. The recombinant protein samples were diluted with running buffer to 1 μM final concentration before injection. The interactions between RNA and the recombinant proteins were analyzed in real time through a sensogram (Fig. 2), in which the RUs were plotted as a function of time. One RU is equivalent to a change in adsorbed mass of 1 pg/mm2 of the sensor surface (BIA Applications Handbook; Biacore). All the data shown in Fig. 2 were corrected based on data obtained from the control (Fc2).
FIG. 2.
SPR analysis of interactions between the TBSV RNA and the recombinant replicase proteins. SPR analysis was carried out using BIACORE X (Biacore), as described in Materials and Methods. A 5′-biotinylated 20-nt RNA derived from the 3′ end of TBSV RNA (the minimal TBSV genomic promoter [34]) was immobilized (440 RU) onto a streptavidin-coated sensor chip. The purified recombinant proteins (1 μM) in a binding buffer were passed over the RNA-coated surface of the chip, and the change in mass due to interaction between RNA and protein that altered the refractive index of the medium was recorded in real time in a sensogram. The time allowed for association and dissociation phases was 180 and 200 to 260 s, respectively, in each protein-RNA interaction assay. Interactions between the target RNA and p33, p92C, p92 (all three were tested as MBP fusion proteins), and MBP are shown in panels A, B, C, and D, respectively. The sensogram data were corrected for nonspecific binding based on data obtained using a control surface (shown as the baseline), which was free of RNA. Note that the RU were not normalized; therefore, they should not be compared directly for proteins of different molecular mass. The small positive response in the MBP test (in panel D) is likely due to loose, nonspecific binding, which was subsequently washed out with the buffer—as is evident from the curve reaching the baseline rapidly during the washing step.
RESULTS
Expression, purification, and RNA binding by recombinant p33 and p92 replicase proteins.
In order to obtain sufficient amounts of soluble TBSV p33 and p92 proteins (Fig. 1A) for biochemical studies, we overexpressed them in E. coli as maltose-binding fusion proteins. This expression strategy allowed for affinity-based purification of the recombinant p33 and p92 proteins, as shown in Fig. 1B. Standard gel mobility shift experiments with a 32P-labeled RNA probe, representing the 82-nt TBSV replication enhancer, termed region III(-) (35; this probe was used in all the experiments unless stated otherwise), demonstrated that both the recombinant p33 and p92 could bind to RNA efficiently (Fig. 1C). In contrast, comparable amounts of bovine serum albumin and the full-length MBP alone (expressed and purified under the same conditions as the recombinant p33 and p92 proteins) did not bind to the RNA probe efficiently (Fig. 1C), thus ruling out the possibility that MBP or any contaminating proteins from E. coli contribute to RNA-binding activity. Interestingly, the N-terminally truncated version of p92 that included the entire unique sequence of p92 (termed p92C; Fig. 1A) also bound to RNA efficiently (Fig. 1C, lane p92C). This observation suggests that p92 contains a minimum of two RNA-binding regions—one in the overlapping region and another in the unique region (see below). Since p33 and p92 have a common RNA-binding region within the overlapping sequence, we used mostly the recombinant p33 in the experiments below (unless stated otherwise).
To test whether the presence of the N-terminal MBP fusion might affect the ability of p33 to bind to RNA and/or affect the migration of the RNA-protein complex in the gel, we compared a purified recombinant p33 preparation, which had the MBP cleaved off by protease factor Xa (sample p33 [cleaved], Fig. 1D), with the uncleaved recombinant MBP/p33 fusion protein preparation (sample MBP/p33, Fig. 1D) in a gel mobility shift assay (Fig. 1E and F). We found that the overall efficiencies of RNA binding by the recombinant p33 and MBP/p33 fusion protein were similar (Fig. 1E and F), thus suggesting that the MBP fusion does not alter the ability of p33—and likely p92—to bind to RNA. Also, the mobility of the RNA-protein complex was the same for the two preparations (Fig. 1E and F). Therefore, we used the maltose-binding fusion proteins in the following experiments.
To corroborate the results obtained from gel mobility shift analysis, we carried out SPR measurements with a Biacore biosensor as described in Materials and Methods. Briefly, the SPR provides data about real-time protein-RNA interactions (11a) by measuring the change in refractive index that takes place between the immobilized RNA and the protein that is being passed in an aqueous buffer over the surface of the chip. The refractive index of the medium changes near the chip surface due to change in mass resulting from RNA-protein interaction (11a). For this study, we fixed a 20-nt-long 5′-biotinylated TBSV RNA, which includes the minimal genomic (i.e., minus-strand initiation) promoter (34) to the surface of a streptavidin-coated chip. The recombinant p33, p92, p92C, and MBP proteins diluted with running buffer were passed separately over the immobilized RNA (Fig. 2). These experiments confirmed that p33 (Fig. 2A), p92 (Fig. 2B), and p92C (Fig. 2C) could bind efficiently and stably to the RNA, while MBP (Fig. 2D) could not. Detailed kinetic measurements on p33/p92 and RNA interactions will be published elsewhere.
Preferential binding of recombinant p33 to single-stranded RNA.
