Abstract
During the immediate-early (IE) phase of reactivation from latency, the Kaposi's sarcoma-associated herpesvirus (KSHV) replication and transcription activator protein (RTA) (or ORF50) is thought to be the most critical trigger that upregulates expression of many downstream viral lytic cycle genes, including the delayed-early (DE) gene encoding the replication-associated protein (RAP) (or K8). RAP physically interacts with and stabilizes the cellular transcription factor CCAAT/enhancer-binding protein-α (C/EBPα), leading to upregulated expression of the cellular C/EBPα and p21CIP-1 proteins followed by G0/G1 cell cycle arrest. Furthermore, RTA also interacts with C/EBPα, and both RAP and RTA cooperate with C/EBPα to activate the RAP promoter through binding to a strong proximal C/EBP binding site that also serves as an RTA-responsive element (RRE). Here we show that C/EBPα also activates the IE RTA promoter in transient-cotransfection reporter gene assays and that addition of either RTA or RAP enhances the effect. Electrophoretic mobility shift assay and deletion analysis revealed three C/EBP binding sites that mediate cooperative transactivation of the RTA promoter by C/EBPα and RTA. Furthermore, chromatin immunoprecipitation assay results showed that the endogenous C/EBPα, RTA, and RAP proteins all associate with RTA promoter sequences in tetradecanoyl phorbol acetate-induced primary effusion lymphoma (PEL) cells. Induction of endogenous KSHV RTA mRNA in PEL cells by exogenously introduced C/EBPα was confirmed by reverse transcription-PCR analysis and by double-label indirect immunofluorescence assays. Reciprocally, expression of exogenous RTA also led to an increase of endogenous C/EBPα expression that could be detected by Western immunoblot assays even in KSHV-negative DG75 cells. Cotransfected RTA also increased positive C/EBPα autoregulation of the C/EBPα promoter in transient-cotransfection reporter gene assays. Finally, C/EBPα proved to strongly activate the promoters of two other KSHV DE genes encoding PAN (polyadenylated nuclear) RNA and MTA (ORF57), which was again mediated by C/EBP binding sites that also contribute to RTA activation. Overall, these results support a model in which the cellular transcription factor C/EBPα and RTA:C/EBPα interactions play important roles both upstream and downstream of the two major KSHV regulatory proteins RTA and RAP during the early stages of lytic cycle reactivation.
Latent infection with Kaposi's sarcoma-associated herpesvirus (KSHV) is detected in the spindle cells of all forms of KS (5), as well as in plasmablast-like cells of AIDS-associated multicentric Castleman's disease and primary effusion lymphoma (PEL) (2, 3, 38). Genome analysis revealed that KSHV belongs to the gamma-2 herpesvirus subfamily and is related to Epstein-Barr virus (EBV) (34). Based on the kinetics of gene expression after viral reactivation, KSHV-encoded genes are classified into four broad categories: latent, immediate-early (IE), delayed-early (DE), and late. Several PEL cell lines carrying multicopy KSHV episomes have been established (2, 3), and virus derived from them can convert dermal microvascular endothelial cells (DMVECs) into KS-like spindle cells (9a). Only latent-state KSHV proteins, including LANA1, v-CYC-D, v-FLIP, and K15 (or LAMP), are expressed in the majority of PEL cells, KS spindle cells, and infected DMVECs (4, 9a, 28, 31). However, treatment with tetradecanoyl phorbol acetate (TPA) or sodium butyrate can disrupt the latency of KSHV infection, especially in PEL cells, and induce lytic viral replication in a subset of the cells (27, 30). Induction of several transcripts, including those from the major RTA (or ORF50), ORF45, and ORF-K4.2 promoters as well as a minor RAP (or K8) promoter of KSHV have been detected within 4 h after lytic cycle induction in the presence of cycloheximide, and therefore these genes have been defined as the likely IE regulatory genes of KSHV (33, 55).
The KSHV-encoded RTA protein is generally considered to function as the principal molecular switch converting infected cells from a latent state into the lytic cycle by specific transcriptional activation of many downstream KSHV promoters (39). The 120-kDa RTA protein (691 amino acids [aa]) is a homologue of the EBV DNA-binding transactivator RTA (or BRLF1), which both play crucial roles as the triggers of complete lytic reactivation from latency. KSHV RTA transactivates various downstream KSHV lytic cycle promoters, including those for RAP (a positional homologue of EBV ZTA), KSHV MTA (or ORF57 a homologue of EBV MTA), vIL6, vMIP (or vCCL), K12 (or Kaposin), vOX2/GPCR, and PAN (polyadenylated nuclear) (or T1.1) RNA (9, 24-26, 35, 37, 44, 45). In at least some target promoters (e.g., PAN, K12, and vIL6), RTA clearly functions by direct DNA binding to specific type II RTA-responsive elements (RREs) (4a, 12a, 37, 46). Although Lukac et al. (24) reported that baculovirus-expressed RTA also binds to the MTA and RAP RRE motifs in electrophoretic mobility shift assay (EMSA) experiments, we observed that in vitro-translated RTA does not bind directly to the type I RAP RRE (46). Instead, RTA activates RAP expression through an interaction with CCAAT/enhancer-binding protein-α (C/EBPα), which itself directly binds to the RRE site and activates the RAP promoter (46). Furthermore, a strong physical interaction was demonstrated between C/EBPα and RTA that requires basic residues in the RTA DNA-binding domain (DBD) between aa 151 and 273. RTA has also been reported to activate target gene expression at another type I RRE within the MTA promoter via a functional interaction with the cellular CBF1 (or RBP-Jκ) DNA binding factor (22), a target of the Notch signaling pathway that also plays an important role in EBNA2-mediated functions during EBV latency (17, 18, 23a).
Previous studies have suggested that KSHV RTA positively autoregulates its own promoter in reporter gene cotransfection assays in 293 cells (12, 32). In addition, a consensus motif at position −220/−213 in the RTA promoter was shown to bind to the OCT1 protein and was interpreted to be important for RTA-mediated autoregulation in 293L cells (32). However, because of our evidence that KSHV RTA physically interacts with C/EBPα (46) and the predicted presence of C/EBP sites in the RTA promoter, we wished to evaluate other potential mechanisms for controlling expression of the RTA lytic cycle trigger protein. C/EBPα was the first identified member of the leucine zipper family of transcription factors, which includes c-JUN, c-FOS, ATF, CREB, and EBV ZTA (6, 20). Members of this family bind to specific DNA elements as homodimers or heterodimers through the conserved bZIP domain located at the C terminus and modulate gene expression through an N-terminal activation domain. Numerous studies have shown that C/EBPα promotes differentiation (11, 21, 47, 53) and inhibits cell proliferation through multiple molecular pathways leading to G1 cell cycle arrest, including elevating the level of p21CIP-1/WAF-1 protein by both directly binding to and activating the p21CIP-1 promoter and stabilizing p21 through a protein-protein interaction (41, 42, 50). C/EBPα also inhibits E2F transcription of genes, including c-MYC, which is involved in DNA synthesis and mitosis through direct interaction with retinoblastoma (Rb) and Rb-like proteins (19, 36), as well as directly inhibiting CDK2 and CDK4 (16, 43).
The 35-kDa KSHV-encoded RAP protein (237 aa) is also a bZIP-like protein (23, 40, 55) that targets to nuclear PML oncogenic domains and seems to play an important role in lytic cycle viral DNA replication (49). Unlike its positional homologue ZTA, the major lytic cycle trigger protein encoded by EBV, RAP alone does not bind to any known downstream KSHV target gene promoters, nor does it act alone as a trigger for KSHV lytic cycle induction (9, 29). However, both RAP and ZTA display similar properties in upregulating levels of the cellular C/EBPα and p21CIP-1 proteins, resulting in G0/G1 cell cycle arrest during progression of the KSHV or EBV lytic cycles, respectively (48, 50). The mechanism of C/EBPα induction was found to involve strong protein-protein interactions between C/EBPα and RAP or C/EBPα and ZTA, leading to both stabilization of C/EBPα protein and cooperative transcriptional upregulation of the cellular C/EBPα and p21 promoters (50, 51). C/EBPα also proved to reciprocally activate viral RAP and ZTA gene expression by directly binding to strong promoter proximal C/EBP binding sites (46, 51; F. Y. Wu et al., unpublished data).
In the present study, we have asked whether C/EBPα might also play a role in activating KSHV IE gene expression and have used EMSA and mutation analysis to identify three C/EBPα binding sites within the proximal 914-bp RTA promoter. Cotransfection of a series of deletion mutant or mutated RTA-LUC target reporter genes in several cell types was used to evaluate both direct and cooperative responsiveness to C/EBPα, RAP, and RTA. A chromatin immunoprecipitation (ChIP) assay was employed to investigate whether C/EBPα, RAP, and RTA associate in vivo with the RTA promoter in KSHV lytically infected PEL cells. We then also tested the ability of C/EBPα to upregulate endogenous RTA in PEL cells and possible cooperative effects of RTA plus C/EBPα in regulating the cellular C/EBPα promoter. The kinetics of induction of C/EBPα mRNA and protein levels in TPA-induced PEL cells were also examined. Finally, we searched for C/EBP binding sites and investigated C/EBPα responsiveness, C/EBPα plus RTA cooperativity, and in vivo ChIP associations in two other downstream KSHV DE promoters that are associated with the PAN and MTA genes and have been shown to respond strongly to RTA.
MATERIALS AND METHODS
Cells and plasmids.
HeLa and 293T cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif.) containing 10% fetal bovine serum in a humidified 5% CO2 incubator at 37°C. KSHV-positive human PEL cells BCBL1 cells, and JSC1 cells, as well as the KSHV-negative DG75 B-lymphoblast cell line, were grown in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum. For KSHV lytic cycle induction, TPA was added to the medium at a final concentration of 20 ng/ml.
