Abstract
Rotaviruses utilize integrins during virus-cell interactions that lead to infection. Cell binding and infection by simian rotavirus SA11 were inhibited by antibodies (Abs) to the inserted (I) domain of the α2 integrin subunit. To determine directly which integrins or other proteins bind rotaviruses, cell surface proteins precipitated by rotaviruses were compared with those precipitated by anti-α2β1 Abs. Two proteins precipitated by SA11 and rhesus rotavirus RRV from MA104 and Caco-2 cells migrated indistinguishably from α2β1 integrin, and SA11 precipitated β1 from α2β1-transfected CHO cells. These viruses specifically precipitated two MA104 cell proteins only, but an additional 160- to 165-kDa protein was precipitated by SA11 from Caco-2 cells. The role of the α2 I domain in rotavirus binding, infection, and growth was examined using CHO cell lines expressing wild-type or mutated human α2 or α2β1. Infectious SA11 and RRV, but not human rotavirus Wa, specifically bound CHO cell-expressed human α2β1 and, to a lesser extent, human α2 combined with hamster β1. Binding was inhibited by anti-α2 I domain monoclonal Abs (MAbs), but not by non-I domain MAbs to α2, and required the presence of the α2 I domain. Amino acid residues 151, 221, and 254 in the metal ion-dependent adhesion site of the α2 I domain that are necessary for type I collagen binding to α2β1 were not essential for rotavirus binding. Rotavirus-α2β1 binding led to increased virus infection and RRV growth. SA11 and RRV require the α2 I domain for binding to α2β1, and their binding to this integrin is distinguishable from that of collagen.
Virus attachment and entry into host cells are multistep processes that influence cellular tropism and can involve sequential recognition of multiple receptors and coreceptors. Rotaviruses, a genus within the Reoviridae family, cause severe gastroenteritis following infection of intestinal enterocytes. The virus spike protein, VP4, which is a major determinant of tropism and receptor binding (4, 20, 51, 58), is proteolytically cleaved by trypsin into VP5* and VP8*, which increases the virus infectivity and internalization rate (1, 14, 28, 29). Several glycoconjugates have been implicated in rotavirus attachment (4, 5, 22, 32, 38, 42, 68, 74, 84). Although a minority of animal rotaviruses, including simian strains SA11 and RRV, can utilize terminal sialic acids (SA) as receptors (12, 13, 22, 32), SA are not essential for infectivity (63). SA-using porcine rotaviruses OSU and CRW-8 appear to use ganglioside- and glycolipid-based receptors, respectively (43, 68). RRV binds sialosides with low affinity via a galectin-like region in VP8* (24, 25).
In searching for rotavirus receptors, Coulson et al. found that VP4 and rotavirus outer capsid protein VP7 contain sequences corresponding to integrin recognition sites (17). Integrins are α/β heterodimeric, transmembrane glycoproteins important for cell surface adhesion and signaling. The RDGE sequence in VP4 at amino acids (aa) 307 to 310 corresponds to the putative α2β1 integrin recognition sequence DGE(A) in type I collagen (75). VP7 contains the αxβ2 integrin ligand sequence, GPR (56), and several potential α4β1 integrin ligand sites (41, 52). Monoclonal antibodies (MAbs) to α2β1 and αxβ2, and peptides containing these integrin ligand sequences, inhibited SA11 and human rotavirus RV-5 infection of MA104 and Caco-2 cells, which were shown to express α2β1 and αxβ2 integrins, by 30 to 90% (15, 17, 41). As Caco-2 cells model small intestinal epithelial cells, this suggested that SA11 could use α2β1 for infection of intestinal cells. Surface expression of α2β1 correlated with susceptibility of MA104, Caco-2, RD, K562, and COS-7 cells to SA11 infection (57). SA11 showed increased levels of binding and growth in α2β1- and α4β1-transfected K562 cells, which were specifically blocked by anti-α2 and anti-α4 MAbs, respectively. From these data, it was concluded that α2β1 and α4β1 can act as SA11 receptors (41). It has been proposed that rotavirus-cell binding can involve initial carbohydrate recognition followed by integrin interaction (41).
More recently, the neuraminidase-resistant RRV mutant nar3 was shown to bind α2β1 (85). Another integrin, αvβ3, has been shown to promote infection by RRV, nar3, and human rotavirus Wa. Rotavirus binding to αvβ3 was not detected (11, 36). It has been confirmed that infection by SA11 and RRV is inhibited by anti-α2 MAbs and DGE-containing peptides (11, 15, 17, 85). The infectivity of several other rotaviruses, including Wa, was also inhibited by anti-α2 MAbs (11, 36). However, binding of RRV to α2β1 could not be detected in two studies (11, 85), and evidence that SA11, Wa, and other rotavirus strains bind to α2β1 was not found by one of these groups (11). A summary of the previously published studies of anti-integrin MAb blockade of rotavirus-cell binding and infection is presented in Table 1.
TABLE 1.
Summary of previously published studies of anti-integrin MAb blockade of rotavirus-cell binding and infection
| Rotavirus strain (cell line) | Integrin subunit | Blockade of given event by Abs specific for the indicated integrin subunit (MAb identity and blockade citation, epitope recognized [if mapped], epitope citation)
|
|
|---|---|---|---|
| Virus-cell binding | Infection | ||
| SA11 | |||
| MA104 | α2 | − (AK7a, I domainb) | + (AK7c; RMAC11c, I domain aa 173-259d; P1E6e, I domain aa 210-211 and 268-273d,f; Gi9e, I domainb) |
| β1 | NDp | + (4B4e, I-like domain aa 207-218g) | |
| β1 | ND | − (P4C10c, aa 207-218g; 8A2c, aa 207-218g; A-IA5c, aa 207-218g; Lia 1/2e, aa 207-218g; MEM-101Ae; MAR4e; CLB-CD29e; K20e, aa 426-587g) | |
| αx | ND | − (99.1.1.1.1c) | |
| β2 | ND | + (MHM23c, I-like domain aa 197-209h; MEM-148e, N-terminal and midregioni; AZN-L18, N-terminal and midregioni; CLB-LFA1e) | |
| α2-K562 | α2 | + (AK7j) | + (AK7j) |
| α4-K562 | α4 | + (P4C2j, aa 108-182k) | + (P4C2j) |
| RRV | |||
| MA104 | α2 | − (AK7a; P1E6l) | − (AK7a; P1E6m) |
| β1 | ND | − (4B7Rm) | |
| αx | ND | − (polyclonal Abm) | |
| β2 | ND | + (polyclonal Abm) | |
| αvβ3 | − (polyclonal Abm) | + (LM609m, αv, and aa 171-184 + aa 187-193 of β3n,o) | |
| Caco-2 | α2 | − (AK7a) | ND |
| CHO α2 | α2 | − (AK7a) | ± (AK7a) |
| CHO α2β1 | α2 | − (AK7a) | + (AK7a) |
| Wa | |||
| MA104 | α2 | − (AK7a) | + (AK7a,m) |
| α2 | − (AK7a) | − (AA10a, I domain aa 173-259d) | |
| β2 | ND | + (MHM23m) | |
| αvβ3 | ND | + (LM609m) | |
| RV-5 | |||
| MA104 | α2 | + (AK7c) | ND |
| Caco-2 | ND | β2 | + (MHM23c) |
Ciarlet et al. (11). In this study, virus-cell binding assays were performed using cell monolayers that had been placed in suspension.
Bergelson et al. (7).
Coulson et al. (17).
Kamata et al. (45).
Coulson (15).
Dickeson et al. (23).
Takada and Puzon (78).
Poloni et al. (66).
Tan et al. (81).
Hewish et al. (41). In this study, virus-cell binding assays were performed on K562 cells that normally grow in suspension. The assays showed specific blockade of SA11 binding to α2β1.
Kamata et al. (46).
Zarate et al. (85). In this study, virus-cell binding assays were performed using cell monolayers that had been placed in suspension. The assays showed no blockade of RRV binding to α2β1.
Guerrero et al. (36). In this study, virus-cell binding assays were performed using cell monolayers that had been placed in suspension. The assays showed no blockade of virus binding to α2β1.
Takagi et al. (80).
Chen et al. (10).
ND, not determined.
To directly investigate the cell surface proteins bound by rotaviruses, we developed a novel virus immunoprecipitation technique designed to maintain integrin ligand-binding function, which included divalent cations, maintenance of the α/β subunit association, and preservation of disulfide bonds. Using this method, we show here that SA11 precipitates surface β1 from α2β1-transfected Chinese hamster ovary (CHO) cells, SA11 and RRV precipitate two surface proteins with the characteristics of α2 and β1 from MA104 cells, and SA11 precipitates these two proteins and a third protein of 160 to 165 kDa from Caco-2 cells.
As SA11 binding and infectivity were inhibited by anti-α2 MAbs which also block α2β1 binding to collagen (15, 17, 41, 45) and as infectivity was inhibited with the collagen-derived peptide sequence DGE (17), it was proposed that SA11 might bind α2β1 similarly to collagen (41). Within α2β1, the upper surface of the inserted (I) domain of α2, including the metal ion-dependent adhesion site (MIDAS), contains all the components required for collagen binding (47, 73). Within the α2 MIDAS, aa residues 151, 221, and 254 are essential for α2β1 binding to collagen (45, 47). Echovirus type 1 binds to α2β1, its cellular receptor, via a region of the α2 I domain partially overlapping that bound by a collagen-derived, triple-helical peptide (50). However, in contrast to collagen, the MIDAS is not involved in echovirus binding to α2β1 (50).
It has been reported that the infectivity of RRV and other rotaviruses was increased in CHO cells following transfection with human α2 or α2β1 but that the level of virus binding was not altered (11) (Table 1). From this study, it was concluded that an anti-α2 MAb inhibited the increased infectivity but had no effect on virus binding to the α2- or α2β1-transfected CHO cells. To analyze the roles of human α2, β1, the α2 I domain, and the α2 MIDAS in rotavirus recognition of α2β1, we utilized integrin-transfected CHO cell lines which had been used previously to map the human α2β1 regions involved in collagen binding (45, 47). We found that infectious SA11 and RRV specifically bound CHO cell-expressed human α2β1 integrin and, to a lesser extent, human α2 combined with hamster β1. This binding was inhibited by anti-α2 I domain MAbs but not by non-I domain MAbs to α2. It required the presence of the α2 I domain but not MIDAS residues 151, 221, and 254. Our experiments showed that the infectivity of SA11 and RRV and the replication of RRV were increased following virus binding to α2β1 integrin on CHO cells.
MATERIALS AND METHODS
Cell lines, viruses, and antibodies.
MA104 and Caco-2 cells were propagated as described before (17, 57). The derivation of CHO K1 cell transfectants expressing human α2 (CHO α2), the human α2 and β1 integrin subunits (CHO α2β1), the human α2 subunit with alanine-swapping mutations (CHO α2 D151A, CHO α2 D254A, and CHO α2 T221A), the human β1 and α2 subunits with an α2 I domain deletion (CHO α2delIβ1), and the PBJ-1 empty vector (CHO K1 PBJ-1) has been described previously (45, 48, 50, 77-79). Cultivation of CHO cell lines was performed as for MA104 cells, except that G418 sulfate (Gibco BRL, Grand Island, N.Y.) at 0.1 to 1 mg/ml was included in transfectant growth medium. Integrin expression on CHO cell lines was monitored regularly by flow cytometry as described previously (41, 57).
Rotaviruses SA11, RRV, and Wa were grown in MA104 cells (16, 41). For purification by glycerol gradient ultracentrifugation, virus was grown in cell cultures on beads (42).
Rabbit antisera to SA11 (anti-SA11) and RRV (anti-RRV) were produced as described before (19) and showed fluorescent focus reduction neutralization assay titers (16) of 1:100,000 and 1:80,000, respectively, against homologous virus. According to the results of enzyme immunoassays (EIA) performed as described previously (18), preimmune rabbit sera (control Ab) showed reciprocal titers of <100 against SA11.