To test whether the recombinant p33 could bind only to single-stranded RNA or to other nucleic acids as well, we used various nucleic acids in template competition experiments, which were evaluated by using gel mobility shift assay. Briefly, the same amounts of 32P-labeled region III(-) RNA probe and purified recombinant p33 were used in the presence of increasing amounts of unlabeled competitors, such as single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), ssDNA, and dsDNA (Fig. 3). All these sequences were derived from the same region of the TBSV genome (minus- or double-stranded region III). These experiments demonstrated that the ssRNA was the best competitor in binding to p33, while ssDNA competed moderately well (Fig. 3B). In contrast, dsRNA and dsDNA templates were poor competitors under the experimental conditions used. Overall, the data suggest that p33 is a single-stranded nucleic acid-binding protein with the highest preference toward ssRNA.
We also tested if the recombinant p33 could preferentially bind to a TBSV-derived ssRNA sequence by using four different but comparably sized ssRNAs in competition experiments as shown in Fig. 4. One competitor was region III(-) [named DI-RIII(-); Fig. 4A], while the other three RNAs were nonviral. One of these competitor ssRNAs consisted of an AU-rich sequence (named AU; Fig. 4A) and another consisted of a GC-rich (named GC; Fig. 4A) artificial sequence (10), while the fourth RNA was tRNA. Comparison of the abilities of these RNAs to compete with the 32P-labeled ssRNA probe in binding to p33 based on the gel mobility shift experiments revealed that the region III(-) was by far the best competitor among the ssRNAs (Fig. 4B and C). The artificial AU-rich RNA was a moderately good competitor, while the artificial GC-rich RNA and the tRNA were poor competitors (Fig. 4B and C). These experiments suggest that p33 preferably binds to the TBSV-derived sequence and, at a lower efficiency, to an AU-rich sequence. Note that the artificial AU and GC competitor RNAs have double- versus single-stranded regions that are comparable in length to those present in DI-RIII(-) (Fig. 4A). Thus, increased binding by p33 to DI-RIII(-) is likely due to the favorable sequence and structure of region III(-).
Cooperative binding of p33 to ssRNA.
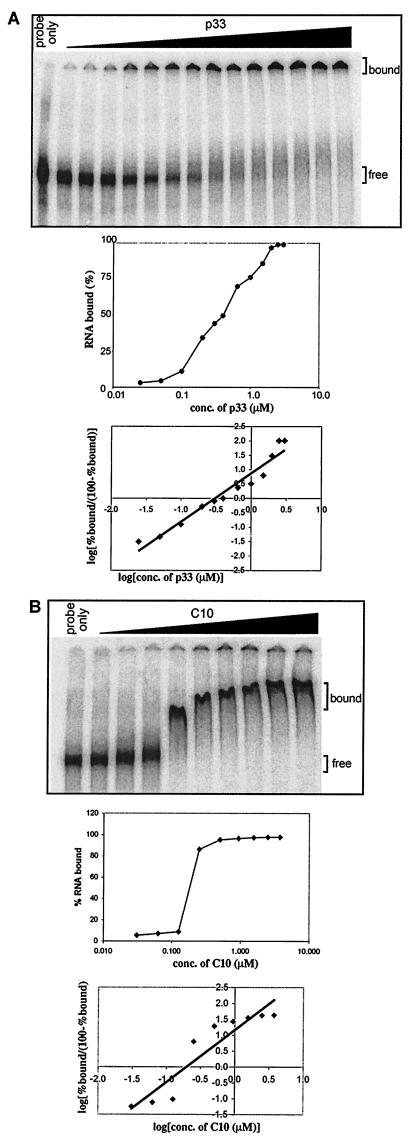
In order to characterize the binding behavior of p33 to TBSV RNA, we incubated progressively increasing amounts of recombinant p33 proteins in the presence of a 32P-labeled probe followed by gel mobility shift assay. In the presence of a small amount of p33 (Fig. 5A), the RNA probe was found mostly in unbound form, while increasing the amount of p33 in the binding reaction resulted in rapid transition of the RNA probe to bound form (samples on right side in Fig. 5A). The absence of intermediately shifted bands, resulting from limited binding of the probe by p33 between the completely bound and the free probe in the gel (Fig. 5A), suggests that most of the RNAs are either coated with p33 or are not bound to p33 at all—depending on the amount of protein present in the RNA-binding reactions. The Hill coefficient of 1.8 for RNA-binding by p33 also supports the supposition that p33 binds to RNA in a cooperative manner (Fig. 5A). Interestingly, when we used a truncated p33 containing only a 60-amino-acid segment of p33 that includes the RNA-binding domain (construct C10, Fig. 5B), then we still observed cooperative RNA binding by this truncated p33, as is clearly evident from the rapid transition from the totally free to completely bound state of probe RNA with a marginal rise in protein concentration (Fig. 5B).
FIG.5.