Plasmids pSEW-C01, pSEW-C02, pJX15, and pCTC581a expressing full-length human C/EBPα, Flag-tagged C/EBPα, KSHV RTA, and Flag-tagged KSHV RAP effector genes driven by CMV or SV2 promoter-enhancers were described previously (46). Plasmid pSEW-R03 encodes the RTA(1-377) DBD segment; plasmid pSEW-R23 encodes RTA(1-691Δ151-167) with an in-frame internal deletion from position 151 to 167 (46). Reporter plasmid pGL3-FL(−914) containing the intact RTA(−914/+34)-LUC reporter gene was kindly provided by Koichi Yamanishi (32). Deletion derivatives of the full-length RTA-LUC reporter gene were generated by PCR using RTA(−914/+34)-LUC as the template to generate RTA(−587/+20)-LUC (in pSEW-RP1), RTA(−554/+20)-LUC (pSEW-RP2), RTA(−261/+20)-LUC (pSEW-RP3), RTA(−241/+20)-LUC (pSEW-RP4), and RTA(−212/+20)-LUC (pSEW-RP5) or by PCR-based mutagenesis using pSEW-RP4 or pSEW-RP1 as the templates to generate mutants RTA(−241/+20 pm−48/−43)-LUC (pSEW-RP6) or RTA(−587/+20 pm−48/−43)-LUC (pSEW-RP7), in which GCAATG at position −48 to −43 has been changed to GATATC (throughout this work, mutated nucleotides are indicated in boldface type). A plasmid encoding the human C/EBPα(−500/+10)-LUC reporter gene (pSEW-CP1) contains the proximal C/EBPα promoter region inserted into a minimal promoter in the pGL3-Basic background, as described previously (50). The PAN(−210/+15)-LUC reporter gene contains the wild-type PAN promoter inserted into the pGL3-Basic background (in plasmid pSEW-PP1) and point mutant derivatives PAN(−190/+15)-LUC (pSEW-PP2), PAN(−190/+15pm2A)-LUC (pSEW-PP3), PAN(−190/+15pm2B)-LUC (pSEW-PP4), PAN(−190/+15pm4A)-LUC (pSEW-PP5), and PAN(−190/+15pm4B)-LUC (pSEW-PP6) (see Fig. 10A) Point mutant reporter gene PAN(−190/+15RREm)-LUC (in plasmid pSEW-PP7) contains five point mutations within the RRE region at position −61 to −52, (GCTAACCTGT to TCTACCCGTG). A set of MTA promoter reporter plasmids expressing MTA(−160/+10)-LUC, MTA(−100/+10)-LUC, and MTA(−60/+10)-LUC in pSEW-MP1, pSEW-MP2, and pSEW-MP3, respectively, were generated by PCR amplification from BCBL1 DNA. Point mutant derivative pSEW-MP4 expressing MTA(−100/+10 pm −94/−90)-LUC in which ACAAT at position −94 to −90 has been changed to TCTAG was generated by a PCR-based method using pSEW-MP2 as the template.
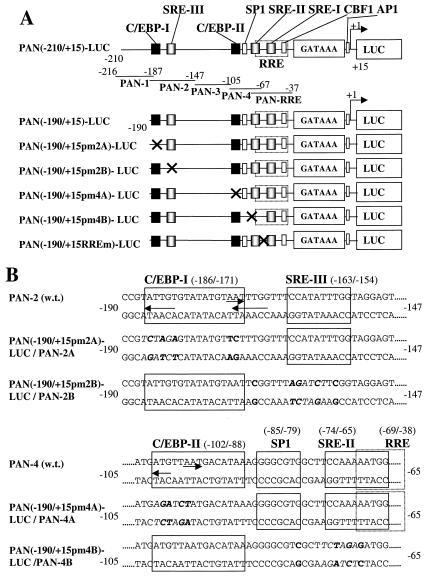
FIG. 10.
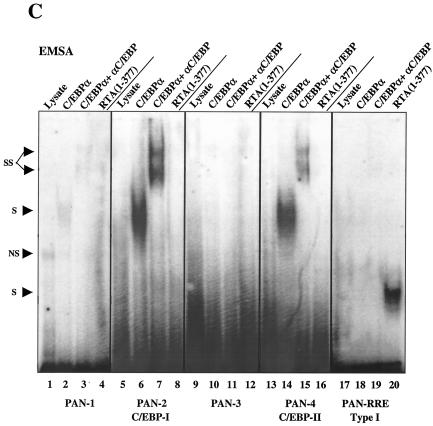
Identification and functionality of two C/EBP binding sites within the PAN promoter. (A) Schematic diagram of the PAN promoter region from position −210 to +15 relative to the defined transcription start site at genomic coordinate 28667, including the previously reported RRE, a putative SP1 site, three potential SRE sites, and the two C/EBP sites identified here. Promoter regions contained in probes PAN-1 to -4 and PAN-RRE and in wild-type and mutant PAN-LUC reporter gene derivatives are represented by solid lines. Mutations (X) are indicated. (B) Nucleotide sequences of six wild-type and mutant PAN EMSA probes and the PAN-LUC reporter genes shown above. Motifs that may serve as C/EBP and other consensus binding sites are delineated by arrows and boxes. Mutated nucleotides are indicated in boldface type. (C) EMSA to test the binding affinity of six different 34- to 44-bp PAN promoter probes to in vitro-translated C/EBPα, C/EBPα plus specific antibody, or the RTA(1-377) DBD protein fragment. Abbreviations: S, shifted bands; SS, antibody supershifts; NS, nonspecific bands. (D) EMSA comparing C/EBPα binding affinity of wild-type and mutant forms of the PAN-2 and PAN-4 probes. (E) Results of transient-expression assays with mutated PAN(−190/+15)-LUC target reporter genes introduced into HeLa cells to test their responsiveness to cotransfected C/EBPα or RTA or both together. Transfections were carried out as described above using 0.1 μg of reporter plasmid DNA and the indicated amounts of effector DNA. Error bars, standard deviations.
DNA transfection and luciferase assay.
Transfection of HeLa cells was performed with 5 × 105 cells/sample in six-well plates using Lipofectamine (Invitrogen) according to the Invitrogen protocol. BCBL1 and DG75 B-lymphoblast cell lines (107 cells/sample) were transfected using the electroporation method described previously (46). Transfected cells were harvested at 48 h posttransfection for luciferase assays as described by Wang et al. (46). All samples also received added CMV-βGAL expression plasmid DNA as an internal control for transfection variability, and the βGAL activity was found not to be significantly affected by cotransfection with either C/EBPα, RTA, or RAP or combinations of them.
Extraction of mRNA and RT-PCR.
BCBL1 cells were transfected by electroporation, and mRNA was extracted at 40 h posttransfection using a GenElute Direct mRNA Miniprep kit (Sigma) according to the product instructions as described previously (46). The purified mRNA samples (1 μg) were incubated with 1 U of DNase I (Invitrogen) and 10 μl of DNase I reaction buffer for 15 min at 20°C. Then, DNase I was inactivated by adding 1 μl of 25 mM EDTA to the mixture and heating for 10 min at 65°C. Reverse transcription (RT) was performed using avian myeloblastosis virus reverse transcriptase (Promega) as previously described (46), and the synthesized cDNA samples were used as templates for PCRs using RTA coding region primers LGH4289 (5′-GAACTACTCGAGATGGTCACTGGACTGTCCTATCC-3′) and LGH4290 (5′-TGACGAAGCTTCAGGTACCAGGTGTCGTGGTCG-3′). The cDNA PCR products (915 bp) were analyzed on a 1.2% agarose gel.
IFA.
Indirect immunofluorescence assay (IFA) was performed 24 h after BCBL1 cells were transfected or induced by TPA using the methanol fixation method. Primary antibodies included rabbit antipeptide antiserum against KSHV RTA (46), mouse anti-Flag monoclonal antibody (MAb) (Sigma, St. Louis, Mo.), and goat (Santa Cruz, Santa Cruz, Calif.) or rabbit anti-C/EBPα polyclonal antibody (PAb). Secondary donkey- or goat-derived fluorescein isothiocyanate (FITC)- or rhodamine-conjugated anti-rabbit or anti-mouse or anti-goat immunoglobulin G (Jackson Pharmaceuticals, West Grove, Pa.) was used to detect the primary antibodies. Mounting solution with DAPI (4′,6′-diamidino-2-phenylindole) (Vector Shield) was used to visualize cellular DNA.
EMSA.
C/EBPα protein samples used for EMSA were in vitro translated using the TNT Quick Coupled Transcription/Translation system (Promega) according to the manufacturer's procedures, and pBBV-C/EBPα plasmid DNA was used as the template. Annealed double-strand oligonucleotides were radiolabeled with [α-32P]dCTP by Klenow DNA polymerase. For EMSA, approximately 50,000 cpm of the 32P-labeled probe and 2 μl of in vitro-translated C/EBPα protein were incubated for 30 min at 20°C in a binding system containing 10 mM HEPES (pH 7.5), 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100, 5% glycerol, and 2 μg of poly(dI-dC). For supershift experiments, 0.5 μl of C/EBPα rabbit antiserum (46) or RTA rabbit antiserum against an RTA epitope between aa 527 and 539 (46) was added to the mixture after 30 min and allowed to incubate for another 30 min before gel loading. Samples were separated on a 4.5% polyacrylamide gel in 1× HEE buffer (10 mM HEPES [pH 7.5], 1 mM EDTA, 0.5 mM EGTA) at 150 V at 4°C as described previously (7). The gels were dried and subjected to autoradiography with Kodak X-ray film.
All oligonucleotides used were purchased from Qiagen Operon. The following RTA promoter probes were used: LGH4341 (5′-GATCTGGCGCTACAGTGGGTGATTTCTTCTACCACGG-3′) and LGH4342 (5′-GATCCCGTGGTAGAAGAAATCACCCACTGTAGCGCCA-3′) annealed to form probe RP-1; LGH4343 (5′-GATCTCATACATTGGTGGCACCCACAGGCCTGTTCCA-3′) and LGH4344 (5′-GATCTGGAACAGGCCTGTGGGTGCCACCAATGTATGA-3′) annealed to form probe RP-2; LGH4345 (5′-GATCGTGGTACCGAATGCCACAATCTGTGCCCTCCAG-3′) and LGH4346 (5′-GATCCTGGAGGGCACAGATTGTGGCATTCGGTACCAC-3′) annealed to form probe RP-3; LGH4347 (5′-GATCACAATTTTCATCTCCAATACCCGGAATTGGGAT-3′) and LGH4348 (5′-GATCATCCCAATTCCGGGTATTGGAGATGAAAATTGT-3′) annealed to form probe RP-4; LGH4349 (5′-GATCCCAGAAACCAGTAGCTGGGTGGCAATGACACGT-3′) and LGH4350 (5′-GATCACGTGTCATTGCCACCCAGCTACTGGTTTCTGG-3′) annealed to form probe RP-5; LGH4363 (5′-GATCCCAGAAACCAGTAGCTGGGTGGATATCACACGT-3′) and LGH4364 (5′-GATCACGTGTGATATCCACCCAGCTACTGGTTTCTGG-3′) annealed to form probe RP-5M.
The entire PAN promoter insert from PAN(−210/+15)-LUC was isolated by cleavage with SacI/HindIII and end labeled to obtain probe PAN-225. The PAN-1 probe was generated by annealing oligonucleotides LGH4971 (5′-GATCATTAATGAAAGTTTATTAATGTTCATCCGT-3′) and LGH4972 (5′-GATCACGGATGAACATTAATAAACTTTCATTAAT-3′); probes PAN-2, PAN-2A, and PAN-2B were generated by annealing LGH4973 and LGH4974, LGH4990 and LGH4991, and LGH4992 and LGH4993, respectively, which contain an additional GATC at the 5′ ends (see Fig. 10); probe PAN-3 was generated by annealing LGH4975 (5′-GATCATGGAGTTTTCTTATGGATTATTAAGGGTCAGCTTGAAGG-3′) and LGH4976 (5′-GATCCCTTCAAGCTGACCCTTAATAATCCATAAGAAAACTCCAT-3′); probes PAN-4, PAN-4A, and PAN-4B were generated by annealing LGH4977 and LGH4978, LGH4994 and LGH4995, and LGH4996 and LGH4997, respectively, which contain an additional GATC at the 5′ ends (see Fig. 10). Probe PAN-RRE (41 bp) was described previously (46).