Hybridoma cells producing the SA11-neutralizing, anti-VP4 MAb 2G4 (59, 71) were provided by M. Estes and J. Burns, Baylor College of Medicine, Houston, Tex. By fluorescent focus reduction neutralization assay, the titer of MAb 2G4 ascitic fluid against SA11 was 1:40,000. The derivation of antirotavirus MAbs RVA (to VP6), 60 (to a VP7 nonneutralization epitope), and RV-3:5 (to a VP7 neutralization epitope) has been described previously (19, 54, 71).
MAbs P4C10, QE2.E5, and P5D2 to human β1 (70, 78) were provided by D. Leavesley and P. Simmons, Royal Adelaide Hospital, Adelaide, South Australia, Australia (P4C10, QE2.E5, and P5D2), and E. Wayner, Fred Hutchinson Cancer Research Center, Seattle, Wash. (P4C10 and P5D2). Anti-human α2 MAbs AK7 (33, 65), which (according to flow cytometry results) binds MA104 cell surface-expressed α2β1 integrin (17, 57) and blocks SA11 infection of MA104 cells by 50 to 60% at 12 to 40 μg/ml (17), and P1E6 were purchased from Pharmingen, San Diego, Calif., and Invitrogen, Burwood, Victoria, Australia, respectively. Anti-human α2 MAb 12F1 and anti-β1 MAb B3B11 were purchased from Chemicon, Temecula, Calif. Purified anti-human α2 MAbs HAS3 and HAS4 were provided by F. Watt, Imperial Cancer Research Fund, London, England. The epitopes recognized by the anti-integrin MAbs used are listed in Table 1. The pan-class I-reactive MAb W6/32 (3) was provided by A. Brooks, Department of Microbiology and Immunology, The University of Melbourne, Melbourne, Victoria, Australia. Negative control MAbs MOPC21 (purchased from Cappel, ICN Pharmaceuticals Inc., Aurora, Ohio) and RV5:2 (specific to RV-5 rotavirus VP4) (18) were matched with test MAbs for isotype and protein concentration. Binding of MAbs to cells was characterized by flow cytometry and expressed as the relative linear median fluorescence intensity (MFI), as described previously (41, 57).
Immunoprecipitation of integrin subunits using virus or anti-integrin antibodies.
Surface proteins on confluent cell monolayers (5 × 107 cells) were biotinylated as described previously (30), and then cells were lysed in 50 mM Tris-HCl buffer (pH 7.5) containing 5 mM iodoacetamide (Sigma, St. Louis, Mo.), 1 mM phenylmethylsulfonyl fluoride (Sigma), 1 mM CaCl2, 1 mM MgCl2, and 0.5% (vol/vol) Triton X-100 (Sigma) for 30 min at 4°C. Lysates were clarified by centrifugation at 10,000 × g for 10 min at 4°C, and lysate protein concentrations were determined using a detergent-compatible protein assay kit (Bio-Rad, Hercules, Calif.).
Purified virus infectivity was activated with porcine trypsin type IX-S (Sigma) (10 μg/ml) for 10 min at 37°C. Activated virus (5 μg; 107 to 108 infectious virions) was mixed with cell lysate (70 μg of protein in 500 μl of phosphate-buffered saline [PBS]) overnight at 4°C. Anti-rotavirus Ab was added to a final dilution of 1:100 for 1 h at 4°C, and Ab complexes were collected onto protein A-Sepharose beads (Amersham Pharmacia Biotech, Uppsala, Sweden) (2 mg in 100 μl) which had been blocked for 1 h at 4°C in PBS containing 5% (wt/vol) skim milk powder (Diploma; Bonlac Foods, Altona, Australia). Beads were collected by centrifugation at 10,000 × g for 10 s and washed extensively with PBS.
For MAb precipitations, cell extract (70 μg of protein) was reacted with 5 to 10 μg of MAb for 1 to 16 h at 4°C. For precipitation of α2β1 from MA104 cell lysates with α2-specific MAbs, MAb-cell protein complexes were collected with protein A-Sepharose beads that had been coated with rabbit anti-mouse immunoglobulins (Ig) (Dako, Germany) (0.2 mg anti-mouse Ig, mainly IgG, per 20 mg of beads) overnight at 4°C and washed extensively with PBS. For all other MAb precipitations, Ab complexes were collected onto blocked protein A-Sepharose beads as described above for virus complexes.
Precipitated proteins were separated from beads by boiling for 5 min in Laemmli's sample buffer (53) containing 5% (vol/vol) β-mercaptoethanol. Proteins and protein molecular weight standards (Novex, San Diego, Calif.) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 7.5% (wt/vol) acrylamide resolving gel at 125 V. Proteins were electrophoretically transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.) for 1 h at 100 V. After blocking in PBS containing 0.1% (vol/vol) Tween 20 (PBS-T) and 5% (wt/vol) skim milk powder for 1 h at 37°C or overnight at 4°C, the membrane was washed extensively with PBS-T. Biotinylated proteins were detected by incubation with streptavidin-conjugated horseradish peroxidase (StAv-HRP) (Silenus, Melbourne, Victoria, Australia), optimally diluted in PBS-T, for 1 h at 20°C. Following washing as described above, bound StAv-HRP was detected by enhanced chemiluminescence (ECL) with ECL Hyperfilm (Amersham Pharmacia Biotech) as specified by the manufacturer, the positions of molecular weight markers were marked, and films were scanned using Adobe Photoshop software.
Virus binding, infectivity, and growth assays.
Assays of infectious rotavirus binding to CHO K1 cell lines were performed using confluent monolayers in 24-well trays (Nunclon, Roskilde, Denmark) as previously described (41). Harvested virus titers were determined by indirect immunofluorescent staining of confluent MA104 cell monolayers inoculated with serial dilutions of virus and were expressed as the number of fluorescent cell-forming units (FCFU) per well (16). For MAb blockade of rotavirus binding, washed cell monolayers were incubated with MAbs (200 μl/well) at 37°C for 1.5 h and then cooled to 4°C prior to MAb removal and virus addition for the binding assay.
Rotavirus infectivity in CHO cell lines was quantitated using confluent monolayers in 96-well trays (Nunclon), as described previously (17, 41). In some experiments, virus was not trypsin activated to maintain cellular integrity. The number of virus-infected CHO cells was determined (using MAb RVA at a 1:2,000 dilution [16]) by indirect immunofluorescent staining.
Rotavirus infection blockade (using anti-integrin MAbs) in MA104 cells was performed using the fluorescent focus reduction assay described previously (17), and growth curves were obtained using confluent MA104 or CHO cells in 24-well trays as described previously (41, 57).
Assay for MAb AK7 binding to the α2 integrin I domain.
Soluble glutathione S-transferase-α2 I domain (GST-α2I) and wild-type GST (wt-GST) were produced and purified as described previously (47), and their binding to MAb AK7 was determined using an EIA adapted from earlier studies (19). Microtiter plates (Immunosorb; Nunclon) were coated with GST-α2I or wt-GST diluted in PBS, at 0 to 500 μg/ml, for 2 h at 37°C. MAbs (10 μg/ml) diluted in PBS containing 2.5% skim milk powder and 0.05% (vol/vol) Tween 20 (Sigma) were reacted overnight at 4°C. Bound MAb was detected using HRP-conjugated anti-mouse Ig (Silenus).
Statistical analysis.
The unpaired, two-tailed Student t test or two-way analysis of variance was used to assess the statistical significance of differences in virus-cell binding and infectivity. Significance was set at the 95% level. Unless otherwise stated, error bars represent the 95% confidence interval and the data presented in each figure were derived from at least three experiments.
RESULTS
Rotaviruses specifically precipitate MA104 and Caco-2 cell surface proteins that migrate indistinguishably from those immunoprecipitated by MAbs to α2β1.
The proteins precipitated by purified SA11 from lysates of surface-biotinylated MA104 cells and collected using anti-SA11 are shown in Fig. 1A. Precipitated proteins ranged in molecular mass from approximately 160 to 30 kDa. Apart from a nonspecifically detected protein migrating at 50 kDa that was present in all precipitations, no proteins were precipitated by control Ab in the presence of virus or by anti-SA11 in the absence of virus.
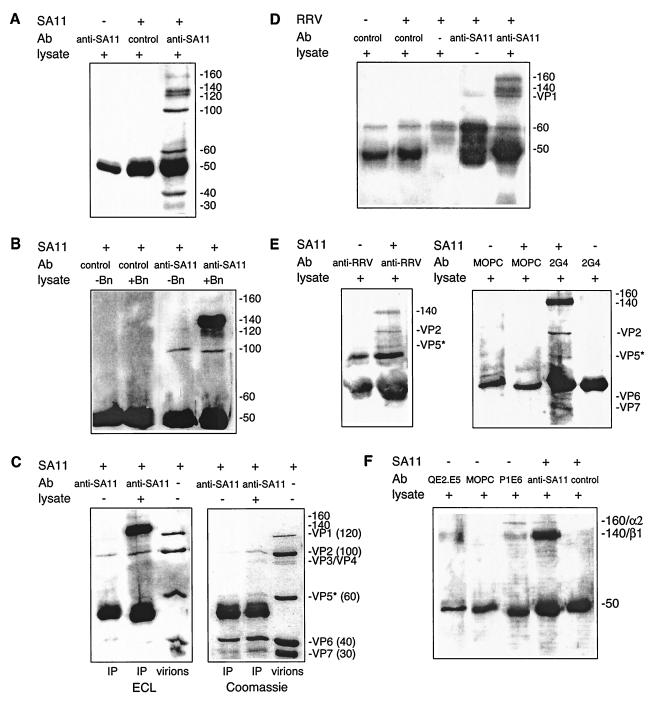
FIG.1.
Comparison of MA104 cell surface proteins precipitated by SA11 and RRV with those precipitated by MAbs to α2β1. (A) SA11 precipitated a range of proteins. Cell lysate proteins were reacted with SA11 (+) or PBS (−) and then collected with anti-SA11 or control Ab. The data represent those obtained in twenty experiments. (B) Of the proteins precipitated by SA11, only the 160- and 140-kDa proteins were biotinylated and derived from the cell surface. SA11-precipitated, mock-biotinylated (−Bn) or biotinylated (+Bn) cell lysate was reacted with anti-SA11 or control Ab. Mock-biotinylated lysates were prepared identically to biotinylated lysates, with the omission of N-hydroxysulfosuccinimide-biotin. (C) The nonbiotinylated proteins in SA11 precipitates were viral in origin. Precipitated proteins were separated in duplicate gels run simultaneously. The biotinylated proteins in one gel (ECL) were detected with StAv-HRP and ECL after transfer to a polyvinylidene difluoride membrane (see Materials and Methods). Proteins in the other gel (Coomassie) were stained at 20°C with 0.1% (wt/vol) Coomassie brilliant blue (Bio-Rad) in 20% (vol/vol) methanol and 20% (vol/vol) acetic acid for 3 h and destained overnight in 40% (vol/vol) methanol and 10% (vol/vol) acetic acid. Cell lysate proteins (+) or PBS (−) were reacted with SA11 and then collected with anti-SA11 and corun in comparison with 2 μg of reduced, purified SA11 proteins (virions). (D) RRV also precipitated two MA104 cell surface proteins of 160 and 140 kDa. Cell lysate proteins (+) or PBS (−) were reacted with RRV (+) or PBS (−) and then collected with anti-SA11, control Ab, or PBS. (E) Rabbit anti-RRV serum and anti-VP4 MAb 2G4 also precipitated the 160- and 140k-Da proteins following SA11 incubation with cell lysate. Lysate proteins precipitated by SA11 (+) or PBS (−) were collected using anti-RRV, 2G4 MAb, or MOPC21 (MOPC) MAb. These data represent those obtained in two experiments. (F) Comparison of the 160- and 140-kDa proteins precipitated by SA11 with those immunoprecipitated by MAbs to α2β1. Lysate proteins were reacted with SA11 and then precipitated with anti-SA11 or control Ab as described previously. These were corun with lysate proteins reactive with anti-β1 MAb QE2.E5, anti-α2 MAb P1E6, or control MAb MOPC21. Complexes in the latter two reactions were collected onto anti-mouse Ig-coated beads (see Materials and Methods). The molecular masses of the major protein bands, calculated from the positions of the molecular mass standards, are indicated in kilodaltons on the right sides of the panels. The identified rotaviral proteins are designated using the “VP” nomenclature, and their molecular masses in kilodaltons are shown in parentheses on the right sides of the panels.