Cooperative RNA binding by the full-length and truncated recombinant p33. (A) Increasing molar concentrations of p33 were incubated with 32P-labeled probe [region III(-); Fig. 1B], and the RNA-protein complex was resolved in a gel mobility shift assay. The middle panel shows the semi-log plot of the percentage of RNA bound versus molar concentration of p33 determined by using a phosphorimager. The bottom panel shows a Hill plot of the experimental data obtained from the binding assay (top of panel A). To deduce the Hill coefficient, we used the points that correspond to the middle of the curve in the semi-log graph (as cooperativeness is negligible at the extremes). (B) The gel mobility shift assay was performed with a truncated recombinant p33, termed C10 (Fig. 7), which contained only a 60-amino-acid-long region, including the RNA binding site in the recombinant p33. The middle and bottom graphs were prepared as described for panel A above.
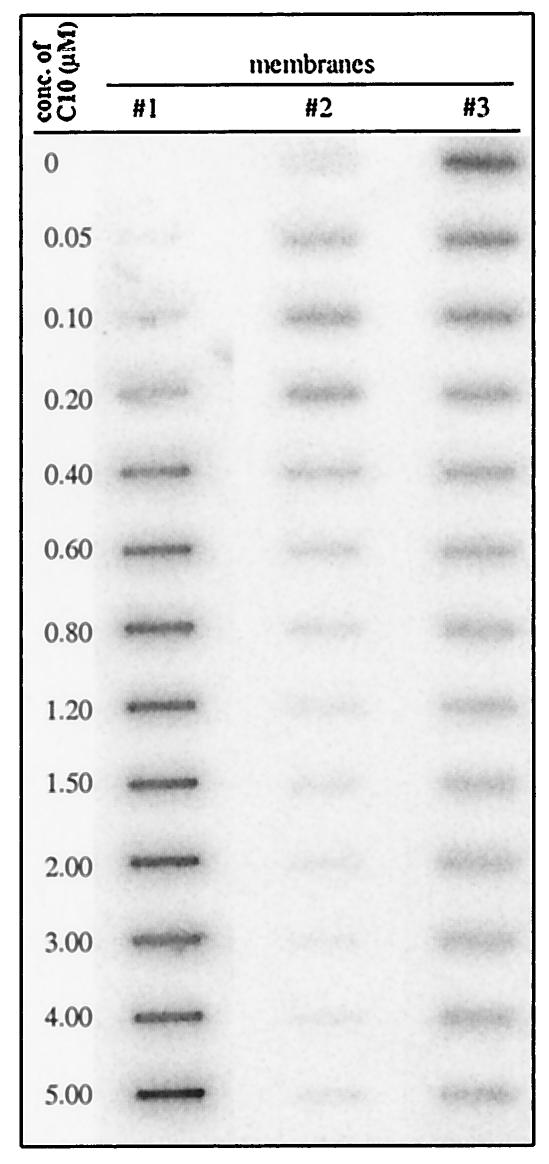
The ability of the truncated p33 to bind cooperatively to RNA was surprising (because of the small size of the protein); therefore, we also used the three-membrane sandwich method as described by Pata et al. (36) to confirm the above finding. Briefly, the first polysulfone membrane, which has low affinity to both RNA and protein, was expected to retain only large RNA-protein complexes (36). The second nitrocellulose membrane can bind to small protein-RNA complexes (i.e., those that have not been retarded on the first membrane), while the third, positively charged membrane retains all unbound RNA. Increasing amounts of C10 protein were added to the same amount of RNA probe, as described in Fig. 6. Aliquots of the binding reactions were filtered through the three-membrane sandwich, followed by detection of the retarded 32P-label on each membrane (Fig. 6). These experiments demonstrated that the RNA probe was retarded as part of a large complex in the presence of elevated amounts of C10 proteins (see membrane 1, Fig. 6), while the formation of small RNA-protein complex was relatively inefficient (Fig. 6, membrane 2). Overall, the shown data are most consistent with the model of cooperative RNA binding, which predicts that most of the RNA is present in a coated form (seen as a large complex) when a sufficient amount of the truncated p33 is present in the binding reaction. The goal of future experiments will be to analyze the structure of the p33-RNA complex.
FIG. 6.
Three-membrane sandwich experiments to demonstrate cooperative RNA binding by a truncated recombinant p33. The purified recombinant C10 protein (Fig. 7; the applied amount is shown on the left side) was incubated with 32P-labeled region III(-) RNA probe for 30 min at 25°C, followed by filtering the reaction products through a three-membrane sandwich (36) in a slot blot apparatus. The membranes were then washed twice with (300 μl) binding buffer, air dried, and analyzed using a phosphorimager. The membranes were used in the following order: polysulfone on the top (column 1), nitrocellulose in the middle (column 2), and Hybond N+ on the bottom (column 3). Note that the unbound label at the highest protein concentration likely represents labeled ribonucleotides (which, in spite of a purification step, are still present in the RNA probe as a carryover from the labeling reaction) that were unbound were to protein C10.
Mapping the RNA-binding site in p33.