Probe MTA-1 was generated by annealing LGH4980 (5′-GATCCTTCATTCCATTAGGGTGAGCGAAGTCACGGTAACACTTATGA-3′) and LGH4981 (5′-GATCTCATAAGTGTTACCGTGACTTCGCTCACCCTAATGGAATGAAG-3′); probe MTA-2 was generated by annealing LGH4982 (5′-GATCGTCAGTGTTTTGCCAGCAAGTGTAACAATAATGTTCCCACGGC-3′) and LGH4983 (5′-GATCGCCGTGGGAACATTATTGTTACACTTGCTGGCAAAACACTGAC-3′); probe MTA-3 was generated by annealing LGH4984 (5′-GATCCCATTTTTCGTTTGTGGTACCTGTGGGACTGGCCAGTTAATCC-3′) and LGH4985 (5′-GATCGGATTAACTGGCCAGTCCCACTGGTACCACAAACGAAAAATGG-3′); probe MTA-2M was generated by annealing LGH5236 (5′-GATCGTCAGTGTTTTGCCAGCAAGTGTATCTAGAATGTTCCCACGGC-3′) and LGH5237 (5′-GATCGCCGTGGGAACATTCTAGATACACTTGCTGGCAAAACACTGAC-3′).
ChIP.
JSC1 or BCBL1 PEL cells were treated with TPA before harvesting at different time points for ChIP assays as described elsewhere (46) using rabbit PAbs against C/EBPα, KSHV RTA, or KSHV RAP or mouse MAb against EBV ZTA or mouse MAb against CHOP10 as controls. Primers LGH4354 (5′-GAACTACTCGAGCTGTGCCCTCCAGCTCTCAC-3′) and LGH4355 (5′-GGACGTAAGCTTACAGTATTCTCACAACAGAC-3′), specific for a 261-bp region in the KSHV RTA promoter from −241 to +20; primers LGH4973 (described for EMSA) and LGH4998 (5′-TGATCTAAGCTTCTGGGCAGTCCCAGTGCT-3′), specific for a 205-bp region in the KSHV PAN promoter from −190 to + 15; and primers LGH4980 (described for EMSA) and LGH4987 (5′-GATCGCGGGCTATTTTGGGAACCTGGCAGCCAGGTTATATAGTG-3′), specific for a 168-bp region in the KSHV MTA promoter from −160 to +8, were used for PCR detection. Primers LGH4930 (5′-TTCGCCTGTTAGACGAAGC-3′) and LGH4929 (5′-GATTCGCAAGCTTCAGTCTCGGAAGTAATTACG-3′), specific for RTA coding region aa 591 to 691, were used as negative control to detect a nonpromoter region. The PCR products were analyzed on a 2% agarose gel.
Extraction of total RNA and Northern blot assay.
BCBL1 cells were induced with TPA, and total RNA was isolated at different time points using Trizol reagent (GIBCO BRL) according to the manufacturer's protocol. RNA samples (30 μg) were separated on a 1.2% formaldehyde agarose gel in 1× morpholinopropanesulfonic acid (MOPS) running buffer and transferred to a Nytran nylon membrane (Schleicher & Schuell). Restriction enzyme-released fragments containing C/EBPα (aa 251 to 358), KSHV RTA (aa 244 to 548) or KSHV RAP (aa 1 to 237), as well as a PCR-generated fragment containing a 286-bp GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA region, were labeled using the Prime-a-Gene labeling system (Promega) in a 50-μl reaction mixture containing 1× labeling buffer, 20 μM concentrations of dATP, dGTP, and dTTP, 25 ng of denatured DNA template, 20 μg of bovine serum albumin, 333 nM [α-32P]dCTP(3,000 Ci/mmol), and 5 U of Klenow DNA polymerase. After end labeling at 20°C for 60 min, the probes were purified with Sephadex G-50, denatured, and incubated with the membrane in a hybridization mixture (5 ml) containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's solution, 20 mM NaPO4 (pH 6.5), 10% dextran sulfate, and calf thymus DNA (100 μg/ml). Sequential hybridizations were carried out overnight at 42°C, and the membrane was washed twice in 2× SSC-0.1% sodium dodecyl sulfate (SDS) at room temperature and then twice in 0.5× SSC-0.1% SDS at 50°C before autoradiography and reuse. For an RNA loading control, the membrane was stripped again and reprobed with GAPDH cDNA.
Protein isolation and Western immunoblot assay.
Approximately 107 BCBL1 cells or 106 293T cells were harvested at different time points after TPA induction or at 48 h posttransfection, respectively; washed one time with phosphate-buffered saline; gently resuspended in 0.2 ml of ice-cold immunoprecipitation buffer (50 mM Tris-Cl [pH 7.9], 50 mM NaCl, 0.1 mM EDTA, 1% glycerol, 0.2% NP-40, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride); and sonicated for 30 s. The samples were then centrifuged at 15,000 × g for 5 min at 4°C, and the supernatants were collected. Western blot analysis was performed after SDS-polyacrylamide gel electrophoresis, using 10 μl of each protein sample. The proteins were detected by immunoblotting with rabbit polyclonal antiserum against C/EBPα, KSHV RTA, or KSHV RAP or mouse MAb against Flag as described previously (46).
RESULTS
C/EBPα activates the RTA IE promoter in cooperation with KSHV RTA and RAP.
Previous reports indicated that cotransfected RTA (ORF50) autostimulates expression from a target RTA promoter by 40-fold in 293T cells using a large RTA(−3000/+20)-LUC reporter gene (12) or by 6-fold in 293L cells using a shorter RTA(−914/+34)-LUC reporter gene (32). In the latter study, OCT1 binding to the RTA promoter was suggested to be important for RTA autoactivation, although OCT1 had little effect on RTA expression when transfected alone. We suspected that C/EBPα might also be involved in the efficient activation of RTA gene expression during the KSHV lytic cycle. Exogenously introduced C/EBPα activates expression of the KSHV DE gene RAP (K8) protein in PELs and cooperates with both RTA and the RAP protein itself to stimulate the RAP promoter in transient-cotransfection assays (46). Those studies also revealed that the C/EBPα and RTA proteins physically interact with each other (as well as with RAP), but whether these two transcription factors regulate each other's expression at the transcriptional level was not known.
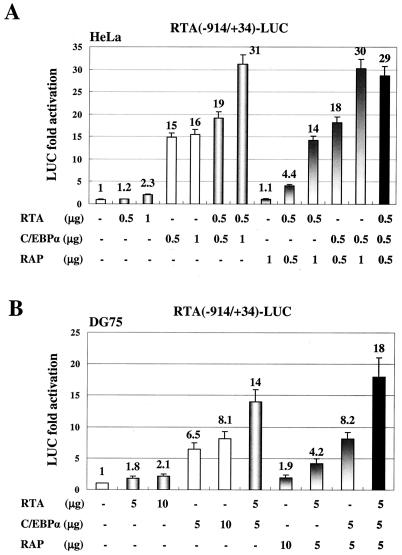
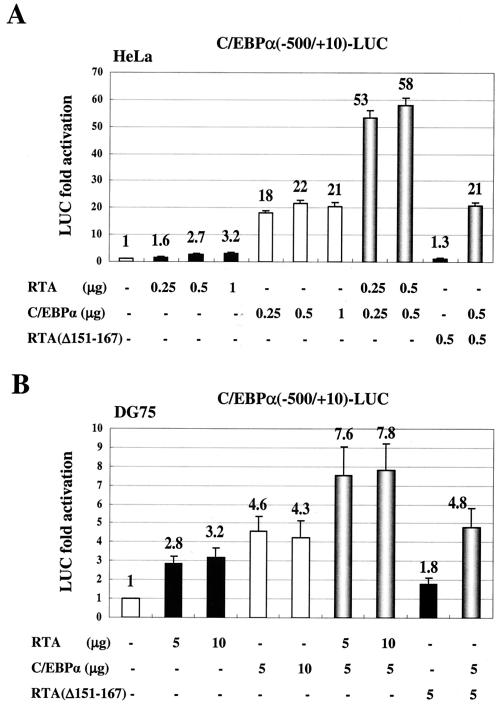
Initially, we transfected the RTA(−914/+34)-LUC target reporter gene into HeLa cells and tested the effects of cotransfected C/EBPα, RTA, and RAP effector genes on RTA promoter expression. Only 2.3-fold autoactivation by RTA alone was observed. However, C/EBPα alone gave a 15-fold stimulation of RTA-LUC activity, and addition of RTA and C/EBPα together resulted in further enhancement up to 31-fold (Fig. 1A). Although RAP had no effect on RTA promoter activity when transfected alone (1.1-fold), cotransfection of RAP and RTA together resulted in up to 14-fold activation in the absence of C/EBPα. When cotransfected with C/EBPα, RAP also elevated the level of transactivation by a factor of 2 up to 30-fold.
FIG. 1.
C/EBPα activates RTA-LUC reporter gene expression in cotransfected cells. (A) HeLa cells were transfected with 0.2 μg of plasmid DNA encoding RTA(−914/+34)-LUC and the indicated amounts of effector plasmid DNA expressing CMV-C/EBPα, CMV-RTA, or SV2-RAP. The total amount of effector plasmid DNA used in each transfection was normalized to 1.5 μg by adding empty CMV promoter vector (pcDNA3.1) DNA. Activation of LUC activity was calculated relative to the basal level control obtained using 1.5 μg of empty CMV promoter vector DNA as the effector. (B) DG75 B-lymphoblast cells were transfected by electroporation with 2 μg of the RTA-LUC target plasmid DNA and the indicated amounts of effector plasmid DNA. The total amount of all plasmid DNA used in each transfection sample was normalized to 15 μg by adding empty CMV promoter vector DNA. Error bars, standard deviations.
To confirm these observations about transactivation of the RTA promoter in the physiologically more relevant B cells, the cotransfection experiments were also carried out in the KSHV- and EBV-negative DG75 lymphoblast cell line by electroporation (Fig. 1B). C/EBPα alone induced RTA-LUC expression up to 8.1-fold (10 μg), but again, although RTA only autoactivated its promoter by 2.1-fold (10 μg), it more than additively increased the activation by C/EBPα, from 6.5-fold up to 14-fold at the 5-μg input DNA level. Addition of RAP also enhanced both C/EBPα and RTA activation levels nearly twofold. Finally, when C/EBPα, RTA, and RAP were all added together to the transfection (5 μg each), the strongest level of activation was obtained (18-fold). Therefore, C/EBPα proved to be the most significant activator of the RTA promoter discovered so far, and just as for the RAP promoter, KSHV RTA and RAP cooperated with C/EBPα to achieve the highest level of activation.
Transfection by C/EBPα increases the level of RTA mRNA in PEL cells.