To confirm that the proteins visualized were biotinylated and derived from the cell surface, proteins precipitated by SA11 and collected using anti-SA11 from lysates of mock-biotinylated MA104 cells were compared with those obtained from cell surface-biotinylated lysates (Fig. 1B). The two proteins of 140 and 160 kDa (present in virus precipitates of biotinylated MA104 cell lysates) were the only proteins not visualized from virus precipitates of mock-biotinylated lysates. Thus, these were the only biotin-accessible proteins on the MA104 cell surface that were specifically precipitated by SA11. The nonbiotinylated proteins of 120 kDa or less proved to be viral in origin, as they migrated in gels indistinguishably from the VP1, VP2, VP5*, VP6, and VP7 proteins solubilized from purified SA11 rotavirus (Fig. 1C). They were precipitated by anti-SA11 only in the presence of virus and were detected in the presence or absence of MA104 cell proteins. Sufficient viral protein, but not 140- and 160-kDa proteins, was precipitated to be visible on Coomassie blue-stained gels, suggesting that ECL was detecting the large amounts of viral proteins present in the precipitations nonspecifically. This nonspecific reaction was not eliminated by StAv-HRP concentration optimization or alternative blocking reagents (data not shown). Streptavidin can bind to peptides of sequences unrelated to biotin (55, 72).
As shown in Fig. 1D, RRV bound to 140- and 160-kDa proteins was precipitated by anti-SA11 from MA104 cell lysates. As SA11 and RRV share VP4 and VP7 serotype specificity, these antisera can be used interchangeably. Anti-SA11 was used rather than anti-RRV, as the anti-SA11 serum showed a higher neutralization and EIA titer to both SA11 and RRV than did the anti-RRV serum. Consistent with the lower titer of anti-RRV against SA11, SA11 that bound to the 140-kDa but not the 160-kDa protein was precipitated by anti-RRV from MA104 cell lysates (Fig. 1E). Anti-VP4 MAb 2G4 precipitated virus complexed with both the 140- and 160-kDa MA104 cell proteins similarly to anti-SA11 and more readily detected the 140-kDa protein (Fig. 1E). MAb 2G4 did not precipitate the 140- and 160-kDa MA104 cell proteins in the absence of virus. Neither protein was detected using MAbs RVA (VP6), 60 (VP7), RV-3:5 (VP7), or MOPC21 (negative control).
The 160- and 140-kDa proteins precipitated by SA11 are comparable in size with the human α2 (approximately 165 kDa) and β1 (approximately 130 kDa) integrin subunits (39) and so might correspond to α2β1. We therefore analyzed the abilities of two anti-α2 MAbs to recognize α2β1 on MA104 cells and inhibit virus infection of these cells. MAb P1E6 bound to MA104 cell surface-expressed α2β1 similarly to MAb AK7 according to flow cytometry, as the histograms obtained with the two MAbs used at the same concentrations were indistinguishable (data not shown). P1E6 and AK7 showed similar ratios of the MFI with anti-integrin MAb to the MFI with control MAb ± range (7.23 ± 0.48 and 6.41 ± 0.57, respectively) (data not shown). MAb P1E6 also blocked SA11 infection of MA104 cells by a mean range of 39% ± 7.1% at 40 μg/ml (P = 0.04). As illustrated in Fig. 1F with data obtained with MAb P1E6, both anti-α2 MAbs AK7 and P1E6 immunoprecipitated two proteins of the expected molecular mass for the α2 and β1 integrin subunits from cell surface-biotinylated MA104 cell extracts which migrated indistinguishably from the 160- and 140-kDa proteins precipitated by SA11.
Although anti-β1 MAbs P4C10, P5D2, and QE2.E5 also immunoprecipitated both the α2 and β1 integrin subunits, these anti-β1 MAbs immunoprecipitated the β1 integrin subunit to a greater extent than the α2 subunit. This is shown in Fig. 1F for MAb QE2.E5. The α2 band precipitated by QE2.E5 is not visible in Fig. 1F, as exposure of the film sufficient to visualize α2 greatly overexposed the β1 band. This is expected, as β1 partners several α subunits, whereas α2 partners only β1. Control MAbs did not precipitate these proteins. Anti-β1 MAb B3B11 recognized a single protein of approximately 140 kDa, according to Western blotting of whole MA104 cell lysate directly applied to the gel (data not shown). As each integrin subunit-specific MAb precipitated both subunits from cell lysates, a significant proportion of the α2 and β1 subunits in the lysates retained their noncovalent association, which is required for efficient ligand interaction (2). Although the intensity of the 140-kDa band tended to be higher than that of the 160-kDa band in SA11 precipitations, the 160- and 140-kDa protein bands precipitated by anti-α2 MAbs were approximately equal in intensity. This suggests that the α2 and β1 proteins were labeled to a similar extent with biotin and that SA11 may have been precipitating more 140- than 160-kDa protein. However, it is also possible that these differences resulted from the nonquantitative nature of the immunoprecipitation and protein visualization techniques. Overall, these data suggest that the 140- and 160-kDa MA104 cell surface proteins bound by SA11 rotavirus are the α2 and β1 integrin subunits. Possibly, SA11 precipitates free β1 integrin, as well as α2β1, from MA104 cell lysates.
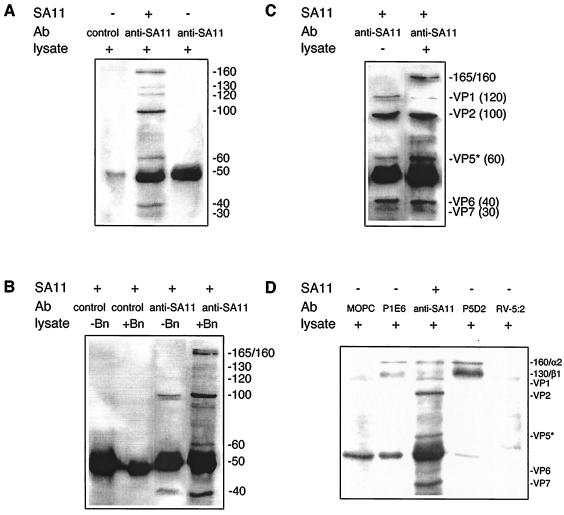
As shown in Fig. 2, SA11 precipitated a similar range of proteins from Caco-2 cell lysates as from MA104 cell lysates. The exceptions were that a 130-kDa Caco-2 protein was precipitated, rather than a 140-kDa protein, and two Caco-2 cell proteins of approximately 165 and 160 kDa were resolved in several experiments. No SA11-bound proteins were precipitated by control Ab (Fig. 2A and B). The origin of the Caco-2 cell proteins visualized was determined by comparison of proteins precipitated by SA11 from mock- and surface-biotinylated cell lysates (Fig. 2B) and in the absence or presence of cell lysate (Fig. 2C). As the proteins of approximately 120 kDa or less precipitated from biotinylated lysates were also present in precipitates from mock-biotinylated Caco-2 cell lysates and in the absence of lysate, only the three proteins at approximately 165, 160, and 130 kDa precipitated by SA11 originated from the Caco-2 cell surface. These SA11-precipitated proteins migrated indistinguishably from the two Caco-2 cell proteins precipitated by anti-β1 MAb P5D2 and anti-α2 MAb P1E6 (Fig. 2D). The 165- and 160-kDa proteins precipitated with virus could not be resolved sufficiently to determine which protein comigrated with the 160-kDa α2 integrin band precipitated by MAb P1E6. Control MAbs RV-5:2 and MOPC21 did not precipitate α2β1. The protein precipitated by RV-5:2 that migrated near β1 did not comigrate with β1 in gels with increased protein separation (data not shown). Thus, the 130- and 160- or 165-kDa Caco-2 cell surface proteins bound by SA11 appear to include α2 and β1. Differences in integrin glycosylation between MA104 and Caco-2 cells might explain the difference in sizes estimated for the β1 subunit in MA104 (140 kDa) and Caco-2 (130 kDa) cells (40). In contrast to the results of MA104 cell studies, SA11 precipitated similar amounts of the 160- or 165- and 130-kDa proteins as judged by band intensity.
FIG. 2.
Comparison of Caco-2 cell surface proteins precipitated by SA11 with those precipitated by MAbs to α2β1. (A) SA11 precipitated a range of proteins. Proteins in cell lysates were reacted with SA11 (+) or PBS (−) and then collected with anti-SA11 or control Ab. (B) Of the proteins precipitated by SA11, only the 165-, 160-, and 130-kDa proteins were biotinylated and derived from the cell surface. SA11-precipitated, mock-biotinylated (−Bn), or biotinylated (+Bn) cell lysate was reacted with anti-SA11 or control Ab. (C) The nonbiotinylated proteins in SA11 precipitates are viral in origin. Cell lysate proteins (+) or PBS (−) were reacted with SA11 and then collected with anti-SA11. The 130-kDa band was not detected in this particular experiment. (D) Comparison of the 165-, 160-, and 130-kDa proteins precipitated by SA11 with proteins immunoprecipitated by MAbs to α2β1. Lysate proteins were reacted with SA11 and then precipitated with anti-SA11 as described previously. These were corun with lysate proteins reactive with anti-β1 MAb P5D2, control MAb RV-5:2, anti-α2 MAb P1E6, or control MAb MOPC21. The estimated protein sizes and identities are indicated as described in the legend to Fig. 1.
No surface-biotinylated Caco-2 cell proteins were visualized in precipitations with purified Wa. As the maximum ratio of infectivity (FCFU) to micrograms of protein obtainable with Wa (6.8 × 105) was 10- to 67-fold less than that obtained with SA11, it appears that the amount of infectious Wa obtainable was insufficient for detection of Wa binding to lysate proteins.
Under standardized assay conditions which included using identical MAbs at the same concentrations, anti-α2 and anti-β1 MAbs precipitated more α2β1 integrin from Caco-2 cells than from MA104 cells, as judged by the intensity of staining of the protein bands. This is consistent with the finding of a higher level of α2β1 expression on Caco-2 cells than on MA104 cells as detected by flow cytometry (57). The amount of α2β1 precipitated by the MAbs also would have been affected by their specificity for human rather than monkey integrins.
N-terminal sequence determination and Western blotting with the anti-β1 MAb B3B11 that detected β1 in whole MA104 cell lysate of the 160- and 140-kDa proteins precipitated by SA11 from cell lysates were unsuccessful due to insufficient amounts of protein for analysis.
Mapping of MAb AK7 to the α2 I domain and flow cytometry characterization of integrin-transfected CHO cells.