In order to identify the RNA-binding domain in p33 and in the corresponding region of p92, we made a series of 13 overlapping constructs with various truncations within the p33 gene (Fig. 7A). The truncated p33 proteins were overexpressed in E. coli, followed by affinity purification of the proteins as MBP fusions (see above) and SDS-PAGE analysis (Fig. 7B). The ability of the obtained truncated p33 proteins to bind to the 32P-labeled probe was tested in gel mobility shift experiments (Fig. 7C). These experiments revealed that the C-terminal segment of p33 contains an RNA-binding domain (Fig. 7C, lane C1), while the N-terminal segment does not bind to RNA under the in vitro conditions (Fig. 7C, lane N). Further testing of C-terminally nested segments of p33 indicated that the central portion of the C-terminal region harbors the RNA-binding domain (Fig. 7C, compare lanes C2 through C4 versus C5 and C6). This was further supported by the observation that deletions of the C-terminal 20 to 56 amino acids in p33 did not affect RNA-binding (Fig. 7C, lanes C7 and C8). Deletion of 103 amino acids from the C terminus, however, abolished RNA binding (Fig. 7C, lane C9). Deletions starting from both the N and C termini confirmed that the shortest p33 derivative that still bound to RNA was 30 amino acids long and covered the central portion of the C-terminal region (Fig. 7C, lane C11). Deletion of an 8-amino-acid portion of construct C11—which has the sequence TGRPRRRP—completely abolished RNA binding (Fig. 7C, lane C12). Indeed, all the p33 derivatives that carried the above arginine- and proline-rich motif bound to the RNA probe efficiently, while those lacking this domain did not bind to the RNA probe (Fig. 7C). Based on these data, we propose that the arginine- and proline-rich motif, which we call the RPR-motif, might be the primary RNA-binding site in p33 and p92.
To confirm that the RNA binding by the truncated p33 derivatives is an inherent feature of these proteins and not due to the presence of a contaminating protein from E. coli, we analyzed the RNA-binding ability of a selected group of short-deletion mutants of p33 in a Northwestern assay (Fig. 7D). Briefly, the full-length p33 and four of the purified recombinant p33 derivatives, C9, C10, C11, and C12 (Fig. 7A), were subjected to SDS-PAGE, followed by blotting to a PVDF membrane. This was followed by probing the membrane with a 32P-labeled RNA probe as described in Materials and Methods. Importantly, we could detect the protein-RNA complex by autoradiography in the portion of the membranes that contained proteins p33 (WT), C10, and C11, but we could not detect RNA-protein complex in the case of C9 and C12 (Fig. 7D, panel Northwestern). Therefore, the Northwestern analysis confirmed the results obtained in the above gel mobility shift experiments, those being that the RPR motif is the core region in RNA binding in p33.
Mapping additional RNA-binding sites in p92.
In order to identify the RNA-binding domains present in the unique segment of p92 (Fig. 1A, p92C), we used a series of deletion derivatives of p92C that were overexpressed and purified from E. coli. Surprisingly, many of the expressed truncated p92C proteins were unstable in E. coli, thereby preventing us from generating large enough numbers of p92C derivatives that could have been useful to pinpoint the RNA-binding sites precisely (data not shown). To this end, we were able to obtain 14 truncated p92C proteins in large enough amounts suitable for biochemical assays (Fig. 8A). Nested truncations of 82 to 133 amino acids from the C terminus of p92C resulted in reduced RNA binding, thus suggesting that this portion of p92C contributes to RNA binding, although the residual RNA binding by these proteins was still significant (Fig. 8C, lanes R1 and R2). Deletion of 434 amino acids from the C-terminal end of p92C abolished RNA binding (Fig. 8C, lane R4). The p92C derivatives carrying segments from the central portion of the protein were found to bind to RNA (Fig. 8C, lanes R5 through R11). In contrast, p92C derivatives, which lacked the above central segment and the C-terminal segment, did not bind efficiently to RNA (Fig. 8C, lanes R12 and R13). Interestingly, protein R14, which contained the 303-amino-acid C-terminal segment of p92C, bound to RNA—though with reduced efficiency when compared to p92C (Fig. 8C, lane R14). Overall, analysis of RNA binding by the above deletion series of p92C revealed that two segments of p92C are involved in RNA binding: the central segment (represented by R6 and R7) and the very C-terminal segment (∼131 amino acids). These two segments can bind to the RNA independently of each other, although each shows somewhat reduced efficiency when compared to p92C.
To further delineate the ∼108-amino-acid-long central RNA-binding site in p92C, we separately introduced five clusters of five to seven alanine and serine mutations into construct R6 (Fig. 8A), as shown schematically in Fig. 8B. We targeted short regions that contained clusters of positively charged amino acids, such as arginine, lysine, and histidine (Fig. 8B, lanes R15 through R19). Surprisingly, all these mutated proteins retained their abilities to bind to RNA (Fig. 8C, lanes R15 through R19), suggesting that either (i) the selected amino acids are not involved in RNA binding or (ii) several amino acids, possibly located at different parts of the central segment of the p92C protein, are brought together by protein folding to form an RNA-binding groove—as is the case for other viral RdRps (5, 16). Therefore, it is possible that the cluster mutagenesis approach was not effective since it did not modify all the important amino acids at once. Therefore, we decided to introduce 27- to 45-amino-acid deletions into construct R6 as shown schematically for constructs R20 through R22 (Fig. 8B). These deletion derivatives bound to RNA poorly (especially protein R22—but compare lanes R20 and R21 of Fig. 8C). Overall, this analysis suggests that an RNA-binding region might be present within the 45-amino-acid-long segment of the central region in p92. Since we had unexpected difficulties in obtaining many other mutants (due to protein stability problems during expression in E. coli; see above), we could not further map the actual amino acids involved in RNA binding for the central segment in p92C using this approach.