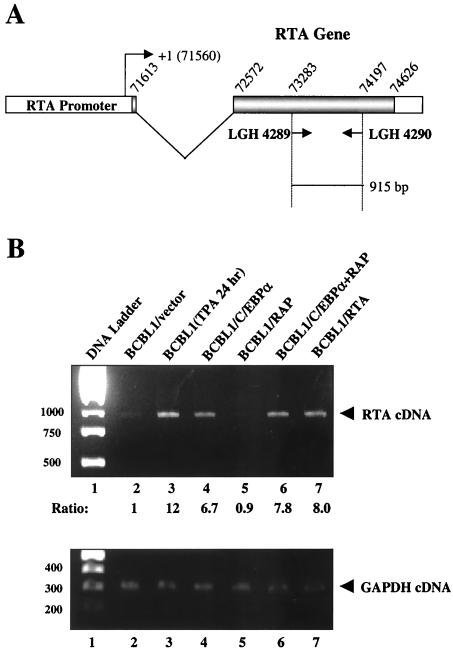
The cotransfection data above strongly suggest that C/EBPα is capable of upregulating KSHV RTA expression at the transcriptional level. To ask whether this effect can be demonstrated to occur in vivo in KSHV-infected PEL cells, we performed RT-PCR in C/EBPα-transfected BCBL1 cells. The mRNA samples were isolated and treated with DNase I to eliminate genomic DNA contamination before RT was carried out. After cDNA synthesis, two primers specific for the RTA gene were used for PCR amplification, giving an expected product of 915 bp (Fig. 2A) In empty vector-transfected control cells, the level of RTA mRNA detected was very low (Fig. 2B, lane 2); however, TPA treatment for 24 h resulted in the activation of KSHV lytic replication in 15% of the cells (as detected by IFA for RAP) and led to a 12-fold increase in the level of RTA mRNA (Fig. 2B, lane 3). In BCBL1 cells electroporated with C/EBPα expression plasmid DNA, the RTA mRNA level increased by 6.7-fold compared to empty vector-transfected cells, whereas there was no effect on RTA expression in RAP-transfected cells (Fig. 2B, lanes 4 and 5). In BCBL1 cells cotransfected with both C/EBPα and RAP, there was a small additive increase in the expression of RTA mRNA (7.8-fold) compared to cells transfected with C/EBPα alone (Fig. 2B, lane 6). Authentication of the expected size of the RT-PCR product was provided using RNA from BCBL1 cells transfected with an RTA cDNA expression plasmid (Fig. 2B, lane 7); however, any effect of RTA on autoactivation here was unable to be detected because both endogenous RTA expression and the transfected RTA plasmid give RT-PCR products of the same size.
FIG. 2.
Induction of endogenous RTA mRNA by transfected C/EBPα in PEL cells. (A) Schematic diagram showing the structure of the RTA gene and the positions of the two primers used for RT-PCR amplification (915 bp). (B) RT-PCR analysis showing that the level of RTA mRNA was elevated in C/EBPα-transfected BCBL1 cells is depicted in the upper panel. Control RT-PCR analysis for cellular GAPDH mRNA levels showing equal loading of all lanes is depicted in the lower panel. Lane 1, DNA size marker; lane 2, negative control RNA for BCBL1 cells transfected with empty pcDNA3.1 vector DNA only; lane 3, positive control RNA for untransfected BCBL1 cells treated with TPA for 24 h; lane 4, CMV-C/EBPα-transfected BCBL1 cell RNA; lane 5, SV2-RAP-transfected BCBL1 cell RNA; lane 6, CMV-C/EBPα− and SV2-RAP-cotransfected BCBL1 cell RNA; lane 7, positive control RNA from BCBL1 cells transfected with CMV-RTA plasmid DNA, used as a size marker for the PCR amplification of RTA cDNA.
Introduction of C/EBPα induces endogenous RTA protein expression in PEL cells.
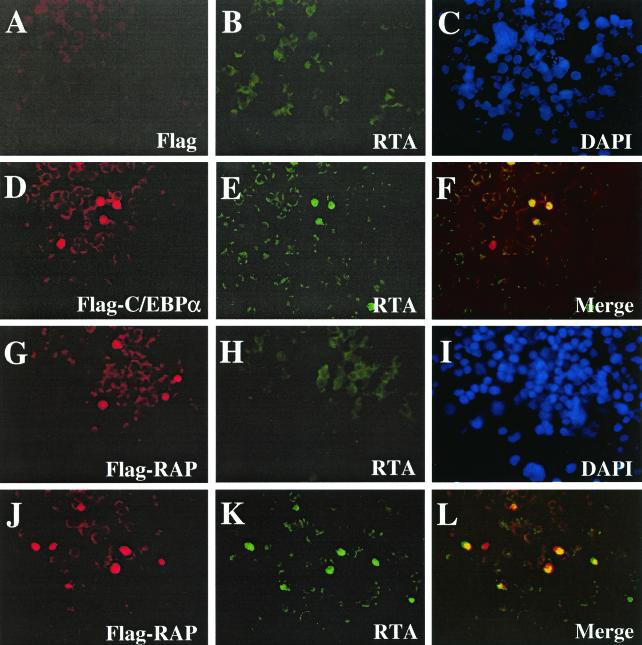
To confirm that RTA protein expression can be induced by C/EBPα, double-label IFA experiments were performed with CMV-Flag-C/EBPα transfected latently infected BCBL1 cells. Expression of endogenous RTA protein was absent in empty vector-transfected control cells (Fig. 3A to C); however, in C/EBPα-transfected cells, 37% of the cells that expressed exogenous Flag-tagged C/EBPα were also now positive for RTA (Fig. 3D to F). In comparison, transfection with SV2-Flag-RAP did not induce RTA protein expression in any of the RAP-positive cells as detected by anti-Flag MAb and anti-RTA PAb (Fig. G to I). Interestingly, in BCBL1 cells doubly transfected with CMV-C/EBPα and SV2-Flag-RAP, 51% of RAP-positive cells became positive for RTA protein expression. Additional IFA experiments confirmed that more than 95% of cells expressing either one of the transfected effector plasmids in the double-transfection experiments were positive for expression of both proteins (data not shown). Therefore, expression of exogenous C/EBPα alone can activate RTA expression from a latent state in some cells and RAP can evidently augment C/EBPα-mediated transactivation of the RTA promoter, although it has no effect alone.
FIG. 3.
Exogenously introduced C/EBPα triggers endogenous RTA protein expression in PEL cells. (A to C) Control double-stain IFA showing lack of background expression of either Flag epitope or the endogenous RTA protein in BCBL1 cells transfected with empty CMV-Flag vector DNA as detected with anti-Flag MAb (A) (rhodamine [red]) or anti-RTA PAb (B) (FITC [green]). (C) DAPI nuclear staining (blue) showing all cells in the same field. (D to F) Expression of Flag-C/EBPα protein (D) (red) in transfected BCBL1 cells induces expression of endogenous RTA protein in the same cells as detected with anti-RTA PAb (E) (green). (F) Merged image. (G to I) Expression of Flag-RAP protein in transfected BCBL1 cells (G) (red) fails to induce endogenous RTA protein expression (H) (green). (I) DAPI (blue) showing the whole cell population. (J to L) Expression of Flag-RAP protein (J) (red) in C/EBPα plus Flag-RAP cotransfected BCBL1 cells and induction of RTA (K) (green) in the same cells. (L) Merged image. Note that spontaneous background RTA expression occurs in approximately 1% of untreated BCBL1 cells.
Identification of three C/EBP binding sites in the RTA promoter.
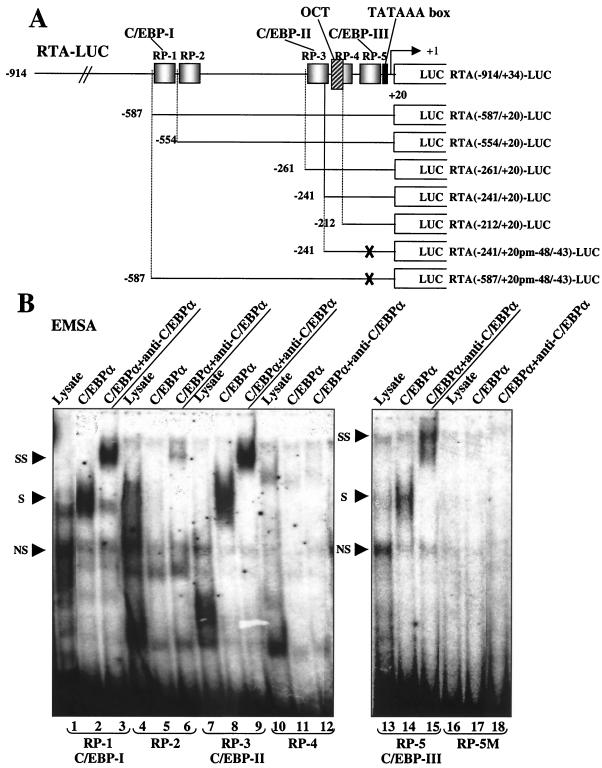
DNA sequence analysis revealed five potential C/EBP binding sites within the RTA promoter region between positions −914 and +20 (Fig. 4A). Complementary oligonucleotide pairs (37 nucleotides long) containing each of these putative sites (designated as RP-1 to RP-5) were synthesized, annealed, and 32P-labeled as probes for EMSA to test their affinity for C/EBPα protein. Strong EMSA bands were observed with probes RP-1 and RP-3 when in vitro-translated C/EBPα protein was added (Fig. 4B, lanes 2 and 8), and the addition of antibody against C/EBPα resulted in the formation of typical slower-migrating supershifted bands (Fig. 4B, lanes 3 and 9). Low-affinity binding to the C/EBPα protein was also observed with probe RP-5 (Fig. 4B, lanes 14 and 15), but neither probe RP-2 nor RP-4 showed any affinity for C/EBPα, and a triple-point mutation introduced into RP-5 M destroyed its ability to bind C/EBPα (Fig. 4B, lanes 17 and 18).
FIG. 4.
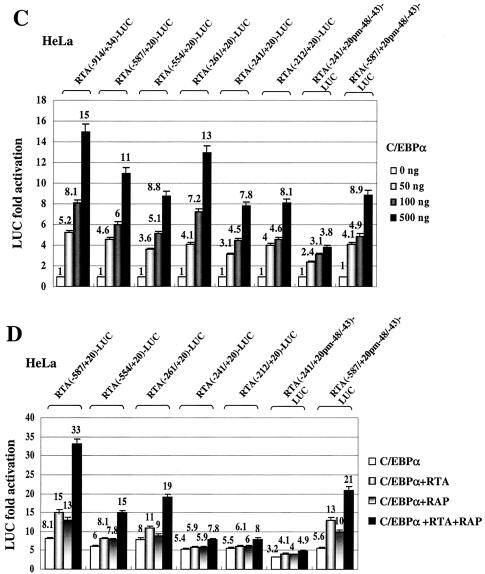
Deletion analysis of the RTA (ORF50) promoter and identification of C/EBP binding sites. (A) Schematic diagram of the RTA promoter region between positions −914 and +20 relative to the mRNA start site at genomic coordinate 71560, including showing the locations of the three C/EBP binding sites defined here. Five oligonucleotide probes encompassing potential C/EBP motifs predicted by sequence analysis are designated RP-1 to RP-5. The OCT site, which has been reported to be important for RTA autoactivation, and the TATA box are also indicated. The promoter region segments retained in a set of RTA-LUC deletion derivative plasmids that sequentially remove motifs RP-1 to RP-5 are designated according to their 5′ boundaries and are represented by solid lines. The X indicates that the C/EBP-III site (RP-5) was destroyed by changing GCAATG (positions 71512 to 71517) to GATATC. (B) EMSA experiment showing the C/EBPα binding ability of each putative C/EBP motif in the RTA promoter. Annealed 34-bp double-stranded oligonucleotide probes containing motifs RP-1 to RP-5, as well as the mutated probe RP-5M, were 32P end labeled and tested for the ability to bind to in vitro-translated C/EBPα protein (lanes 2, 5, 8, 11, 14, and 17). A sample of the unprogrammed reticulocyte lysate was used as control for nonspecific shifted bands (lanes 1, 4, 7, 10, 13, and 16). Rabbit polyclonal antiserum against C/EBPα was used to generate specific supershifted bands (lanes 3, 6, 9, 12, 15, and 18). Abbreviations: S, C/EBPα-specific shifted bands; SS, antibody supershifts; NS, nonspecific bands. (C) Responsiveness of seven different RTA-LUC deletion mutant derivatives to C/EBPα transactivation in cotransfected HeLa cells. Transfections were carried out using 200 ng of target reporter promoter DNA and 0-, 50-, 100-, or 500-ng input doses of C/EBPα effector expression plasmid DNA as described in the legend to Fig. 1A. Error bars, standard deviations. (D) Cooperative activation of seven different RTA-LUC deletion derivatives by C/EBPα, RTA, and RAP in HeLa cells. Transfection was carried out using 200 ng of target reporter plasmid DNA, 250 ng of C/EBPα expression plasmid DNA and 500 ng of either RTA or RAP expression plasmid DNA alone or a combination of both as indicated. Error bars, standard deviations.