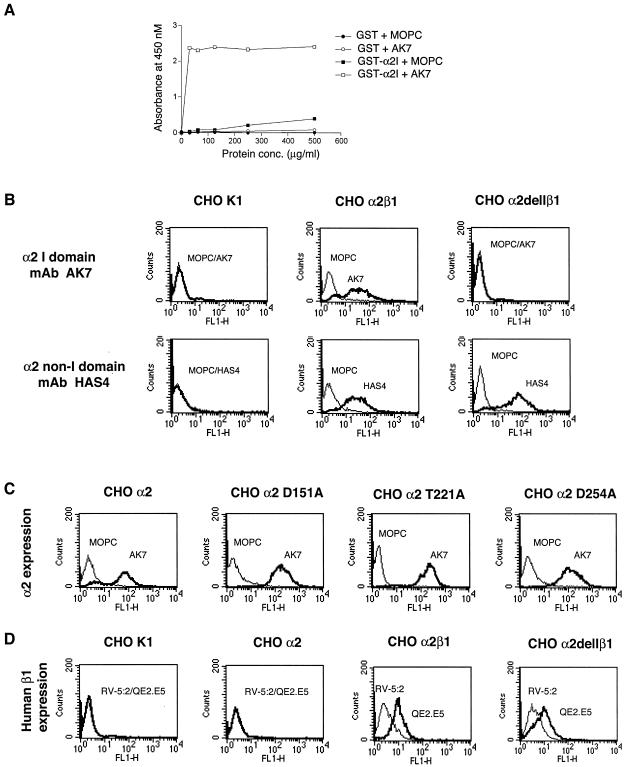
As shown in Fig. 3A, AK7, the most effective α2-specific MAb tested for blockade of SA11 cell binding and infectivity (17, 41; P. Halasz and B. S. Coulson, unpublished data), bound to GST-α2I as determined by EIA, confirming previous data showing that AK7 maps to the α2 I domain (7). Flow cytometry analysis using MAb HAS4 (which maps to the N-terminal region of α2 outside the I domain [7, 47]) and MAb AK7 showed that CHO K1 cells lacked human α2, CHO α2β1 cells expressed both I domain and non-I domain epitopes, and CHO α2delIβ1 cells lacked the I domain but expressed non-I domain epitopes similarly to CHO α2β1 cells (Fig. 3B). MAb HAS3 maps to the same α2 region as MAb HAS4 (7, 47) and showed the same results as MAb HAS4 (data not shown). CHO α2, CHO α2 D151A, CHO α2 T221A, and CHO α2 D254A cells all expressed the α2 I domain AK7 epitope (Fig. 3C). Human β1 (combined with human α2) was present on CHO α2β1 and CHO α2delIβ1 cells but was absent from CHO K1 and CHO α2 cells, as expected (Fig. 3D). We calculated from the flow cytometry histograms in Fig. 3 that approximately 40% of the CHO α2β1 cells expressed human α2 dimerized with hamster β1. These histograms show that approximately 55% of the CHO α2β1 cells expressed human β1, as detected using MAb QE2.E5, which is specific for human β1 and does not recognize hamster β1 (Fig. 3D). Thus, the remaining 45% of these cells expressed hamster β1 only. As detected using MAbs AK7 and HAS4 and determined from the data presented in Fig. 3B, 86% of CHO α2β1 cells expressed human α2. So, on average, approximately 90% of the CHO α2β1 cells that expressed hamster β1 also expressed human α2 and, overall, approximately 40% (calculated as 90% of 55%) of all the CHO α2β1 cells expressed human α2 dimerized with hamster β1. As the N and C termini of the I domain are closely associated in the three-dimensional structure and the I domain is inserted as an additional region sitting above the α subunit in a subset of integrins (2), deletion of the I domain is expected not to affect the function of the remaining regions of α2 or β1, and this appears to be the case from our studies.
FIG. 3.
Mapping of MAb AK7 to the α2 I domain and flow cytometry analysis of α2β1 integrin expression on CHO K1 cell lines. (A) MAb AK7 binds by EIA to GST-α2I but not to wt-GST control. (B, C, and D) Detection by flow cytometry of surface expression of human α2 and β1 on CHO cell lines used in this study. (B) The I domain deletion from CHO α2delIβ1 cells and its presence on CHO α2β1 cells were confirmed using MAb AK7, and the presence of the human α2 subunit on CHO α2β1 and CHO α2delIβ1 cells was demonstrated using MAb HAS4. (C) The presence of the α2 subunit, including the I domain, on CHO α2, CHO α2 D151A, CHO α2 T221A, and CHO α2 D254A cells was confirmed using MAb AK7. (D) Human β1 expressed on CHO α2β1 and CHO α2delIβ1 cells, but not on CHO α2 cells, was detected using MAb QE2.E5.
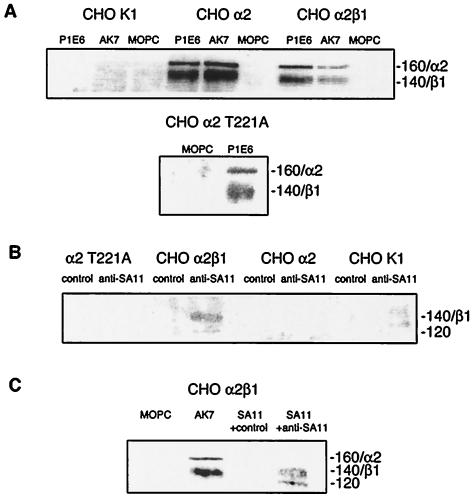
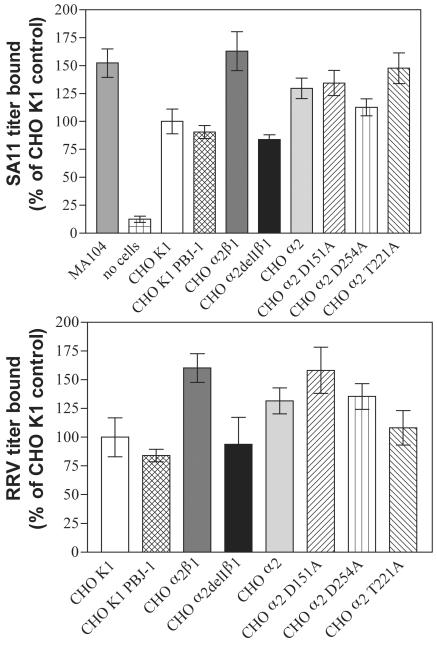
SA11 rotavirus precipitates a CHO α2β1 cell surface protein that migrates indistinguishably from β1 and is not precipitated by SA11 from CHO K1, CHO α2, CHO α2delIβ1, or CHO α2 T221A cell lysates.
Anti-α2 MAbs immunoprecipitated two proteins from CHO α2 and CHO α2β1 cell lysates (AK7 and P1E6) and from CHO α2 Τ221Α lysates (P1E6) which migrated at the expected molecular masses for α2β1 integrin (approximately 120 and 140 kDa) (Fig. 4A). No proteins were immunoprecipitated from CHO K1 lysates (Fig. 4A) and CHO α2delIβ1 cell lysates (data not shown) by these MAbs. According to the results of Western blotting of CHO α2β1 and CHO α2delIβ1 cell lysates, anti-β1 MAb B3B11 recognized a single protein, of approximately 140 kDa that was not detected in CHO K1 or CHO α2 lysates (data not shown). SA11 weakly precipitated two proteins, of approximately 140 and 120 kDa (Fig. 4B and C), from CHO α2β1 cell lysates which were not precipitated from CHO K1, CHO α2, and CHO α2 Τ221Α lysates (Fig. 4B) or CHO α2delIβ1 lysates (data not shown). As shown in Fig. 4C, the 140-kDa protein precipitated by SA11 from CHO α2β1 cells migrated indistinguishably from the β1 integrin subunit immunoprecipitated in conjunction with the α2 subunit by MAb AK7. As CHO α2 and CHO α2β1 cells differ only in the presence of human β1, this shows that SA11 precipitated the β1 integrin from CHO α2β1 cell lysates. The inability of SA11 to precipitate α2 or β1 from CHO α2delIβ1 cells suggests that the β1 precipitation from CHO α2β1 cell lysates was dependent on the presence of the α2 I domain. The identity of the 120-kDa protein precipitated by virus from CHO α2β1 cell lysates (Fig. 4B and C) is unclear. It may have been associated with β1 or may have been viral in origin.
FIG. 4.
SA11 immunoprecipitated β1 and a 120-kDa protein from CHO α2β1 cells. (A) The precipitation of cell surface α2 and β1 by anti-α2 MAbs AK7 and P1E6 from CHO α2, CHO α2 T221A, and CHO α2β1 cells, but not from CHO K1 cells, is shown. (B) SA11 precipitated 140- and 120-kDa surface proteins from CHO α2β1 cells but not from CHO α2, CHO α2 T221A, and CHO α2β1 cells. (C) The 140-kDa protein was identified as β1 by comigration with the β1 that was coprecipitated with α2 by MAb AK7. The estimated protein sizes and identities are indicated as described in the legend to Fig. 1.
As shown in Fig. 4A, the β1 band precipitated by anti-α2 MAbs P1E6 and AK7 appears more intense than the α2 band. This suggests that in contrast to results with MA104 cells, the β1 may be more heavily biotinylated than the α2 in CHO cells. This could explain the detection of SA11 precipitation of β1, but not α2, from CHO α2β1 cell lysates.
Binding of SA11 and RRV to α2β1 and infectivity of SA11 in MA104 cells are blocked by an anti-α2 I domain MAb but not by MAbs directed to a region of α2 outside the I domain.
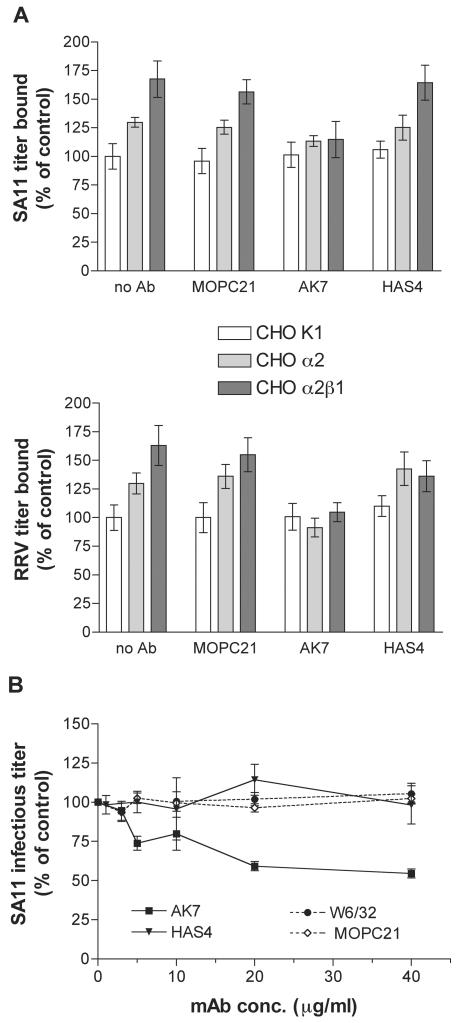
At a multiplicity of infection (MOI) of 5 (calculated from MA104 infectious cell titers) and in the absence of any Ab treatment, titers of SA11 and RRV bound to CHO α2β1 and CHO α2 cells were significantly higher than those of SA11 and RRV bound to CHO K1 or CHO K1 PBJ-1 cells (P < 0.001) (Fig. 5A and 6). CHO α2β1 and CHO α2 cells bound 1.6- to 1.7-fold- and 1.3-fold-higher titers of SA11 and RRV, respectively, than did CHO K1 or CHO K1 PBJ-1 cells. CHO α2β1 cells bound significantly higher virus titers than did CHO α2 cells (P < 0.01), suggesting that the β1 subunit plays a role in facilitating virus binding to α2β1 integrin. As shown in Fig. 5A, the increase in RRV binding to CHO α2β1 and CHO α2 cells was abolished by cellular treatment with MAb AK7 but not with MAbs HAS4 and MOPC21. Similarly, the increased SA11 binding to CHO α2β1 and CHO α2 cells was reduced to approximately 110% of binding to CHO K1 cells by treatment with MAb AK7 but not by treatment with MAbs HAS4 and MOPC21. MAb HAS3 had the same effect as MAb HAS4 on SA11 and RRV binding to MA104 cells (data not shown). Thus, SA11 and RRV bind human α2β1 expressed on CHO K1 cells to a higher level than to that of human α2 dimerized with hamster β1 and this binding involves the α2 I domain. At the maximum MOI achievable (an MOI of 1), CHO K1, CHO α2, and CHO α2β1 cells bound Wa at similarly low titers (data not shown). Thus, no binding of Wa to human α2β1 or human α2 dimerized with hamster β1 was detected using integrin-transfected CHO cells.