DISCUSSION
Binding of the replicase proteins to the viral RNA is predicted to be important during many steps of the viral infectious cycle, including the recruitment of the viral RNA to the site of replication, recognition of cis-acting elements during replication, cRNA synthesis, etc. (1, 6, 7, 19). To gain insights into the replication process of tombusviruses, we have in this paper characterized the ability of p33 and p92 replicase proteins to bind to RNA. Although we found that binding of p33 to ssRNA was the strongest, ssDNA was also bound by p33 with moderate efficiency (Fig. 3). In contrast, binding to dsRNA or dsDNA was poor, suggesting that p33 is a single-stranded nucleic acid binding protein. Interestingly, the ability of p33 replicase protein to bind single-stranded nucleic acids is similar to other plant viral proteins, such as movement proteins and coat proteins (2, 3, 11-13, 18, 22, 30, 31, 39, 40, 48). In spite of its ability to bind to viral as well as nonviral RNAs, p33 showed preference for the TBSV-derived sequence, which was the best substrate among four different similar-sized ssRNA templates (Fig. 4). An artificial AU-rich template was also bound by p33 moderately well. In contrast, a GC-rich RNA and tRNA bound poorly to p33 (Fig. 4). Overall, the observed selectivity of p33 in RNA binding may not be enough for p33 (and possibly p92) to bind to only TBSV-related RNAs in infected cells. It is possible that other factors, or a combination of factors, are needed to achieve such levels of selectivity in template use.
Based on p33-RNA binding experiments, we propose that that both p33 (Fig. 5) and p92 (data not shown) bind RNA in a cooperative manner. Results (Fig. 5A) show typical “all or none” behavior characteristic of cooperative binding (15, 24, 25). The RNA bound by the recombinant p33, which was either fused to MBP (Fig. 5A) or was cleaved off the MBP domain (Fig. 1F), stayed in the well, probably due to the large size of the complex. In contrast, the p33 or p33 fused to MBP did migrate into the gel in the absence of RNA, as shown in Fig. 1D. In addition, a 60-amino-acid-long truncated p33 (Fig. 7A, protein C10) also bound to RNA in a cooperative manner (Fig. 5B). Indeed, detection of large complexes between the truncated p33 and the RNA probe in the three-membrane sandwich assay (Fig. 6) is also consistent with cooperative RNA binding by this truncated p33.
The proposed ability of p33 to bind RNA in a cooperative manner suggests that, after the initial binding by the first p33 to the RNA, subsequent binding of additional p33 molecules to the same RNA is greatly facilitated not only by the stabilizing effect coming from binding to the RNA but also by p33-p33 (protein-protein) interactions. This may lead to complete coating of the RNA with p33 molecules. In support of this model, we found p33-p33 interactions in vitro (K. S. Rajendran and P. D. Nagy, unpublished data).
Binding to RNA in a cooperative manner can be advantageous for both the viral RNA and the replicase proteins, since this may increase the stability of RNA-protein complexes inside the infected cells. Therefore, it is not surprising that many viral proteins, including 2D of poliovirus, NS5B RdRp of hepatitis C virus (HCV) (50), viral coat proteins, and plant viral movement proteins (2, 3, 11-13, 18, 22, 30, 31, 39, 40, 48), were found to bind viral RNAs in a cooperative manner. The functional significance of cooperative binding by p33 and p92 is currently not known. It is possible that p33 can coat the viral ssRNAs in infected cells, which may be beneficial during template recruitment and/or replication. The p33-coated viral RNAs may be more resistant to nucleases and less accessible to host-mediated gene silencing than are free viral RNAs (49, 53). Since cooperative binding depends on the amounts of replicase proteins and viral RNAs present in the cells, it is likely that this feature may be important for the function of p33, which is 20-fold more abundant than the p92 replicase protein in the infected cells (45). The ability to bind cooperatively to RNA, possibly in combination with p33, may also be important for the function of p92 during replication. For example, cooperative binding between p92 and p33 may facilitate recruitment of the less abundant p92 proteins to the viral RNA in infected cells. Alternatively, binding of p92 to the viral RNA templates in a cooperative manner may enhance its RdRp activity, as it has been shown for the poliovirus 2D (RdRp) protein that binds RNA cooperatively (17, 36). In addition, it has been shown that the poliovirus 2D protein forms large complexes that might be involved in virus replication (23).
Deletion analysis revealed that p33 has one high-affinity RNA-binding site in its C-terminal region. This RNA-binding region includes the RPR motif, which has been shown to be essential for RNA binding (Fig. 7C, compare proteins C11 and C12). Mutagenesis of the RPR motif in p33 has also revealed that this domain is essential for the replication of a tombusvirus and an associated DI RNA in vivo (35a). Mutations within the RPR motif have affected subgenomic RNA synthesis as well (35a). These observations demonstrate that the RPR motif plays a central role in viral RNA synthesis and metabolism.