The three putative C/EBP sites that proved to interact with C/EBPα in vitro were designated C/EBP-I (within probe RP-1), C/EBP-II (within probe RP-3), and C/EBP-III (within probe RP-5). To examine the contribution of these sites to the transactivation of the RTA promoter by C/EBPα, we constructed a series of deletion mutants with 5′ boundaries at positions −587, −554, −261, −241, and −212 that sequentially removed the C/EBP-I and C/EBP-II sites from the full-length RTA(−914/+34)-LUC reporter gene, as illustrated in Fig. 4A. In addition, the same mutated version of C/EBP-III that was generated in RP-5M was introduced into both RTA(−587/+20)-LUC and RTA(−241/+20)-LUC. The responsiveness of each of these truncated RTA reporter genes to cotransfection with increasing doses of C/EBPα effector plasmid DNA was measured, and the results are summarized in the histogram shown in Fig. 4C. Basal activity across the series did not change by more than 50% and is not included in the data. Only the mutant version of RTA(−241/+20)-LUC without any of the three identified C/EBP sites remaining showed a fourfold reduction, with all of the others giving less than twofold reduction in responsiveness to C/EBPα alone. Therefore, evidently both the C/EBP-II and C/EBP-III sites alone are sufficient for direct C/EBPα activation.
We also examined the role of the C/EBP binding sites in cooperative activation by C/EBPα plus RTA and RAP. Deletion of C/EBP-I in RTA(−554/+20)-LUC led to partial impairment of enhanced C/EBPα activation by both RTA and RAP, whereas when C/EBP-II was also deleted in RTA(−241/+20)-LUC, cooperativity between RTA and RAP with C/EBPα was nearly abolished (Fig. 4D). However, although mutation of C/EBP-III in RTA(−241/+20 pm−48/−43)-LUC significantly impaired direct C/EBPα activation, the cooperative enhancement by RTA and RAP was not affected. Therefore, either one or both of the C/EBP-I and -II sites that bind strongly to C/EBPα protein in EMSA appear to be required for the cooperative activation by C/EBPα, RTA, and RAP, with C/EBP-II alone being sufficient and C/EBP-III not contributing.
In parallel experiments, neither deletion nor point mutation of the OCT1 site (−220/−213) described by others (32) had any significant effect on basal activity or on C/EBPα or RTA transactivation in our studies in HeLa or DG75 cells (data not shown). A recent study by Saveliev et al. (33) suggests that the KSHV RTA promoter region may initiate a second set of IE transcripts in a bidirectional fashion from near position −100 and that in addition a minor IE version of RAP mRNA may initiate from position −250 in the KSHV RAP promoter. If so, the C/EBP-III site described here would still represent an upstream regulator for both the spliced RTA (ORF50) and ORF48/19 IE transcripts, but C/EBP-I and -II would be downstream and possible negative regulators of the ORF48/29 leftward spliced IE transcript. Similarly, the overlapping C/EBP-RRE motif in the RAP promoter would be downstream of the minor IE RAP transcription start site and would likely have negative regulatory effects compared to the positive effects observed on the predominant DE version of the RAP promoter that we worked with previously (46).
C/EBPα, RTA, and RAP all associate with the RTA promoter in vivo by ChIP assay.
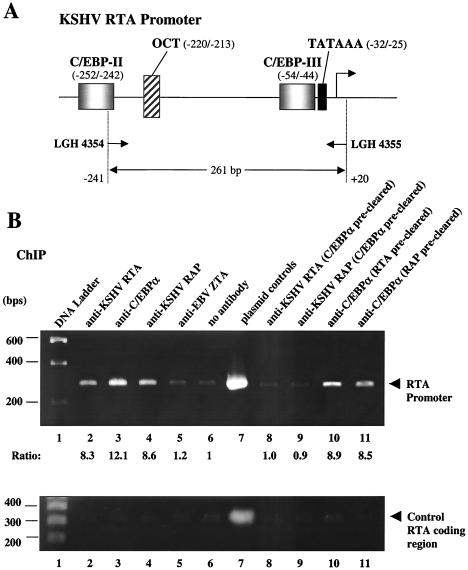
ChIP assays provide a powerful method to investigate the association between proteins and specific DNA regions in vivo. For example, we showed previously that addition of either in vitro-translated intact RTA or RAP proteins to C/EBPα in EMSA experiments interferes with the C/EBPα:DNA-shifted bands but fails to produce a supershifted band (46, 51). That result suggested but did not prove that RTA and RAP can piggyback bind onto the DNA-bound C/EBPα protein. However, the use of chromatin immunoprecipitation assays with PEL cell extracts confirmed that both RTA and RAP do indeed associate with the RAP promoter in a C/EBPα-dependent fashion in vivo, despite their inability to bind to it directly in vitro (46, 51). To conduct a similar experiment with the RTA promoter, we incubated a cross-linked cell extract obtained from KSHV-infected JSC1 cells after 40 h of TPA treatment with antibodies against C/EBPα, RTA, or RAP followed by immunoprecipitation. The presence of RTA-promoter DNA fragments within the immunoprecipitated samples was detected by PCR amplification using specific primers encompassing a 261-bp region in the RTA promoter (Fig. 5A). The recovery of PCR products of this size on an agarose gel revealed that all three antibodies were able to precipitate complexes containing RTA-promoter DNA fragments (Fig. 5B, upper panel, lanes 2 to 4). In contrast, antibody against EBV ZTA or a negative control using no antibody failed to recover any RTA-promoter DNA (lanes 5 and 6). Importantly, these same immunoprecipitates did not display any preferential association with nonpromoter DNA as judged by parallel PCR analysis with RTA-coding region primers (Fig. 5B, lower panel).
FIG. 5.
ChIP assay showing that the C/EBPα, RTA, and RAP proteins all associate with RTA promoter DNA in vivo. (A) Schematic diagram of the 261-bp region of the RTA promoter that was targeted for PCR amplification. (B) Results of ChIP assays carried out with samples from JSC1 cells after TPA treatment for 40 h are shown in the upper panel. Lane 1, DNA size marker; lanes 2 to 6, ChIP assay PCR products from immunoprecipitates obtained using antibodies against either RTA, C/EBPα, RAP, or EBV ZTA or with no antibody; lane 7, size control for 261-bp PCR amplification product from the RTA(−914/+34)-LUC reporter plasmid; lanes 8 and 9, ChIP assay PCR products obtained from immunoprecipitates using antibodies against RTA or RAP with cell extracts that had been precleared with C/EBPα antibody; lanes 10 and 11, ChIP assay PCR products obtained from immunoprecipitates using C/EBPα antibody and cell extracts that had been precleared with antibodies against RTA or RAP. Control PCR amplification of RTA coding region sequences (300 bp) from the same ChIP immunoprecipitates are shown in the lower panel. These results demonstrate that the binding is specific for promoter region sequences.
Both KSHV RTA and RAP have been shown to physically interact with C/EBPα by immunoprecipitation and in vitro glutathione S-transferase binding assays (46), but neither bound directly to any of the identified C/EBP site probes from the RTA promoter in EMSA experiments (data not shown). Therefore, to investigate whether the association of RTA and RAP with the RTA promoter depends on indirect interaction with C/EBPα bound to C/EBP binding sites, we next precleared the cell extract with C/EBPα antibody before performing the ChIP assay on immunoprecipitates using antibodies against RTA or RAP. After removing C/EBPα, neither of these two antibodies was able to precipitate any RTA promoter region target DNA (Fig. 5B, lanes 8 and 9). However, C/EBPα antibody was still able to recover RTA promoter DNA after preclearing with antibody against RTA or RAP (Fig. 5B, lanes 10 and 11). This suggests a model involving the formation of transcriptional transactivating complexes on the RTA promoter in which C/EBPα binds directly to its target sites, whereas KSHV RTA and RAP only associate more indirectly with this promoter mediated through piggyback protein-protein interactions with C/EBPα.
Expression of C/EBPα is induced during the early phase of KSHV reactivation.
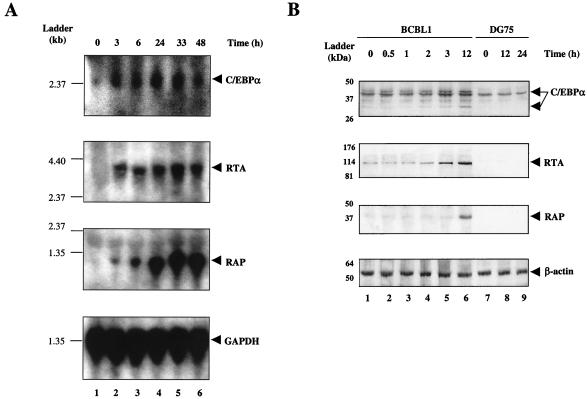
The KSHV IE gene RTA is likely to be the first viral gene expressed after primary infection as well as during reactivation from latency. We have shown above that exogenous C/EBPα can activate RTA expression both in latently infected PEL cells and in cotransfection reporter gene assays; however, whether C/EBPα is a factor that naturally triggers RTA early expression or vice versa remains to be determined. To investigate the kinetics of C/EBPα gene expression compared with that of RTA and RAP during KSHV reactivation in PEL cells, we performed a Northern blot experiment using total RNA samples extracted from BCBL1 cells treated with TPA for 0, 3, 6, 24, 33, or 48 h. A 32P-labeled C/EBPα DNA probe was used to detect the 2.5-kb C/EBPα mRNA. Expression of C/EBPα mRNA was limited in uninduced BCBL1 cells (TPA 0 h) but was elevated by 3 h after TPA induction, when the 3.2-kb RTA mRNA was also first observed, and it appeared to reach a peak at 24 h somewhat earlier than RTA (Fig. 6A). In contrast, although a low level of the 1.1-kb RAP mRNA was induced at 3 h, the major stimulation occurred significantly later and peaked between 33 and 48 h.
FIG. 6.