FIG. 5.
Inhibition by anti-I domain MAb AK7, but not by non-I domain MAb HAS4, of infectious SA11 (upper panel) and RRV (lower panel) binding to human α2β1 and to human α2 combined with hamster β1 on CHO cells (A) and of SA11 infection of MA104 cells (B). MOI = 5. MAb W6/32 is directed to MHC class I. MOPC21 is an isotype-matched negative control MAb for AK7, W6/32, and HAS4. Bars represent 95% confidence intervals (A) or standard deviations (B).
FIG. 6.
Titers of SA11 (upper panel) and RRV (lower panel) bound to CHO K1 cells and their transfectants show that the α2 I domain is essential for virus binding to α2β1 and that point mutations in the MIDAS do not affect virus binding. MOI = 5.
MAbs HAS4 (Fig. 5B) and HAS3 (data not shown) had no effect on SA11 infectivity in MA104 cells. In contrast, MAb AK7 inhibited SA11 infectivity in a dose-dependent fashion, as reported previously (17). According to the results of flow cytometry, the major histocompatibility complex (MHC) class I-specific MAb W6/32, which has been used previously as a negative control for anti-α2 MAb blockade of echovirus 1 cell binding (7), bound 99.5% of MA104 cells with an MFI of 20.1 ± 0.5, but this binding did not affect SA11 infectivity, further demonstrating the specificity of the inhibition by MAb AK7. W6/32 also had no effect on the infectivity of RRV and Wa (data not shown). Thus, the interaction of SA11 with monkey α2β1 on MA104 cells is specific for α2 and involves the α2 I domain.
A comparison of the abilities of anti-α2 MAbs to inhibit rotavirus, echovirus 1, and collagen binding is presented in Table 2. Echovirus and collagen both bind the α2 I domain (45, 47, 50, 73). As the patterns of MAb inhibition are very similar between echovirus, collagen, and rotavirus and only anti-I domain MAbs block the interaction of these three ligands with α2, these MAb data strongly suggest that rotavirus also binds the α2 I domain.
TABLE 2.
Ability of anti-α2 MAbs to inhibit cellular interactions of rotaviruses, echovirus 1, and collagen
| Anti-α2 MAb | Region (domain) of α2 recognizeda | Inhibition by MAb of:
|
||
|---|---|---|---|---|
| Rotavirus binding and/or infectivityb | Echovirus 1 bindingc | Collagen bindingd | ||
| AK7 | I | + | + | + |
| RMAC11 | I | + | NDh | + |
| P1E6 | Ie | + | + | + |
| 12F1 | If | + | + | − |
| AA10 | I | − | + | −/±g |
| Gi9 | I | + | − | + |
| HAS3 | Non-I | − | − | − |
| HAS4 | Non-I | − | − | − |
Published by Coulson et al. (17) (AK7 and RMAC11), Coulson (15) (Gi9), and Ciarlet et al. (11) (AA10) and in this paper (P1E6, 12F1, HAS3, and HAS4). MAb 12F1 inhibited RRV infectivity in a dose-dependent fashion to 16.9% ± 0.2% at 20 μg/ml. At this concentration, according to flow cytometry results, 12F1 bound to 91.4% of MA104 cells with an MFI of 5.1 ± 0.2.
Published by Bergelson et al. (7).
Residues 210 to 211 and 268 to 273 within the I domain required for P1E6 MAb binding to α2 (23).
Residues 199 to 203 within the I domain required for 12F1 binding to α2 (23).
No inhibition of collagen binding to CHO α2 cells was detected (7), whereas partial inhibition of binding to RD cells transfected with α2 was found (6).
ND, not determined.
The binding of SA11 and RRV rotaviruses to α2β1 is eliminated by deletion of the α2 I domain but not by MIDAS point mutations.
As shown in Fig. 6, titers of SA11 and RRV bound to CHO α2delIβ1 cells were significantly (1.6- to 1.7-fold) lower than those of SA11 and RRV bound to CHO α2β1 cells (P < 0.001), whereas titers of SA11 and RRV bound to CHO α2delIβ1, CHO K1, and CHO K1 PBJ-1 cell lines were indistinguishable (P > 0.05). Thus, deletion of the α2 I domain eliminated virus binding to α2β1. CHO α2β1 cells bound levels of SA11 similar to those of permissive MA104 cells (P > 0.05). The CHO α2, CHO α2 D151A, CHO α2 D254A, and CHO α2 T221A cell lines all bound similar levels of SA11 and RRV (P > 0.05), so binding of these viruses to human α2 dimerized with hamster β1 is not affected by mutagenesis of α2 I domain amino acid residues in the MIDAS region (D151, T221, and D254) that are critical for the ligation of collagen (45, 47).
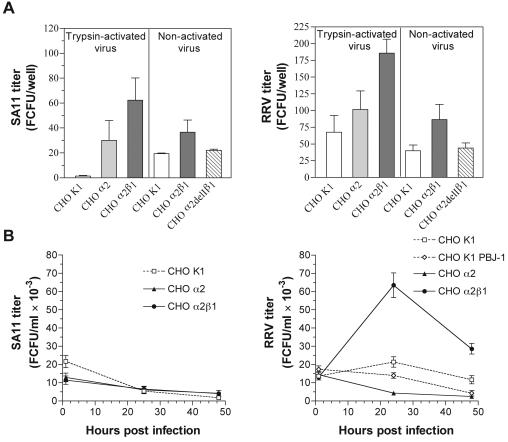
The infectivity of SA11 and RRV mediated via α2β1 is eliminated by deletion of the α2 I domain.
Rotavirus infectivity was measured as the number of cells infected after a single round of virus replication at 16 h after infection. Consistent with previous studies (11), CHO K1 cells were highly resistant to infection with SA11 and RRV. As shown in Fig. 7A, at a MOI of 20 (calculated using MA104 cells), SA11 infected only 1 to 20 of 50,000 (≤0.04%) cells and RRV infected only 40 to 70 (≤0.1%) cells. Trypsin-activated virus infectivity levels were higher in CHO α2 cells than in CHO K1 cells. SA11 titers in CHO α2 cells were 90-fold higher (P = 0.03) and RRV titers were 1.5-fold higher (P < 0.05) than those in CHO K1 cells. Greater increases in infectivity of trypsin-activated virus were measured in CHO α2β1 cells than in CHO K1 cells (190-fold for SA11 [P = 0.02] and 2.8-fold for RRV [P < 0.001]). Although SA11 and RRV infected significantly more CHO α2β1 cells than CHO K1 cells, the infected cells represented only 0.1 to 0.4% of the cells expressing α2β1. For each CHO cell line, the relative levels of virus infectivity closely mirrored the bound virus titers.
FIG. 7.
The increased infectivity of SA11 (left panels) and RRV (right panels) in CHO α2β1 cells compared to that seen with CHO K1 cells was abolished by deletion of the α2 I domain (A), and the growth of RRV, but not SA11, was increased in CHO α2β1 cells compared to that seen with CHO K1 and CHO α2 cells (B). (A) Panels labeled “Trypsin-activated virus” show data for virus grown in the presence of trypsin; panels labeled “Non-activated virus” show data for virus grown in the absence of trypsin. MOIs of 20 (A) and 5 (B) were used.
SA11 and RRV that were not trypsin activated infected fewer CHO α2β1 cells than did fully activated virus (Fig. 7A), but levels of trypsin-activated and nonactivated virus bound relative to CHO K1 cells were similar. Nonactivated SA11 infected 1.9-fold more CHO α2β1 cells (P < 0.05) than CHO K1 cells, and nonactivated RRV infected 2.2-fold more CHO α2β1 cells (P < 0.001) than CHO K1 cells. CHO α2delIβ1 cells were highly sensitive to trypsin and lost adhesion readily, apparently due to their loss of the ability to bind extracellular matrix by using the α2 I domain, so they were infected with nonactivated viruses only. Consistent with the relative abilities of SA11 and RRV to bind CHO α2delIβ1 and CHO K1 cells, nonactivated SA11 and RRV each infected similar numbers of these cells (P > 0.05), showing that the infectivity of nonactivated SA11 and RRV mediated by α2β1 depends on the presence of the α2 I domain. No Wa-infected CHO K1, CHO α2, or CHO α2β1 cells were detected using virus at a MOI of 1 (data not shown).
The growth of RRV, but not SA11 or Wa, in CHO K1 cells is enhanced by transfection with α2β1.
As shown in Fig. 7B, RRV growth was barely detectable in CHO K1 cells, with a 1.6-fold increase in titer at 24 h postinfection (p.i.) and a reduced titer at 48 h p.i. RRV titers in CHO K1 PBJ-1 and CHO α2 cells continuously declined. Thus, the process of transfection and/or the requirement for G418 selection for maintenance of transfectants rendered the cells completely resistant to production of infectious RRV and this resistance was not overcome by α2 transfection. Others have shown that G418 exposure of CHO cells carrying empty vector selected for endogenous Bcl-2 expression (82) and that Bcl-2 expression limited reovirus growth (67), suggesting that G418 selection might have been responsible for limiting RRV growth. However, CHO α2β1 cells supported a sixfold increase in RRV titer at 24 h p.i., showing that α2β1 transfection allowed completion of the RRV replicative cycle and production of infectious RRV. By 48 h p.i., the increase was reduced to twofold, suggesting that only a single cycle of RRV replication had occurred. In all CHO cell lines tested, net yields of SA11 (Fig. 7B) and Wa (data not shown) declined.
A summary of all the experiments described in this paper, their outcomes, the MAbs used, and the MAb epitopes is presented in Table 3.
TABLE 3.