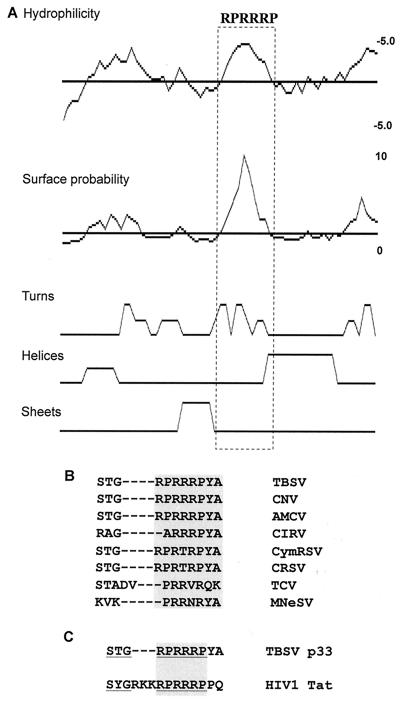
The RPR motif is highly conserved among the replicase proteins of tombusviruses and the related turnip crinkle virus (TCV) (Fig. 9B). Accordingly, a recombinant TCV p88 protein, which is the RdRp protein similar to p92 of TBSV, contains an RPR motif in its N-terminal portion and has been shown to bind RNA efficiently in vitro (38). Moreover, the RPR motif in TBSV is similar to the previously proposed ARM motif (i.e., arginine-rich motif), which is present in several viral- and host RNA-binding proteins (14), including the trans-activator protein (Tat) of human immunodeficiency virus type 1 (Fig. 9C) (4, 9). The structure of the ARM motif, however, is rather different in several RNA-binding proteins. Therefore, it is not known if the ARM motif functions similarly in different proteins (14).
FIG. 9.
Primary and secondary structure analysis of the RNA-binding region in p33. (A) SEQWeb version 1.1 from GCG Wisconsin package was used to predict the secondary structure, surface probability, and hydrophilicity of the RNA-binding region in p33. The arginine- and proline-rich motif involved in RNA binding is boxed. (B) Sequence alignment of the arginine- and proline-rich motifs present in the p33-like replicase proteins of different tombusviruses and the related proteins in carmoviruses. The conserved motif is boxed. The following abbreviations were used: CNV, cucumber necrosis virus; AMCV, Artichoke mottled crinkle virus; CIRV, Carnation Italian ringspot virus; CymRSV, Cymbidium ringspot virus; CRSV, Carnation ringspot virus (a dianthovirus); TCV, turnip crinkle virus (a carmovirus); MNeSV, maize necrotic streak virus; and HIV-1, human immunodeficiency virus type 1. (C) Alignment of the arginine- and proline-rich motif in TBSV p33 and arginine-rich motif in HIV-1 Tat, which is involved in RNA binding (4, 9). The residues important for RNA binding in the Tat protein are underlined, while the arginine-rich sequence is boxed.
Protein analysis software predicts that the RPR motif in TBSV p33 constitutes a hydrophilic pocket, and it is exposed to solvent (Fig. 9A). This structural prediction is compatible with the proposed function of the RPR motif in RNA binding. The secondary structure analysis predicts that the RPR motif itself has mainly turns (Fig. 9A), suggesting that this motif may take stable conformation upon binding to RNA. Indeed, the ARM motif in the human immunodeficiency virus type 1 Tat protein (37, 46) constitutes flexible turns that can adopt two different conformations upon binding to two different viral RNAs (46).
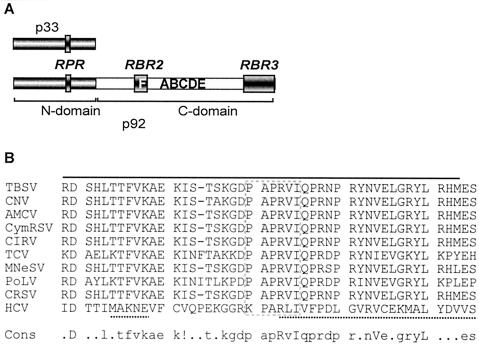
In addition to its role in p33 functions, the RPR motif is likely functional in p92 as well, since its N-terminal sequence (the so-called “overlapping domain”) is identical to p33. We could not test this in vitro, however, since p92 has additional RNA-binding sites—one in the central part (named the RBR2 region, located in the vicinity of the RdRp signature motifs; Fig. 10A) and another in the C-terminal segment (named RBR3; Fig. 10A), which can facilitate binding to RNA in the absence of the RPR motif (Fig. 1B, construct p92C). Nevertheless, separate mutagenesis of the RPR motif in p33 and p92, followed by testing virus replication in the protoplast, revealed that the RPR motif in p92 is essential for tombusvirus replication (35a). Therefore, based on (i) sequence identity between p33 and the overlapping domain of p92 and (ii) the in vivo requirement of the wild-type RPR motif in p92 for tombusvirus replication, we propose that the RPR motif in p92 is a functional RNA-binding site. However, the actual function(s) of the RPR motif in p92 will need further studies.
FIG. 10.