Enhanced expression of C/EBPα mRNA and protein at very early stages during the KSHV lytic cycle. (A) Kinetics of C/EBPα and KSHV early gene mRNA synthesis during TPA induction in PEL cells. Northern blotting was carried out using total RNA extracted from BCBL1 cells at different time points after TPA induction (0, 3, 6, 24, 33, and 48 h, as indicated). Upper panels show membranes that were sequentially hybridized with denatured 32P-labeled DNA fragment probes detecting mRNA for C/EBPα (2.5-kb), RTA (3.2-kb), or RAP (1.1-kb). The lower panel shows a probe for GAPDH (1.3-kb) that was used as loading control for the different samples. Positions of an RNA size marker ladder are indicated at the left of each panel. (B) Examination of C/EBPα protein levels at early times during the KSHV lytic cycle in PEL cells. Protein samples were isolated at 0, 0.5, 1, 2, 3, and 12 h after TPA induction from BCBL1 cells and at 0, 12, and 24 h from DG75 cells as indicated. The upper three panels show Western immunoblots that were performed using rabbit antibodies against C/EBPα, RTA, or RAP. In the lower panel, antibody against β-actin was used as a loading control. The positions of protein size markers are indicated at the left of each panel.
To evaluate the time course of C/EBPα induction at the protein level, a Western blot experiment was carried out using BCBL1 lysates from cultures treated with TPA for 0, 0.5, 1, 2, 3, and 12 h (Fig. 6B). An increase in both C/EBPα (42 and 30-kDa) and RTA (120-kDa) protein levels was first observed at 3 h, whereas the RAP (35-kDa) protein could not be detected until 12 h. Therefore, evidence at both the RNA and protein levels indicated that expression of C/EBPα is elevated at the very early stages of KSHV reactivation in PEL cells in parallel with RTA and even before RAP is synthesized.
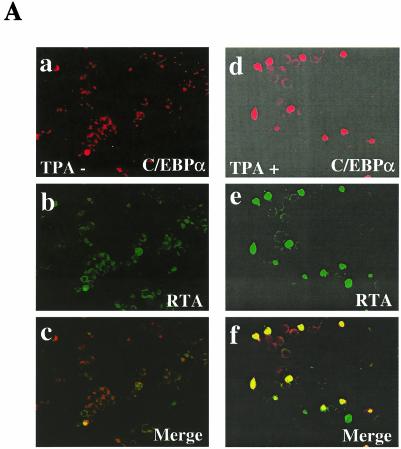
To address how many and which PEL cells were induced to express endogenous C/EBPα and KSHV RTA proteins, double-label IFA experiments were carried out in BCBL1 cells after 24 h in the presence or absence of TPA treatment. Neither protein was detectable in more than 1% of the cells before induction, but both became positive in approximately 15% of the cells after induction. Importantly, 79% of the RTA-positive cells at 24 h after TPA induction were also positive for C/EBPα, and 82% of the C/EBPα-positive cells were also positive for RTA (Fig. 7A). Again, we emphasize that TPA treatment did not induce detectable C/EBPα protein levels in the majority of PEL cells that did not also proceed into the lytic cycle. The observed correlation in expression of both proteins in the same cells during the lytic cycle further supports our in vitro findings that C/EBPα can upregulate the RTA promoter and raised the question of whether RTA might in turn upregulate the C/EBPα promoter.
FIG. 7.
Evidence that RTA expression enhances C/EBPα protein levels. (A) Expression of endogenous C/EBPα protein and that of RTA proteins are both induced in the same subpopulation of BCBL1 cells after TPA treatment. Double-stain IFA shows the absence of C/EBPα and RTA expression in uninduced BCBL1 cells (a to c), compared to high-level expression of both endogenous C/EBPα and RTA in BCBL1 cells after treatment with TPA for 24 h (d to f). Goat anti-C/EBPα PAb (a and d) (rhodamine [red]) and rabbit anti-RTA PAb (b and e) (FITC [green]) wereused for IFA detection. (c and f) Merged images. (B) Immunoblot detection of increased endogenous C/EBPα protein (42- and 30-kDa forms) in KSHV-negative B cells transfected with RTA. DG75 cells were transfected with either empty vector DNA (lane 1), two different input doses of CMV-RTA (lanes 2 and 3), or the CMV-C/EBPα (lane 4) expression plasmids as indicated. Western blotting was performed using antibodies against C/EBPα (upper panel), RTA (middle panel), or β-actin (lower panel). (C) Immunoblot detection of increased cotransfected C/EBPα protein in the presence of RTA. 293T cells were cotransfected with CMV-Flag-C/EBPα together with either the empty CMV promoter vector DNA (lane 1), wild-type CMV-RTA(1-691) expression plasmid (lane 2), or mutant CMV-RTA(1-691Δ151-167) expression plasmid (lane 3). Western immunoblotting was performed using Flag MAb (to detect exogenous C/EBPα) (upper panel), RTA PAb (middle panel), or β-actin MAb (lower panel). Positions of protein size markers are indicated to the left of each panel.
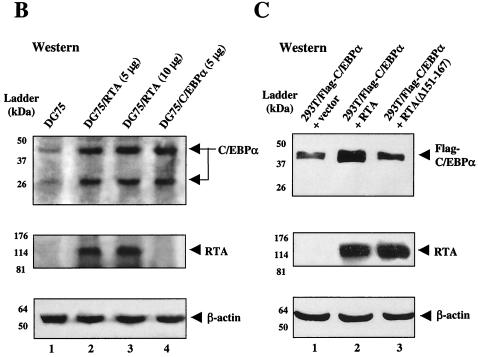
The expression of C/EBPα is not induced by TPA in KSHV-negative DG75 cells, as judged by both IFA (48) and Western immunoblotting (Fig. 6B). Therefore, virus-encoded factors seem likely be responsible for even the earliest induction of C/EBPα expression in TPA-treated BCBL1 cells. Because RTA and C/EBPα mRNA and proteins were both expressed as early as 3 h in TPA-induced BCBL1 cells, well before RAP was detected, we examined the effect of exogenous RTA expression on endogenous C/EBPα protein levels in uninfected DG75 lymphoblast cells. Western immunoblotting performed to detect the 42-kDa (and 30-kDa) forms of C/EBPα showed that (despite only a 20% transfection efficiency) their overall level of expression in the culture was indeed elevated severalfold by addition of the vector DNA expressing RTA (120 kDa) alone, and in fact reached the same level as that attained after addition of the C/EBPα expression vector alone (Fig. 7B).
RTA stimulates activation of the C/EBPα promoter cooperatively with C/EBPα.
To investigate the mechanism by which RTA induces C/EBPα expression, we first tested the effect of cotransfected RTA on a target C/EBPα-LUC reporter gene in HeLa cells. RTA alone activated the C/EBPα promoter by only 2.7- (0.5 μg) to 3.2-fold (1 μg), compared to C/EBPα alone, which had an 18-fold effect (Fig. 8A). However, when RTA was cotransfected together with C/EBPα, the combined effect was elevated more than additively to more than 53-fold. In contrast, the RTA(Δ151-167) mutant, which is unable to interact with the C/EBPα protein (46), had little activity on its own and failed to enhance the activation by C/EBPα. Similarly, in DG75 lymphoblast cells, cotransfection with RTA, but not with RTA(Δ151-167), again enhanced C/EBPα activation from fourfold to nearly eightfold (Fig. 8B). These results suggest that RTA can in turn stimulate the C/EBPα promoter cooperatively with C/EBPα, presumably because of an increase in transcriptional activity by DNA-bound RTA:C/EBPα complexes.
FIG. 8.
RTA activates the C/EBPα promoter cooperatively with C/EBPα in transient cotransfection assays. (A) HeLa cells were transfected with 0.2 μg of C/EBPα-LUC reporter gene plasmid and the indicated amounts of effector plasmids expressing either C/EBPα, wild-type RTA(1-691) or mutant RTA(Δ151-167), or both together. The total amount of effector plasmid DNA used in each transfection was normalized to 1.2 μg by adding empty CMV promoter vector DNA. Activation of LUC activity was calculated based on the control transfection using 1.2 μg of empty CMV vector DNA, representing the basal level. (B) DG75 B-lymphoblast cells were electroporated with 2 μg of C/EBPα-LUC target plasmid and the indicated amounts of effector plasmids. The total amount of effector plasmids used in each transfection was normalized to 15 μg by adding empty vector DNA. Error bars, standard deviations.
To address the possibility that the cooperative enhancement of C/EBPα transactivation by RTA might in addition involve an effective increased stabilization of the level of C/EBPα protein, we compared the amounts of each protein present in the single or combined transfected extracts (Fig. 7C). Because cotransfection with RTA had no effect on the high constitutive levels of CMV-βGAL expression in similar experiments (data not shown), we anticipated that there would be no transcriptional effects on the CMV-C/EBPα target promoter used here. Nevertheless, the results clearly revealed that cotransfection with wild-type RTA(1-691), but not with mutant RTA(1-691Δ151-167), led to a threefold increase in Flag-C/EBPα (42-kDa) protein levels over those obtained with the C/EBPα expression plasmid alone. Therefore, the cooperativity of RTA:C/EBPα complexes, like that of RAP:C/EBPα complexes, may involve a stabilization effect on the relatively short half-lived C/EBPα protein, as well as a transcriptional component.
C/EBPα transactivates the KSHV PAN promoter by specifically binding to two C/EBP sites.
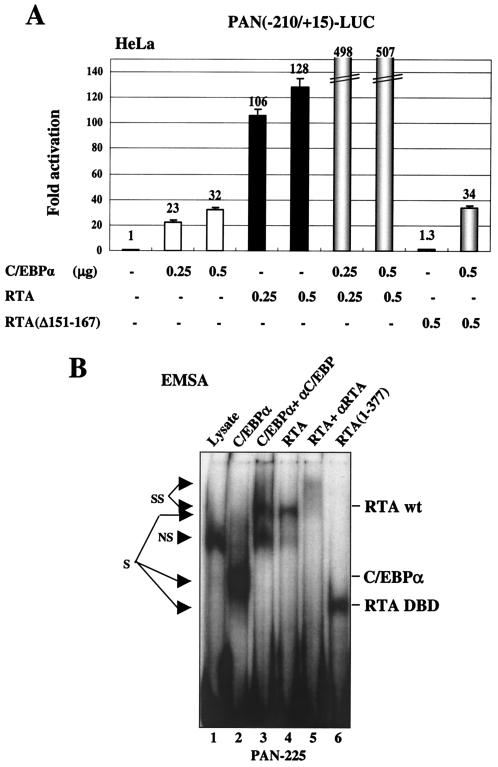
RTA is also known to transactivate expression of the KSHV DE gene encoding PAN RNA by binding directly to a type II RRE in the proximal region of the PAN promoter (4a, 37, 46). To investigate whether C/EBPα may also play a role in regulating the PAN promoter, the effects of cotransfected C/EBPα and RTA, either alone or in combination, were tested on a target PAN(−210/+15)-LUC reporter gene in HeLa cells. C/EBPα alone proved to activate the intact wild-type PAN target promoter up to 32-fold, whereas RTA alone produced up to 128-fold activation (Fig. 9A). Moreover, the combination of C/EBPα and RTA together led to a further dramatic increase in the activation level up to 500-fold. In comparison, the RTA(Δ151-167) deletion mutant, which has lost both its direct DNA binding activity and the ability to interact with C/EBPα (46), neither activated the PAN promoter when transfected alone nor cooperated with C/EBPα when cotransfected.