Outline of experiments performed and results obtained in this study
| Cell line and MAb(s) (specificity) | Flow cytometry | Immunoprecipitation of indicated integrin
|
Comigration of virally precipitated proteins with indicated immunoprecipited integrin
|
Change in SA11 and RRV binding, infectivity, and/or growth compared to that of control cellsl | ||
|---|---|---|---|---|---|---|
| α2 | β1 | α2 | β1 | |||
| MA104 | ||||||
| W6/32 (class I MHCa); MOPC21 | − | − | − | − | − | Inf unaffected (SA11) |
| P1E6 (α2 I domain aa 210-211, 268-73b,c | + | + | + | + | + | Inf↓ (SA11) |
| AK7 (α2 I domaind) | + | + | + | + | + | Inf↓ (SA11) |
| HAS3; HAS4 (both to α2 non-I domaind) | NDk | ND | ND | ND | ND | Inf unaffected (SA11) |
| P4C10 (β1 I-like domain aa 207-218e) | +f | + | + | + | + | Inf unaffected (SA11)g |
| P5D2 (β1 I-like domain aa 207-218e,h) | ND | + | + | + | + | ND |
| QE2.E5 (β1 aa 426-587i) | ND | + | + | + | + | ND |
| Caco-2 | ||||||
| MOPC21 control | − | − | − | − | − | ND |
| P1E6; P5D2 | ND | + | + | + | + | ND |
| CHO K1 | ||||||
| MOPC21; RV-5:2 (RV-5 VP4j); no Ab | − | − | − | − | − | Bd, Inf unaffected |
| AK7 | − | − | − | − | − | Bd unaffected |
| P1E6 | − | − | − | − | − | ND |
| HAS3; HAS4 | − | ND | ND | ND | ND | Bd unaffected |
| QE2.E5 | − | ND | ND | ND | ND | ND |
| CHO PBJ-1 | ||||||
| No Ab | ND | ND | ND | ND | ND | Bd, Gr unaffected (SA11), Bd unaffected, Gr↓ (RRV) |
| CHO α2 | ||||||
| MOPC21; RV-5:2 (negative control MAbs) | − | − | − | − | − | Bd↑, Inf↑ |
| No Ab | − | − | − | − | − | Bd↑, Inf↑, Gr↓ |
| AK7 | + | + | + | − | − | Bd↓ |
| P1E6 | + | + | + | − | − | ND |
| HAS3; HAS4 | ND | ND | ND | ND | ND | Bd unaffected |
| QE2.E5 | − | ND | ND | ND | ND | ND |
| CHO α2T221A | ||||||
| MOPC21 (negative control MAb) | − | − | − | − | − | Bd↑, Inf↑ |
| AK7 | + | + | + | − | − | ND |
| P1E6 | ND | + | + | − | − | ND |
| CHO α2D151A and D254A | ||||||
| MOPC21 (negative control MAb) | − | − | − | − | − | Bd↑, Inf↑ |
| AK7 | + | ND | ND | ND | ND | ND |
| CHO α2β1 | ||||||
| MOPC21; RV-5:2 (negative control MAbs) | − | − | − | − | − | Bd⇈, Inf⇈ |
| No Ab | − | − | − | − | − | Bd⇈, Inf⇈, Gr↓ (SA11), Gr↑ (RRV) |
| AK7 | + | + | + | − | + | Bd⇊ |
| P1E6 | + | + | + | − | + | ND |
| HAS3; HAS4 | + | ND | ND | ND | ND | Bd unaffected |
| QE2.E5 | + | ND | ND | ND | ND | ND |
| CHO α2de1Iβ1 | ||||||
| MOPC21; RV-5:2 (negative control MAbs) | − | − | − | − | − | Bd unaffected |
| AK7 | − | − | − | − | − | ND |
| HAS3; HAS4; QE2.E5 | + | ND | ND | ND | ND | ND |
Barnstable et al. (3).
Dickeson et al. (23).
Kamata et al. (45).
Bergelson et al. (7).
Takada and Puzon (78).
Londrigan et al. (57).
Coulson et al. (17).
Seltzer et al. (70).
Faull et al. (31).
Coulson et al. (18).
ND, not determined.
Bd, binding; Inf, infectivity; Gr, growth. Designation in parentheses represents rotavirus strain. Control cells were MA104 with no Ab for MA104 cells and CHO K1 with no Ab for CHO cell lines.
DISCUSSION
In the present study, rotaviruses SA11 and RRV were shown to precipitate surface proteins migrating identically with α2β1 from highly permissive monkey kidney and colonic epithelial cell lines and to precipitate surface β1 from CHO cells transfected with human α2β1. On transfected CHO cells, these rotaviruses bound to human α2β1 and human α2 combined with hamster β1. Virus binding to human α2β1 required the presence of the α2 I domain and was facilitated by the presence of the homologous β1 subunit. Virus binding to human α2β1 and to human α2 combined with hamster β1 was inhibited by anti-I domain MAbs but not by anti-α2 MAbs mapping outside the I domain. This is strong evidence that SA11 and RRV bind α2β1 and that this binding requires the α2 I domain. In confirmation of these studies, Graham et al. have shown that that the α2 ligand peptide DGEA and anti-α2 MAb AK7 specifically inhibit SA11, RRV, and Wa binding to MA104 and α2-transfected K562 cells and that as determined by EIA, Escherichia coli-expressed, purified α2 I domain binds to RRV VP5*-GST (34a).
In an earlier study, it was reported that levels of RRV binding to CHO cells transfected with human α2 or α2β1 were similar to those of RRV binding to a range of other cells, including CHO K1 and MA104 cells, and that anti-α2 MAb AK7 did not affect RRV binding to CHO cells transfected with human α2 or α2β1 (11). It was concluded that, with the exception of the nar strain, rotaviruses do not bind to cells via α2β1 (13, 85). Our ability to demonstrate SA11 and RRV binding to α2β1 on CHO cells may have been due to methodological differences between the binding assays used in the cited negative study and that used in our work. Although Ciarlet et al. tried several different binding assay protocols (11), they unfortunately did not use the method which Hewish et al. had found earlier to be successful in the detection of SA11 binding to α2β1 on K562 cells (41) and which had been used by two groups (51, 58) in the mapping of rotavirus cell attachment to VP4. The modifications made by Ciarlet et al. to this method included measurement of virus binding to trypsinized cells in suspension. Removal of cells from the extracellular matrix produces changed expression and activation of cell adhesion molecules, including integrins (9), alterations in activation of signal transduction pathways, and anoikis (69, 76), which could affect rotavirus binding. Exposed sites on suspended cells that are not normally accessible on cells in monolayers could lead to increased levels of nonspecific virus binding. Ciarlet et al. demonstrated rotavirus infectivity blockade by anti-α2 MAb AK7 in monolayers of integrin-transfected CHO cells (11), which has been confirmed in this study, so the rationale for measuring virus binding to cells in suspension is unclear.
Using flow cytometry, the specificity of the virus-cell binding assay used here was demonstrated with three MAbs that bound α2 on CHO α2 and CHO α2β1 cells. The anti-α2 I domain MAb AK7 inhibited SA11 and RRV binding to α2β1 on these cells, whereas virus binding was not affected by two MAbs which mapped to α2 outside the I domain. Binding assay specificity was also shown by the increase in titers of virus bound to CHO α2 and CHO α2β1 cells, but not to CHO α2delIβ1 cells, over those of controls. The inability of anti-class I MAb W6/32 to inhibit virus infectivity, even though this MAb bound 100% of MA104 cells as determined by flow cytometry, further demonstrates the specificity of this binding assay. It has been shown elsewhere that MAb W6/32 does not affect RRV and SA11 binding to MA104 cells, whereas MAb AK7 at 20 μg/ml inhibits binding of these viruses by 30 and 35%, respectively (34a).
Echovirus 1 binding to α2β1 was reduced by mutations at aa 199 to 201, 212, 214, and 216 in the α2 I domain and required N289 (23, 50). In contrast, type I collagen binding required D151, T221, and T254, which provide coordinating side chains for the metal ion in the MIDAS region (27, 45), and collagen binding was reduced by mutations in the α2 I domain at N154, N190, D219, E256, H258, D259, D292, and E299 (49, 73). As SA11 and RRV rotavirus binding to α2 I domain was not affected by alanine mutagenesis of D151, T221, and T254, these MIDAS residues do not appear to be critical for virus binding and rotavirus binding to the α2 I domain is distinguishable from that of type I collagen. More detailed mapping of the regions and residues involved in rotavirus binding to the α2 I domain would be needed to determine whether rotaviruses and echovirus 1 have similar binding requirements. Interestingly, Graham et al. have shown elsewhere that binding of SA11 to α2β1 on K562 cells is increased following α2β1 activation (34a). In contrast, echovirus 1 binding to α2β1 on K562 cells is not affected by α2β1 activation (6). This suggests that α2β1 binding by rotaviruses, echoviruses, and collagen involves distinct α2β1 residues.
The 140-kDa protein precipitated by SA11 from CHO α2β1 cells was identified as the β1 integrin subunit by its comigration with the β1 coprecipitated (using anti-α2 MAbs) with α2 and by the inability of virus to precipitate this protein from parental CHO cells, CHO cells transfected with α2 alone, or CHO cells transfected with α2β1 lacking the α2 I domain. This supports the conclusion that the 130- and 140-kDa proteins, migrating indistinguishably from β1, which were precipitated from Caco-2 and MA104 cells by SA11 and/or RRV, respectively, are likely to be β1. The β1 subunit displays differing degrees of mobility in different cell types due to differing degrees of glycosylation (40). The detection of SA11 precipitation of β1 from CHO α2β1, but not CHO α2, cells is consistent with the levels of binding of infectious SA11 and RRV to α2β1 being highest on CHO α2β1 cells.
The precipitation of β1 but not α2 from CHO α2β1 cells was unexpected. As the β1 subunit appeared to be more heavily biotinylated than the α2 subunit in lysates of these cells, it is possible that precipitated α2 was not detected due to a lack of assay sensitivity. Alternatively, SA11 may be capable of binding β1 epitopes directly. The precipitation of an apparently larger amount of β1 than α2 from MA104 cell lysates by SA11 is consistent with the latter possibility. An additional explanation is that these differences in virus-precipitated protein band intensity result from interexperimental variation rather than true biological differences.
MAb 2G4 binds to the putative fusion region of VP5* (59, 60), which consists of 90 aa in the linear sequence from the DGE tripeptide sequence implicated in α2β1 binding. As MAb 2G4 precipitated SA11 complexed with proteins indistinguishable from α2β1, some 2G4 recognition sites were available for binding after the complexes were formed.
Previous studies of cellular proteins bound by rotaviruses have used virus overlay protein blot assays (VOPBA), in which cellular proteins were reduced and dissociated before being reacted with virus (42, 43). MA104 cell proteins of 66 to 97 kDa bound by SA11 in VOPBA (42, 43) and octyl glucoside-extracted MA104 cell proteins, including heat shock cognate protein 70 (35), which blocked rotavirus infection (37), were not precipitated in our studies. Two proteins were recognized by SA11 in VOPBA through a galactose moiety (42) and so may not have bound virus with sufficient affinity for detection by precipitation.
SA11 and RRV did not precipitate αxβ2 or αvβ3 from MA104 cell lysates, as the only cellular proteins specifically precipitated by virus migrated indistinguishably from α2β1 at 160 and 140 kDa. The αx, β2, αv, and β3 integrin subunits migrate at approximately 150, 95, 150, and 110 kDa (36, 48). Consistent with this, RRV, Wa, and nar3 rotaviruses do not bind β3 on these cells (34a, 36). However, the unidentified Caco-2 cell surface protein of approximately 160 to 165 kDa precipitated by SA11 might be αx or αv. It is also possible that this 160- to 165-kDa protein was associated with α2β1 rather than precipitated by virus directly. It is significant that proteins with the characteristics of α2β1 were precipitated by SA11 from Caco-2 cells, as these cells share some characteristics with human intestinal enterocytes. In microarray analysis of gene expression 16 h after RRV infection of Caco-2 cells, α2 and β1 expression were increased 3.1-fold and 2.5-fold, respectively, and it was suggested that infection might upregulate this receptor on infected or nearby uninfected cells (21).
CHO cell transfection with α2 or α2β1 increased cell binding by SA11 and RRV to similar degrees, and both viruses used α2β1 to infect the cells. However, RRV infected larger numbers of cells and only RRV infection produced detectable infectious virus (in CHO α2β1 cells only) in growth assays. Also, only a small fraction of the SA11 and RRV bound to α2β1 entered CHO K1 cells, as measured by the number of infected cells at 16 h after infection. This low level of infectivity was shown previously for RRV (11) (Table 1). These observations suggest that these cells have a block in rotavirus entry after attachment via α2β1 (and an additional barrier to production or release of infectious virions following viral protein synthesis) and that these blocks affected SA11 to a greater extent than RRV. The existence of a block in rotavirus cell entry is supported by the report that liposome-mediated infection of CHO K1 cells with double-layered rotavirus particles, which are noninfectious when added exogenously to cells and lack VP4 or VP7, resulted in levels of virus infection similar to those seen with MA104 cells (11). It would be interesting to determine whether CHO K1 cells expressing α2β1, αxβ2, and αvβ3 allow more efficient entry of SA11 and RRV, as αxβ2 and αvβ3 have a role in rotavirus entry, and when a mixture of α2β1 and αxβ2 ligand peptides (17) or combinations of MAbs to α2β1, αxβ2, and αvβ3 (11, 17, 36) are used, blockade of rotavirus infectivity in permissive cells is additive.