Locations of the three RNA-binding regions in TBSV p92. (A) Schematic representation of the three mapped RNA-binding regions—RPR, RBR2, and RBR3—in p92. The N-terminal portion of p92, which overlaps with p33, is indicated as a dark box, while the unique C-terminal segment is shown as a light box. The signature motifs of RdRps are marked with letters A through F (29). The names of the RNA-binding regions are shown above the filled boxes. (B) Sequence alignment of the RBR2 region in TBSV and in other replicase proteins of tombusviruses and related viruses as well as the RNA-binding region in NS5B of HCV. The RBR2 region in TBSV is indicated by a solid line above the sequence, while the RNA-binding sequence in NS5B of HCV (5) is marked with a dotted line beneath the sequence. The conserved motif F is boxed. The consensus (cons) sequence is also shown at the bottom. The abbreviations used for viruses are the same as in the legend to Fig. 9, except that we also included Pothos latent virus (PoLV; Tombusviridae) and HCV.
Sequence comparison of the central RBR2 RNA-binding domain within the unique p92C segment of TBSV with other tombusviruses and related viruses revealed that this 45-amino-acid region contains several highly conserved amino acids, including motif F (Fig. 10B). Interestingly, an RNA-binding region, which includes the motif F, has also been defined for the NS5B RdRp protein of HCV (5), as shown in Fig. 10B. This region has been shown for HCV RdRp to include some of the basic amino acids that line up the RNA-binding groove and bind to RNA template during viral replication (5). In addition, the highly conserved arginine and isoleucine or leucine in motif F have been found to bind to ribonucleoside triphosphates (rNTPs) (21). These observations suggest that this 45-amino-acid-long region in TBSV p92 possesses the amino acids responsible for binding to both rNTPs as well as to the template RNA. Based on its similarity in location to the RNA-binding region in the HCV NS5B, the function of the RBR2 RNA-binding region in p92 of TBSV may be to “channel” the RNA template toward the active site in the RdRp. The significance and possible function(s) of the C-terminal RBR3 RNA-binding region in TBSV p92 is currently under further investigation.
In summary, our in vitro studies defined the RNA-binding regions in p33 and p92 replicase proteins of TBSV, which likely play a major role(s) in tombusvirus RNA replication and metabolism. These results will open the way for future studies on the functions of these proteins in particular and on tombusvirus replication in general.
Acknowledgments
We thank Janice D. Pata for suggestions regarding cooperative binding experiments and Judit Pogany and Tadas Panavas for critical reading of the manuscript and for very helpful suggestions. We also thank Murali Kanakasabai for helping to fit the experimental data. We also thank H. Scholthof for providing construct pHS-175.
This work was supported by NSF (MCB0078152) and by the Kentucky Tobacco Research and Development Center at the University of Kentucky, awarded to P.D.N.
Footnotes
Publication 03-12-003 of the Kentucky Agricultural Experiment Station.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Ansel-McKinney, P., and L. Gehrke. 1998. RNA determinants of a specific RNA-coat protein peptide interaction in alfalfa mosaic virus: conservation of homologous features in ilarvirus RNAs. J. Mol. Biol. 278:767-785. [DOI] [PubMed] [Google Scholar]
- 3.Baer, M. L., F. Houser, L. S. Loesch-Fries, and L. Gehrke. 1994. Specific RNA binding by amino-terminal peptides of alfalfa mosaic virus coat protein. EMBO J. 13:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, P., M. Kraft, A. Ejchart, M. Westendorp, R. Frank, and P. Rosch. 1995. Structural studies of HIV-1 Tat protein. J. Mol. Biol. 247:529-535. [DOI] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, K. W. 1999. Replication of tobacco mosaic virus RNA. Philos. Trans. R. Soc. Lond. B 354:613-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgyan, J., L. Rubino, and M. Russo. 1996. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J. Gen. Virol. 77:1967-1974. [DOI] [PubMed] [Google Scholar]
- 9.Calnan, B. J., B. Tidor, S. Biancalana, D. Hudson, and A. D. Frankel. 1991. Arginine-mediated RNA recognition: the arginine fork. Science 252:1167-1171. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, C.-P., J. Pogany, and P. D. Nagy. 2002. Mechanism of DI RNA formation in tombusviruses: dissecting the requirement for primer extension by the tombusvirus RNA-dependent RNA polymerase in vitro. Virology 304:460-473. [DOI] [PubMed] [Google Scholar]
- 11.Citovsky, V., D. Knorr, G. Schuster, and P. Zambryski. 1990. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60:637-647. [DOI] [PubMed] [Google Scholar]
- 11a.Crouch, R. J., M. Wakasa, and M. Haruki. 1999. Detection of nucleic acid interactions using surface plasmon resonance, p. 143-160. In S. Haynes (ed.), Methods in molecular biology, vol. 118. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 12.Donald, R. G., and A. O. Jackson. 1996. RNA-binding activities of barley stripe mosaic virus gamma b fusion proteins. J. Gen. Virol. 77:879-888. [DOI] [PubMed] [Google Scholar]