FIG. 9.
C/EBPα binds to and transactivates the KSHV PAN promoter. (A) HeLa cells were cotransfected with 0.1 μg of PAN(−210/+15)-LUC reporter plasmid DNA and indicated amounts of effector plasmids expressing C/EBPα, wild-type RTA(1-691), or mutant RTA(Δ151-167). Error bars, standard deviations. (B) EMSA with probe PAN-225 showing that in vitro-translated C/EBPα, intact RTA, and the RTA DBD protein segment all bind independently to the PAN(−210/+15) promoter region. Abbreviations: S, shifted bands; SS, antibody supershifts; NS, nonspecific bands.
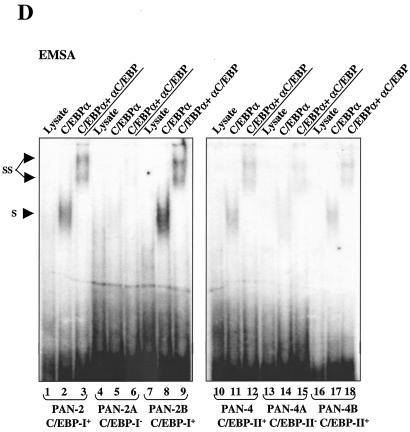
In EMSA experiments, the in vitro-translated C/EBPα protein and either the intact RTA(1-691) protein or the RTA(1-377) DBD protein segment were all able to bind independently to a 225-bp PAN DNA probe encompassing the RRE (Fig. 9B). Evidently, in this case, both C/EBPα and RTA probably activate the PAN promoter through direct binding to their respective target recognition sites. However, additional more complex interactions remained plausible to explain the functional cooperativity demonstrated above. Therefore, to map the number and position of C/EBP target sites within the PAN promoter, we examined the C/EBPα binding ability of a series of overlapping 34- to 44-bp EMSA probes (Fig. 10A) referred to as PAN-1 to -4 that span the whole 180-bp PAN promoter segment upstream of the TATA box. Two of these probes, PAN-2 and PAN-4, were able to form specific shifted complexes with the C/EBPα protein, but probes PAN-1, PAN-3, and the PAN-RRE probe did not do so (Fig. 10C).
To further define the location of these C/EBP sites, we introduced multiple adjacent point mutations into several motifs that resembled either C/EBP or other known recognition motifs within the PAN-2 and PAN-4 probes, as indicated in Fig. 10B. The EMSA results indicated that both the PAN-2A and PAN-4A probes lost the ability to bind to C/EBPα, whereas the mutations that inactivated putative SRE or SP1 sites within the PAN-2B and PAN-4B probes did not significantly affect C/EBPα binding (Fig. 10D). Therefore, the C/EBP-I site maps around position −186 to −171 and the C/EBP-II site maps around position −102 to −88. As expected, the in vitro-translated RTA(1-377) DBD protein fragment bound only to the PAN-RRE probe from position −77 to −37, which encompasses the previously defined type II RRE, but not to any of the other upstream PAN probes. Quantitation of the relative strengths of C/EBP-I and C/EBP-II showed that the C/EBP-I site had an affinity for C/EBPα two- to threefold higher than that for the C/EBP-II site (data not shown).
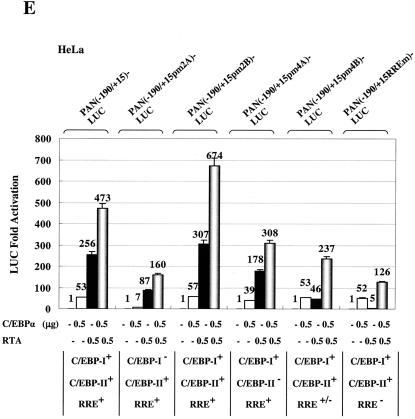
When the mutated versions of the PAN-LUC reporter gene were used in cotransfection experiments (Fig. 10E), the C/EBP-I mutated PAN(−190/+15pm2A)-LUC gene showed an 8-fold decrease in C/EBPα responsiveness and 2.5-fold reduced RTA responsiveness relative to the wild-type version (panel 2), whereas the SRE mutated PAN(−190/+15pm2B)-LUC gene was unaffected (panel 3). However, the C/EBP-II mutated PAN(−190/+15pm4A)-LUC showed only a 30% decrease in both C/EBPα and RTA responsiveness (panel 4), and all three mutant probes retained similar twofold levels of RTA plus C/EBPα cooperativity (Fig. 10E). In contrast, although the SRE and SP1 point mutations in PAN(−190/+15pm4B)-LUC did not affect C/EBPα transactivation or RTA augmentation (panel 5), there was a fivefold decrease in direct RTA-responsiveness, presumably because the first base pair within the defined type II PAN-RRE motif was also altered (Fig. 10B). To evaluate the response to C/EBPα in the total absence of the direct RTA binding motif, we also performed the cotransfection experiments using a mutant reporter gene PAN(−190/+15RREm)-LUC, which contains the same multiple point mutations that completely abolished both RTA binding and RTA responsiveness of the PAN promoter as reported by Song et al. (37). Indeed, only 5-fold RTA-activation remained, but normal levels of C/EBPα activation (52-fold) were observed (panel 6). Importantly, this RRE-knocked-out version of the PAN target promoter still retained some C/EBPα plus RTA cooperativity (giving 126-fold activation). These results indicate that, although RTA alone is sufficient to activate the PAN promoter by binding directly to the proximal type II RRE motif, C/EBPα also activates the PAN promoter primarily through the C/EBP-I site. Furthermore, an additional level of cooperative transactivation also occurs that is mediated by RTA:C/EBPα complexes binding to one or both of the C/EBP-I and C/EBP-II sites independently of the RRE itself.
The KSHV MTA promoter is also transactivated by C/EBPα through a C/EBP site adjacent to the RRE.
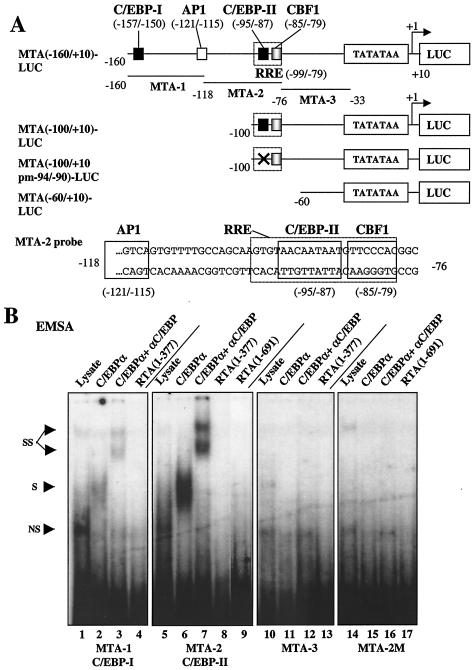
Both the KSHV MTA and RAP (K8) promoters have previously been reported to contain partially homologous versions of a different type of RRE referred to as a palindromic type I RRE (24, 44). However, we showed previously that C/EBPα transactivates the KSHV RAP promoter through a strong C/EBP binding site at position −73 to−67 that overlaps with the identified type I RRE motif (46). Furthermore, RTA did not bind directly to the RAP type I RRE at all, and instead the activation was mediated by interaction with C/EBPα bound to this C/EBP site. Liang et al. (22) have also recently shown that RTA binds indirectly to the MTA type I RRE via interaction with the cellular CBF1 (or RBP-Jκ) protein. Nevertheless, we suspected that the MTA promoter might also use the same indirect mechanism for RTA regulation as does the RAP promoter, because a similar potential C/EBP-like motif at position −95/−87 also forms part of the core of the defined type I RRE sequence within the MTA promoter (Fig. 11A).
FIG. 11.
C/EBPα transactivates the KSHV MTA promoter by binding to specific C/EBP sites. (A) Schematic diagram of the MTA promoter region from positions −160 to +10 relative to the defined transcription start site at genomic coordinate 82003, including the previously reported RTA response element (RRE), CBF1 site, a putative AP1 site and the C/EBP sites identified here. Positions of the probes used for EMSA as well as the sequence for probe MTA-2 are indicated. Promoter regions contained in different derivatives are represented by solid lines. The mutated C/EBP site in MTA(−100/+10 pm−94/−90)-LUC is indicated (X). (B) EMSA to test the binding affinity of three different MTA promoter probes to either unprogrammed reticulocyte lysate (lanes 1, 5, 10, and 14) or in vitro-translated C/EBPα (lanes 2, 6, 11, and 15), C/EBPα plus specific antibody (lanes 3, 7, 12, and 16), full-length RTA(1-691) protein (lanes 9 and 17) or the RTA(1-377) DBD protein segment (lanes 4, 8, and 13). Abbreviations: S, shifted bands; SS, antibody supershifts; NS, nonspecific bands. (C) Results of transient reporter gene expression assays with deleted or mutated MTA-LUC target reporter genes introduced into HeLa cells to test their responsiveness to cotransfected C/EBPα (open bars), wild-type RTA(1-691) (black bars), or both together (shaded bars). Transfections were carried out using 0.2 μg of reporter plasmid DNA and the indicated amounts of effector DNA. Error bars, standard deviations.
To examine the C/EBPα binding activity of this site and to search for other potential C/EBP sites within the MTA promoter, we performed EMSA experiments using three 43-bp probes spanning the proximal 130-bp upstream MTA promoter region. Probe MTA-2 from position −116 to −74 encompassing the putative C/EBP site within the RRE proved to bind strongly to C/EBPα, whereas probe MTA-1 showed only relatively weak binding affinity for C/EBPα and probe MTA-3 failed to bind (Fig. 11B). Importantly, none of these probes, including that containing the RRE, bound directly to either the in vitro-translated intact RTA protein or to the RTA(1-377) DBD fragment (Fig. 11B).
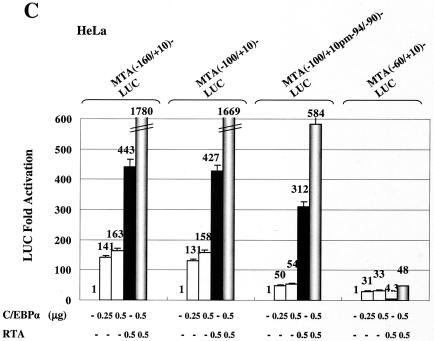
To further test the functions of these C/EBP sites, we carried out cotransfection experiments using a set of MTA-LUC reporter mutants with 5′ truncations at positions −160, −100, and −60 (Fig. 11A). Reporter plasmid MTA(−160/+10)-LUC contains both of the C/EBP-I and C/EBP-II sites detected by EMSA, whereas MTA(−100/+10)-LUC retains only the strong C/EBP-II site within the RRE, and MTA(−60/+10)-LUC lacks both C/EBP sites as well as the overlapping type I RRE motif. C/EBPα alone proved to activate both MTA(−160/+10)-LUC and MTA(−100/+10)-LUC by 160-fold, whereas RTA alone gave 430-fold up-regulation. In both cases, C/EBPα and RTA together cooperated to achieve an enhanced activation of up to 1,700-fold (Fig. 11C). In contrast, the C/EBPα responsiveness of MTA(−60/+10)-LUC was reduced 5-fold and the RTA responsiveness was virtually abolished (down 100-fold). To further define the location of the strong C/EBP site within the EMSA probe MTA-2, we introduced multiple adjacent point mutations into the putative C/EBP-II site AACAATAAT from −95 to −87. The EMSA results indicated that probe MTA-2M containing the mutated C/EBP-II site had lost the ability to bind to C/EBPα (Fig. 11B). Cotransfection experiments using MTA(−100/+10pm−94/−90)-LUC, which contains the same mutated C/EBP-II site as in the probe MTA-2M, gave a threefold decrease in direct C/EBPα activation. Furthermore, it retained threefold-augmented C/EBPα plus RTA responsiveness, but it gave only a 30% decrease in direct RTA responsiveness (Fig. 11C). The surprisingly high residual responsiveness (30- to 50-fold) of both MTA(−60/+10)-LUC and MTA(−100/+10pm−94/−90)-LUC to C/EBPα alone implies that there may still be another more proximal as yet unidentified C/EBP site present within the MTA promoter (perhaps at position −40).