It is noteworthy that the elevated binding of SA11 and RRV to α2β1 in the presence of homologous, rather than heterologous, β1 led to increased SA11 and RRV infectivity and increased RRV growth. The presence of human β1 may have stabilized α2β1 in a higher activation state. It has been shown that activation of human β1 on K562 cells expressing α2β1 results in increased levels of infectious SA11 binding to α2β1 (34a). A rotavirus preference for homologous integrin could help explain species-specific restrictions in virus tropism.
In contrast to the results seen with SA11 and RRV, Wa neither bound α2β1 on CHO cells nor infected CHO cells, a result in line with those of previous studies (11). However, anti-α2 MAb blockade of Wa infectivity in MA104 cells has been demonstrated (11, 36). Also, Graham et al. have shown Wa binding to α2β1 on K562 cells, MAb AK7 blockade of Wa binding to these cells, and AK7 blockade of Wa binding to MA104 cells (34a). Thus, an accessory molecule or additional receptor required for Wa, but not SA11 or RRV, binding to α2β1 appears to be lacking from CHO cells but is present on K562 and MA104 cells. This provides a basis for further studies to determine the full requirements for human rotavirus cell attachment.
Rotavirus binding to α2β1, and interaction with αxβ2 and αvβ3 during cell entry, is consistent with the proposed mechanisms of rotavirus cell entry. Integrin activation following ligand binding leads to formation of focal adhesions in which integrins are clustered and cytoskeletal and signaling proteins are recruited to the integrin cytoplasmic domains (34). Activated integrins can facilitate endocytosis through direct interaction with protein kinase Cα and caveolin (34, 64). Although the pathway of rotavirus cell entry is not defined, there is evidence for both endocytosis and direct membrane penetration (8, 26, 44). Adenovirus recognition of αvβ3 via the penton base leads to both clathrin-dependent endocytosis and macropinocytosis of virus (62), whereas echovirus 1 binding to α2β1 results in virus entry through caveolae (61). Thus, even though both these viruses use integrins which can be associated with caveolin (83), they enter cells through different pathways. Therefore, rotavirus cell entry pathway(s) cannot be predicted from the viral integrin usage. It will be important to determine whether rotavirus is internalized in a complex with integrin following binding or whether the virus-integrin interaction is transient and serves to increase the proximity of the VP5* fusion region to the cell membrane. Induction of signaling cascades by rotavirus-integrin interactions could facilitate cell entry and might also enhance virus replication by modifying cell functions such as survival and differentiation.
The studies reported here have helped establish the role of α2β1 in rotavirus attachment and entry. On CHO cells, SA11 and RRV bind to α2β1, resulting in a low level of cellular infection. This binding requires the α2 I domain and is facilitated by the presence of homologous β1. As SA11 and RRV do not require the same MIDAS residues as type I collagen for binding to the α2 I domain, human rotavirus binding to α2β1 also may be distinguishable from that of collagen. The development of therapeutic agents capable of selectively inhibiting rotavirus binding to α2β1 might therefore be feasible. It will be important to determine whether rotavirus-neutralizing antibodies (induced by natural infection or candidate rotavirus vaccines) act by inhibition of virus binding to α2β1.
Acknowledgments
We are most grateful to F. Watt for provision of MAbs HAS3 and HAS4; M. Estes and J. Burns for MAb 2G4; D. Leavesley, E. Wayner, and P. Simmons for anti-β1 integrin MAbs; and A. Brooks for MAb W6/32. We thank I. H. Holmes, M. Hewish, M. Anders, and L. Harrison for helpful discussions, and the students taking part in the Rotavirus Medical Microbiology Project in 2000 assisted in producing the P1E6 MAb blockade results.
This work was supported by project grants 980635 and 208900 from the National Health and Medical Research Council of Australia (NHMRC). B.S.C. is a Senior Research Fellow of the NHMRC.
REFERENCES
- 1.Arias, C. F., P. Romero, V. Alvarez, and S. Lopez. 1996. Trypsin activation pathway of rotavirus infectivity. J. Virol. 70:5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaout, M. A. 2002. Integrin structure: new twists and turns in dynamic cell adhesion. Immunol. Rev. 186:125-140. [DOI] [PubMed] [Google Scholar]
- 3.Barnstable, C. J., W. F. Bodmer, G. Brown, G. Galfre, C. Milstein, A. F. Williams, and A. Ziegler. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens—new tools for genetic analysis. Cell 14:9-20. [DOI] [PubMed] [Google Scholar]
- 4.Bass, D. M., E. R. Mackow, and H. B. Greenberg. 1991. Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology 183:602-610. [DOI] [PubMed] [Google Scholar]
- 5.Bastardo, J. W., and I. H. Holmes. 1980. Attachment of SA-11 rotavirus to erythrocyte receptors. Infect. Immun. 29:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergelson, J. M., B. M. Chan, R. W. Finberg, and M. E. Hemler. 1993. The integrin VLA-2 binds echovirus 1 and extracellular matrix ligands by different mechanisms. J. Clin. Investig. 92:232-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergelson, J. M., N. F. St. John, S. Kawaguchi, R. Pasqualini, F. Berdichevsky, M. E. Hemler, and R. W. Finberg. 1994. The I domain is essential for echovirus 1 interaction with VLA-2. Cell Adhes. Commun. 2:455-464. [DOI] [PubMed] [Google Scholar]
- 8.Chemello, M. E., O. C. Aristimuño, F. Michelangeli, and M.-C. Ruiz. 2002. Requirement for vacuolar H+-ATPase activity and Ca2+ gradient during entry of rotavirus into MA104 cells. J. Virol. 76:13083-13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D., V. Magnuson, S. Hill, C. Arnaud, B. Steffensen, and R. J. Klebe. 1992. Regulation of integrin gene expression by substrate adherence. J. Biol. Chem. 267:23502-23506. [PubMed] [Google Scholar]
- 10.Chen, P., C. Melchior, N. H. Brons, N. Schlegel, J. Caen, and N. Kieffer. 2001. Probing conformational changes in the I-like domain and the cysteine-rich repeat of human beta 3 integrins following disulfide bond disruption by cysteine mutations: identification of cysteine 598 involved in αIIβ3 activation. J. Biol. Chem. 276:38628-38635. [DOI] [PubMed] [Google Scholar]
- 11.Ciarlet, M., S. E. Crawford, E. Cheng, S. E. Blutt, D. A. Rice, J. M. Bergelson, and M. K. Estes. 2002. VLA-2 (α2β1) integrin promotes rotavirus entry into cells but is not necessary for rotavirus attachment. J. Virol. 76:1109-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. [DOI] [PubMed] [Google Scholar]
- 13.Ciarlet, M., J. E. Ludert, M. Iturriza-Gómara, F. Liprandi, J. J. Gray, U. Desselberger, and M. K. Estes. 2002. Initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 76:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark, S. M., J. R. Roth, M. L. Clark, B. B. Barnett, and R. S. Spendlove. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J. Virol. 39:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulson, B. S. 1997. Effects of workshop monoclonal antibodies on rotavirus infection of cells, p. 391-393. In T. Kishimoto, H. Kikutani, A. E. G. K. von dem Borne, S. M. Goyert, D. Y. Mason, M. Miyasaka, L. Moretta, K. Okumura, S. Shaw, T. A. Springer, K. Sugamura, and H. Zola (ed.), Leucocyte typing VI. Garland Publishing, Inc., New York, N.Y.
- 16.Coulson, B. S., K. J. Fowler, R. F. Bishop, and R. G. Cotton. 1985. Neutralizing monoclonal antibodies to human rotavirus and indications of antigenic drift among strains from neonates. J. Virol. 54:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coulson, B. S., S. L. Londrigan, and D. J. Lee. 1997. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc. Natl. Acad. Sci. USA 94:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulson, B. S., J. M. Tursi, W. J. McAdam, and R. F. Bishop. 1986. Derivation of neutralizing monoclonal antibodies to human rotaviruses and evidence that an immunodominant neutralization site is shared between serotypes 1 and 3. Virology 154:302-312. [DOI] [PubMed] [Google Scholar]
- 19.Coulson, B. S., L. E. Unicomb, G. A. Pitson, and R. F. Bishop. 1987. Simple and specific enzyme immunoassay using monoclonal antibodies for serotyping human rotaviruses. J. Clin. Microbiol. 25:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford, S. E., M. Labbe, J. Cohen, M. H. Burroughs, Y. J. Zhou, and M. K. Estes. 1994. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J. Virol. 68:5945-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuadras, M. A., D. A. Feigelstock, S. An, and H. B. Greenberg. 2002. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 76:4467-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delorme, C., H. Brussow, J. Sidoti, N. Roche, K. A. Karlsson, J. R. Neeser, and S. Teneberg. 2001. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol. 75:2276-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickeson, S. K., N. L. Mathis, M. Rahman, J. M. Bergelson, and S. A. Santoro. 1999. Determinants of ligand binding specificity of the α1β1 and α2β1 integrins. J. Biol. Chem. 274:32182-32191. [DOI] [PubMed] [Google Scholar]
- 24.Dormitzer, P. R., Z. Y. Sun, O. Blixt, J. C. Paulson, G. Wagner, and S. C. Harrison. 2002. Specificity and affinity of sialic acid binding by the rhesus rotavirus VP8* core. J. Virol. 76:10512-10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dormitzer, P. R., Z. Y. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowling, W., E. Denisova, R. LaMonica, and E. R. Mackow. 2000. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 74:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley, J., C. G. Knight, R. W. Farndale, M. J. Barnes, and R. C. Liddington. 2000. Structural basis of collagen recognition by integrin α2β1. Cell 101:47-56. [DOI] [PubMed] [Google Scholar]
- 28.Espejo, R. T., S. Lopez, and C. Arias. 1981. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J. Virol. 37:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evander, M., I. H. Frazer, E. Payne, Y. M. Qi, K. Hengst, and N. A. McMillan. 1997. Identification of the α6 integrin as a candidate receptor for papillomaviruses. J. Virol. 71:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faull, R. J., J. Wang, D. I. Leavesley, W. Puzon, G. R. Russ, D. Vestweber, and Y. Takada. 1996. A novel activating anti-β1 integrin monoclonal antibody binds to the cysteine-rich repeats in the β1 chain. J. Biol. Chem. 271:25099-25106. [DOI] [PubMed] [Google Scholar]
- 32.Fukudome, K., O. Yoshie, and T. Konno. 1989. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell adsorption. Virology 172:196-205. [DOI] [PubMed] [Google Scholar]
- 33.Gamble, J. R., L. J. Matthias, G. Meyer, P. Kaur, G. Russ, R. Faull, M. C. Berndt, and M. A. Vadas. 1993. Regulation of in vitro capillary tube formation by anti-integrin antibodies. J. Cell Biol. 121:931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giancotti, F. G., and E. Ruoslahti. 1999. Integrin signaling. Science 285:1028-1032. [DOI] [PubMed] [Google Scholar]