- 13.Donald, R. G., D. M. Lawrence, and A. O. Jackson. 1997. The barley stripe mosaic virus 58-kilodalton βb protein is a multifunctional RNA binding protein. J. Virol. 71:1538-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Draper, D. E. 1999. Themes in RNA-protein recognition. J. Mol. Biol. 293:255-270. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari, M. E., W. Bujalowski, and T. M. Lohman. 1994. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J. Mol. Biol. 236:106-123. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 16a.Hearne, P. Q., D. A. Knorr, B. I. Hillman, and T. J. Morris. 1990. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177:141-151. [DOI] [PubMed] [Google Scholar]
- 17.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, C. Y., Y. L. Huang, M. Meng, Y. H. Hsu, and C. H. Tsai. 2001. Sequences at the 3′ untranslated region of bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. J. Virol. 75:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 20.Kollar, A., and J. Burgyan. 1994. Evidence that ORF 1 and 2 are the only virus-encoded replicase genes of cymbidium ringspot tombusvirus. Virology 201:169-172. [DOI] [PubMed] [Google Scholar]
- 21.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 22.Li, Q., and P. Palukaitis. 1996. Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology 216:71-79. [DOI] [PubMed] [Google Scholar]
- 23.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 24.McGhee, J. D., and P. H. von Hippel. 1974. Theoretical aspects of DNA-protein interactions: cooperative and non-cooperative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86:469-489. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, K. P., V. Bhakuni, D. Xie, and E. Freire. 1992. Molecular basis of cooperativity in protein folding. III. Structural identification of cooperative folding units and folding intermediates. J. Mol. Biol. 227:293-306. [DOI] [PubMed] [Google Scholar]
- 26.Nagy, P. D., C. D. Carpenter, and A. E. Simon. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. USA 94:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. D., C. Zhang, and A. E. Simon. 1998. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 17:2392-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of cucumber necrosis virus and tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276:279-288. [DOI] [PubMed] [Google Scholar]
- 29.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 30.Osman, T. A., R. J. Hayes, and K. W. Buck. 1992. Cooperative binding of the red clover necrotic mosaic virus movement protein to single-stranded nucleic acids. J. Gen. Virol. 73:223-227. [DOI] [PubMed] [Google Scholar]
- 31.Osman, T. A., P. Thommes, and K. W. Buck. 1993. Localization of a single-stranded RNA-binding domain in the movement protein of red clover necrotic mosaic dianthovirus. J. Gen. Virol. 74:2453-2457. [DOI] [PubMed] [Google Scholar]
- 32.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263-274. [DOI] [PubMed] [Google Scholar]
- 34.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Internal initiation by the cucumber necrosis virus RNA-dependent RNA polymerase is facilitated by promoter-like sequences. Virology 296:275-287. [DOI] [PubMed] [Google Scholar]
- 35.Panavas, T., and P. D. Nagy. 2003. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 77:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Panaviene, Z., J. M. Baker, and P. D. Nagy. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191-205. [DOI] [PubMed] [Google Scholar]
- 36.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 37.Puglisi, J. D., R. Tan, B. J. Calnan, A. D. Frankel, and J. R. Williamson. 1992. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 257:76-80. [DOI] [PubMed] [Google Scholar]
- 38.Rajendran, K. S., J. Pogany, and P. D. Nagy. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao, A. L., and G. L. Grantham. 1996. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology 226:294-305. [DOI] [PubMed] [Google Scholar]
- 40.Richmond, K. E., K. Chenault, J. L. Sherwood, and T. L. German. 1998. Characterization of the nucleic acid binding properties of tomato spotted wilt virus nucleocapsid protein. Virology 248:6-11. [DOI] [PubMed] [Google Scholar]
- 41.Rubino, L., and M. Russo. 1998. Membrane-targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 42.Rubino, L., F. Weber-Lotfi, A. Dietrich, C. Stussi-Garaud, and M. Russo. 2001. The open reading frame 1-encoded (“36K”) protein of Carnation Italian ringspot virus localizes to mitochondria. J. Gen. Virol. 82:29-34. [DOI] [PubMed] [Google Scholar]
- 43.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Scholthof, K. B., H. B. Scholthof, and A. O. Jackson. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365-369. [DOI] [PubMed] [Google Scholar]
- 46.Smith, C. A., V. Calabro, and A. D. Frankel. 2000. An RNA-binding chameleon. Mol. Cell 6:1067-1076. [DOI] [PubMed] [Google Scholar]
- 47.Soldevila, A. I., W. M. Havens, and S. A. Ghabrial. 2000. A cellular protein with an RNA-binding activity copurifies with viral dsRNA from mycovirus-infected Helminthosporium victoriae. Virology 272:183-190. [DOI] [PubMed] [Google Scholar]
- 48.Tsai, M. S., Y. H. Hsu, and N. S. Lin. 1999. Bamboo mosaic potexvirus satellite RNA (satBaMV RNA)-encoded P20 protein preferentially binds to satBaMV RNA. J. Virol. 73:3032-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants—defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White, K. A., and T. J. Morris. 1994. Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc. Natl. Acad. Sci. USA 91:3642-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, K. A., and T. J. Morris. 1999. Defective and defective interfering RNAs of monopartite plus-strand RNA plant viruses. Curr. Top. Microbiol. Immunol. 239:1-17. [DOI] [PubMed] [Google Scholar]
- 53.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]