- 34a.Graham, K. L., P. Halasz, Y. Tan, M. J. Hewish, Y. Takada, E. R. Mackow, M. K. Robinson, and B. S. Coulson. Integrin-using rotaviruses bind α2β1 integrin α2 I domain via VP4 DGE sequence and recognize αXβ2 and αVβ3 by using VP7 during cell entry. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 35.Guerrero, C. A., D. Bouyssounade, S. Zarate, P. Isa, T. Lopez, R. Espinosa, P. Romero, E. Mendez, S. Lopez, and C. F. Arias. 2002. Heat shock cognate protein 70 is involved in rotavirus cell entry. J. Virol. 76:4096-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerrero, C. A., E. Mendez, S. Zarate, P. Isa, S. Lopez, and C. F. Arias. 2000. Integrin αvβ3 mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97:14644-14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerrero, C. A., S. Zarate, G. Corkidi, S. Lopez, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo, C. T., O. Nakagomi, M. Mochizuki, H. Ishida, M. Kiso, Y. Ohta, T. Suzuki, D. Miyamoto, K. I. Hidari, and Y. Suzuki. 1999. Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J. Biochem. (Tokyo) 126:683-688. [DOI] [PubMed] [Google Scholar]
- 39.Hemler, M. E., C. Huang, and L. Schwarz. 1987. The VLA protein family. Characterization of five distinct cell surface heterodimers each with a common 130,000 molecular weight beta subunit. J. Biol. Chem. 262:3300-3309. [PubMed] [Google Scholar]
- 40.Hemler, M. E., J. G. Jacobson, and J. L. Strominger. 1985. Biochemical characterization of VLA-1 and VLA-2. Cell surface heterodimers on activated T cells. J. Biol. Chem. 260:15246-15252. [PubMed] [Google Scholar]
- 41.Hewish, M. J., Y. Takada, and B. S. Coulson. 2000. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jolly, C. L., B. M. Beisner, and I. H. Holmes. 2000. Rotavirus infection of MA104 cells is inhibited by ricinus lectin and separately expressed single binding domains. Virology 275:89-97. [DOI] [PubMed] [Google Scholar]
- 43.Jolly, C. L., B. M. Beisner, E. Ozser, and I. H. Holmes. 2001. Non-lytic extraction and characterisation of receptors for multiple strains of rotavirus. Arch. Virol. 146:1307-1323. [DOI] [PubMed] [Google Scholar]
- 44.Kaljot, K. T., R. D. Shaw, D. H. Rubin, and H. B. Greenberg. 1988. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J. Virol. 62:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamata, T., W. Puzon, and Y. Takada. 1994. Identification of putative ligand binding sites within I domain of integrin α2β1 (VLA-2, CD49b/CD29). J. Biol. Chem. 269:9659-9663. [PubMed] [Google Scholar]
- 46.Kamata, T., W. Puzon, and Y. Takada. 1995. Identification of putative ligand-binding sites of the integrin α4β1 (VLA-4, CD49d/CD29). Biochem J. 305:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamata, T., and Y. Takada. 1994. Direct binding of collagen to the I domain of integrin α2β1 (VLA-2, CD49b/CD29) in a divalent cation-independent manner. J. Biol. Chem. 269:26006-26010. [PubMed] [Google Scholar]
- 48.Kamata, T., R. Wright, and Y. Takada. 1995. Critical threonine and aspartic acid residues within the I domains of β2 integrins for interactions with intercellular adhesion molecule 1 (ICAM-1) and C3bi. J. Biol. Chem. 270:12531-12535. [DOI] [PubMed] [Google Scholar]
- 49.Kapyla, J., J. Ivaska, R. Riikonen, P. Nykvist, O. Pentikainen, M. Johnson, and J. Heino. 2000. Integrin α2I domain recognizes type I and type IV collagens by different mechanisms. J. Biol. Chem. 275:3348-3354. [DOI] [PubMed] [Google Scholar]
- 50.King, S. L., T. Kamata, J. A. Cunningham, J. Emsley, R. C. Liddington, Y. Takada, and J. M. Bergelson. 1997. Echovirus 1 interaction with the human very late antigen-2 (integrin α2β1) I domain. Identification of two independent virus contact sites distinct from the metal ion-dependent adhesion site. J. Biol. Chem. 272:28518-28522. [DOI] [PubMed] [Google Scholar]
- 51.Kirkwood, C. D., R. F. Bishop, and B. S. Coulson. 1998. Attachment and growth of human rotaviruses RV-3 and S12/85 in Caco-2 cells depend on VP4. J. Virol. 72:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komoriya, A., L. J. Green, M. Mervic, S. S. Yamada, K. M. Yamada, and M. J. Humphries. 1991. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J. Biol. Chem. 266:15075-15079. [PubMed] [Google Scholar]
- 53.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 54.Lazdins, I., B. S. Coulson, C. Kirkwood, M. Dyall-Smith, P. J. Masendycz, S. Sonza, and I. H. Holmes. 1995. Rotavirus antigenicity is affected by the genetic context and glycosylation of VP7. Virology 209:80-89. [DOI] [PubMed] [Google Scholar]
- 55.Lindqvist, Y., and G. Schneider. 1996. Protein-biotin interactions. Curr. Opin. Struct. Biol. 6:798-803. [DOI] [PubMed] [Google Scholar]
- 56.Loike, J. D., B. Sodeik, L. Cao, S. Leucona, J. I. Weitz, P. A. Detmers, S. D. Wright, and S. C. Silverstein. 1991. CD11c/CD18 on neutrophils recognizes a domain at the N terminus of the A alpha chain of fibrinogen. Proc. Natl. Acad. Sci. USA 88:1044-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Londrigan, S. L., M. J. Hewish, M. J. Thomson, G. M. Sanders, H. Mustafa, and B. S. Coulson. 2000. Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 81:2203-2213. [DOI] [PubMed] [Google Scholar]
- 58.Ludert, J. E., N. Feng, J. H. Yu, R. L. Broome, Y. Hoshino, and H. B. Greenberg. 1996. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol. 70:487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mackow, E. R., R. D. Shaw, S. M. Matsui, P. T. Vo, M. N. Dang, and H. B. Greenberg. 1988. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc. Natl. Acad. Sci. USA 85:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mackow, E. R., M. Y. Yamanaka, M. N. Dang, and H. B. Greenberg. 1990. DNA amplification-restricted transcription-translation: rapid analysis of rhesus rotavirus neutralization sites. Proc. Natl. Acad. Sci. USA 87:518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marjomaki, V., V. Pietiainen, H. Matilainen, P. Upla, J. Ivaska, L. Nissinen, H. Reunanen, P. Huttunen, T. Hyypia, and J. Heino. 2002. Internalization of echovirus 1 in caveolae. J. Virol. 76:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meier, O., K. Boucke, S. V. Hammer, S. Keller, R. P. Stidwill, S. Hemmi, and U. F. Greber. 2002. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 158:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mendez, E., C. F. Arias, and S. Lopez. 1993. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J. Virol. 67:5253-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng, T., D. Shima, A. Squire, P. I. Bastiaens, S. Gschmeissner, M. J. Humphries, and P. J. Parker. 1999. PKCα regulates β1 integrin-dependent cell motility through association and control of integrin traffic. EMBO J. 18:3909-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Connell, P. J., R. Faull, G. R. Russ, and A. J. F. D'Apice. 1991. VLA-2 is a collagen receptor on endothelial cells. Immunol. Cell Biol. 69:103-110. [DOI] [PubMed] [Google Scholar]
- 66.Poloni, F., P. Puddu, F. Moretti, M. Flego, G. Romagnoli, M. Tombesi, I. Capone, A. Chersi, F. Felici, and M. Cianfriglia. 2001. Identification of a LFA-1 region involved in the HIV-1-induced syncytia formation through phage-display technology. Eur. J. Immunol. 31:57-63. [DOI] [PubMed] [Google Scholar]
- 67.Rodgers, S. E., E. S. Barton, S. M. Oberhaus, B. Pike, C. A. Gibson, K. L. Tyler, and T. S. Dermody. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 71:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rolsma, M. D., T. B. Kuhlenschmidt, H. B. Gelberg, and M. S. Kuhlenschmidt. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 72:9079-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruoslahti, E., and J. C. Reed. 1994. Anchorage dependence, integrins, and apoptosis. Cell 77:477-478. [DOI] [PubMed] [Google Scholar]
- 70.Seltzer, J. L., A. Y. Lee, K. T. Akers, B. Sudbeck, E. A. Southon, E. A. Wayner, and A. Z. Eisen. 1994. Activation of 72-kDa type IV collagenase/gelatinase by normal fibroblasts in collagen lattices is mediated by integrin receptors but is not related to lattice contraction. Exp. Cell Res. 213:365-374. [DOI] [PubMed] [Google Scholar]
- 71.Shaw, R. D., P. T. Vo, P. A. Offit, B. S. Coulson, and H. B. Greenberg. 1986. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology 155:434-451. [DOI] [PubMed] [Google Scholar]
- 72.Skerra, A., and T. G. Schmidt. 2000. Use of the Strep-Tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326:271-304. [DOI] [PubMed] [Google Scholar]
- 73.Smith, C., D. Estavillo, J. Emsley, L. A. Bankston, R. C. Liddington, and M. A. Cruz. 2000. Mapping the collagen-binding site in the I domain of the glycoprotein Ia/IIa (integrin α2β1). J. Biol. Chem. 275:4205-4209. [DOI] [PubMed] [Google Scholar]
- 74.Srnka, C. A., M. Tiemeyer, J. H. Gilbert, M. Moreland, H. Schweingruber, B. W. de Lappe, P. G. James, T. Gant, R. E. Willoughby, R. H. Yolken, et al. 1992. Cell surface ligands for rotavirus: mouse intestinal glycolipids and synthetic carbohydrate analogs. Virology 190:794-805. [DOI] [PubMed] [Google Scholar]
- 75.Staatz, W. D., K. F. Fok, M. M. Zutter, S. P. Adams, B. A. Rodriguez, and S. A. Santoro. 1991. Identification of a tetrapeptide recognition sequence for the α2β1 integrin in collagen. J. Biol. Chem. 266:7363-7367. [PubMed] [Google Scholar]
- 76.Stupack, D. G., and D. A. Cheresh. 2002. Get a ligand, get a life: integrins, signaling and cell survival. J. Cell Sci. 115:3729-3738. [DOI] [PubMed] [Google Scholar]
- 77.Takada, Y., and M. E. Hemler. 1989. The primary structure of the VLA-2/collagen receptor alpha 2 subunit (platelet GPIa): homology to other integrins and the presence of a possible collagen-binding domain. J. Cell Biol. 109:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takada, Y., and W. Puzon. 1993. Identification of a regulatory region of integrin beta 1 subunit using activating and inhibiting antibodies. J. Biol. Chem. 268:17597-17601. [PubMed] [Google Scholar]
- 79.Takada, Y., J. Ylanne, D. Mandelman, W. Puzon, and M. H. Ginsberg. 1992. A point mutation of integrin β1 subunit blocks binding of α5β1 to fibronectin and invasin but not recruitment to adhesion plaques. J. Cell Biol. 119:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takagi, J., T. Kamata, J. Meredith, W. Puzon-McLaughlin, and Y. Takada. 1997. Changing ligand specificities of αvβ1 and αvβ3 integrins by swapping a short diverse sequence of the beta subunit. J. Biol. Chem. 272:19794-19800. [DOI] [PubMed] [Google Scholar]
- 81.Tan, S. M., M. K. Robinson, K. Drbal, Y. van Kooyk, J. M. Shaw, and S. K. Law. 2001. The N-terminal region and the mid-region complex of the integrin beta 2 subunit. J. Biol. Chem. 276:36370-36376. [DOI] [PubMed] [Google Scholar]
- 82.Tey, B. T., R. P. Singh, L. Piredda, M. Piacentini, and M. Al-Rubeai. 2000. Influence of bcl-2 on cell death during the cultivation of a Chinese hamster ovary cell line expressing a chimeric antibody. Biotechnol. Bioeng. 68:31-43. [DOI] [PubMed] [Google Scholar]
- 83.Wary, K. K., A. Mariotti, C. Zurzolo, and F. G. Giancotti. 1998. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell 94:625-634. [DOI] [PubMed] [Google Scholar]
- 84.Willoughby, R. E. 1993. Rotaviruses preferentially bind O-linked sialylglycoconjugates and sialomucins. Glycobiology 3:437-445. [DOI] [PubMed] [Google Scholar]
- 85.Zarate, S., R. Espinosa, P. Romero, C. A. Guerrero, C. F. Arias, and S. Lopez. 2000. Integrin α2β1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50-54. [DOI] [PubMed] [Google Scholar]