Overall, the type I RRE overlapping with the C/EBP-II site in the MTA promoter shows some similar properties to that in the RAP promoter by being partially dependent upon RTA forming piggyback complexes with C/EBPα. However, unlike the RRE motif in the RAP promoter, the presumed interaction of RTA with CBF1 within the adjacent segment of the MTA RRE clearly produces a much greater effect.
Association of C/EBPα, RTA, and RAP proteins with both IE and DE KSHV promoters during early lytic reactivation.
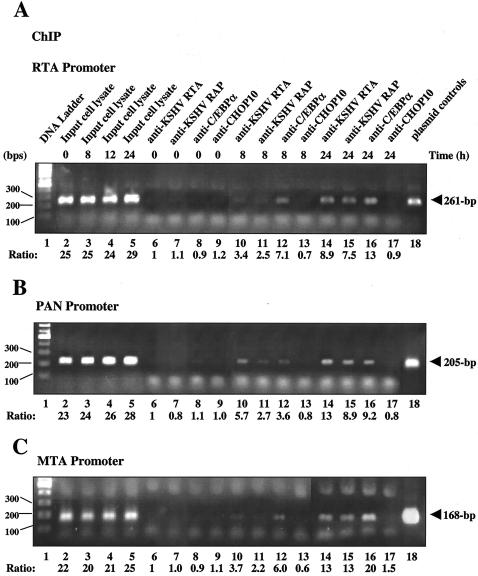
The above-mentioned experiments demonstrated that C/EBPα plays an important role in transactivating the KSHV RTA, PAN, and MTA promoters and that its own expression is elevated by RTA as early as 3 h after TPA treatment in BCBL1 cells. To investigate when C/EBPα, RTA, and RAP can first be detected associating with these three viral promoters during the early stages of lytic reactivation in infected cells, we carried out a time course ChIP assay experiment using BCBL1 cell lysates at 0, 4, 8, 12, and 24 h after TPA treatment. Antibodies against RTA, RAP, C/EBPα, or CHOP10 (negative control) were used to immunoprecipitate the associated DNA fragments (Fig. 12). In the PCR amplification, DNA regions representing all three promoters (RTA, PAN, and MTA) were detected as early as 8 h after TPA treatment in all three (RTA, RAP, and C/EBPα) immunoprecipitates, but not in those obtained with CHOP10 antibody (Fig. 12A to C), whereas no protein-bound DNA fragments were detected in the uninduced BCBL1 cell extracts (0 h) or in cell lysates induced by TPA for only 4 h (data not shown). At 24 h after TPA induction, increased amounts of promoter DNA were precipitated by the RTA, RAP, and C/EBPα antibodies, presumably reflecting increased expression of all three proteins, and the 12-h samples gave intermediate effects (data not shown). Interestingly, for the RTA and MTA promoters, C/EBPα was able to precipitate higher levels of promoter DNA than did RTA and RAP at both the 8- and 24-h time points (Fig. 12A and C), whereas for the PAN promoter, RTA had the strongest association with the promoter DNA (Fig. 12B). This is consistent with the in vitro evidence that RTA strongly and directly binds to PAN-RRE but only indirectly associates with the RTA and MTA promoters through the piggyback interactions with C/EBPα or CBF1.
FIG. 12.
Time course ChIP assays showing the association of C/EBPα, KSHV RTA, and RAP proteins with target viral promoters during early KSHV lytic reactivation. At different time points after TPA induction, aliquots of the BCBL1 cell lysates were precipitated with antibodies against KSHV RTA, RAP, C/EBPα, or CHOP10. Associations between proteins and target promoters were detected by PCR using primers specific for promoter regions of KSHV RTA (A), PAN (B), and MTA (C). Lane 1, DNA size marker; lanes 2 to 5, ChIP assay PCR products from either uninduced input BCBL1 cell lysates (0 h, lanes 6 to 9) or those at 8 h (lanes 10 to 13) and 24 h (lanes 14 to 17) after TPA induction; lanes 6, 10, and 14, PCR products from immunoprecipitates obtained using antibodies against RTA; lanes 7, 11, and 15, PCR products from immunoprecipitates obtained using antibodies against RAP; lanes 8, 12, and 16, PCR products from immunoprecipitates obtained using antibodies against C/EBPα; lanes 9, 13, and 17, PCR products from immunoprecipitates obtained using antibodies against CHOP10; lane 18, size control PCR amplification products from the corresponding promoter reporter plasmids including RTA(−914/+34)-LUC, PAN(−210/+15)-LUC and MTA(−160/+10)-LUC reporter plasmids.
Comparison of four different types of RTA responsiveness.
The relative responsiveness to both C/EBP and RTA transactivation varies greatly among the five promoters that we have studied extensively both here and previously. Table 1 compares and summarizes the average level of enhancement of LUC reporter gene activity from the cellular C/EBPα and KSHV RTA, RAP, PAN, and MTA promoters under parallel conditions in HeLa cells. Interestingly, the properties of the IE RTA promoter here most closely resemble those of the C/EBPα promoter, whereas the three DE promoters are more similar to one another, especially with regard to direct RTA response levels. All five promoters contain one or more relatively high- affinity C/EBP binding sites, and we suggest that perhaps all C/EBP sites of at least moderate affinity act as weak RREs by mediating severalfold cooperative effects through C/EBPα:RTA complex formation. Both the C/EBPα and RTA promoters appear to have only this type of RRE. In contrast, the three DE viral promoters each contain in addition a single more powerful and specific RRE. However, our interpretation of the present evidence indicates that all three DE RREs operate by distinctly different mechanisms.
TABLE 1.
Different types of RTA responsiveness
| Promoter | Basal activitya | Activation (fold) by:
|
Known binding site(s) | ||
|---|---|---|---|---|---|
| C/EBPα | RTA | C/EBPα + RTA | |||
| RTA | 5 × 104 | 15 | 2 | 30 | C/EBP (n = 3) (OCT? [n = 1]) |
| C/EBPα | 2 × 104 | 20 | 3 | 55 | C/EBP (n = 1) |
| RAPd | 1 × 104 | 30 | 70 | 250 | C/EBPe (n = 2) |
| PANb | 5 × 104 | 30 | 110 | 500 | C/EBP (n = 2), RTA (n = 1) |
| MTAc | 3 × 103 | 140 | 440 | 1,700 | C/EBP (n = 2), CBF1 (n = 1) |
Relative luciferase activities of 200 ng of target plasmid DNA in HeLa cells.
Type II RRE.
Type IA RRE.
Type IB RRE.
Presumed additional unknown cellular DNA binding factor.
In the case of the PAN promoter, the mechanism involves classical direct RTA binding to a type II RRE recognition motif. This may also be true for the K12 and vIL6 RREs (12a), although there is no nucleotide sequence homology between the identified vIL6 motif and the other two type II RTA binding sites. For MTA and RAP, the partially homologous type I RREs both encompass C/EBP binding sites and both fail to bind directly to RTA. However, based on recent studies from Lukac et al. (24) and Liang et al. (22), it now seems clear that a CBF1 binding motif lies directly adjacent to the C/EBP site and mediates indirect piggyback RTA binding to the MTA RRE. In contrast, for the RAP RRE, although the C/EBP site is essential for RTA responsiveness (46), this particular site apparently represents a special highly responsive C/EBP motif, presumably because of some other unknown adjacent feature (but it does not resemble a CBF1 site). We suggest that the indirectly responding MTA and RAP sites be referred to as type IA and type IB RREs, respectively. Finally, there remains the question as to why both the C/EBPα and RTA responses of the MTA promoter are so much higher than those of the other promoters tested. One possible explanation may reflect the relatively low basal activity of the MTA promoter, which is nearly 20-fold lower than that of the PAN promoter (Table 1) and might be attributable to the presence of the strong binding site for the CBF1 repressor.
DISCUSSION
Early lytic cycle gene expression by both KSHV and EBV is driven primarily by three virus-encoded nuclear regulatory proteins—namely, in KSHV by RTA (ORF50), MTA (ORF57), and RAP (K8) or in EBV by RTA (BRLF1), MTA (BMLF1), and ZTA (BZLF1), respectively. These proteins from the two subclasses of human gammaherpesviruses have diverged enormously, with only the two RTA DNA-binding transcriptional transactivators retaining as much as 15% overall amino acid identity, whereas the two MTA posttranscriptional RNA shuttle proteins retain only small scattered motifs in common, and the two bZIP family proteins resemble one another only at the structural level and in their splicing patterns. Therefore, there are many questions about the overall degree of similarity or differences in their functional activities.
Genetic knockout experiments in EBV have clearly shown that there are considerable overlap and redundancy in the functional roles of RTA and ZTA (13), although little in the way of any common mechanisms has been described. The ability of both constitutively expressed EBV RTA and ZTA to individually trigger full lytic cycle expression in latently EBV-infected B cells or epithelial cells is highly illustrative of this redundancy (10, 15, 29a, 52). Nevertheless, for KSHV this property has only been ascribed to RTA but not to RAP (9, 14, 24, 39). For EBV RTA-mediated transactivation, almost as many different mechanisms have been proposed as there are identified target promoters, including direct binding to specific DNA-recognition elements, and indirect mechanisms involving interactions with CBP, USF, Rb/E2F, SP1/3, or activation of JN kinase and mitogen-activated protein kinase. Similarly, for the KSHV version, although the target DNA recognition motif is different, both a direct promoter DNA-binding mechanism and several indirect mechanisms involving CBP, OCT1, SP1, AP1, and CBF1 (RBP-Jκ) have all been proposed by others (8, 12a, 22, 32, 54).
We have been focusing instead on another indirect cellular regulatory pathway involving C/EBPα, which unlike many of the others, is evidently targeted by both KSHV and EBV and also includes a physical interaction between C/EBPα and RAP or ZTA, as well as with KSHV RTA (48, 50, 51). For RAP and ZTA, these C/EBPα interactions lead to p21-mediated G1 cell cycle arrest in both viruses (48, 50) and also contribute to complex positive autoregulatory loops that apparently serve to strongly and coordinately upregulate levels of the RAP, ZTA, C/EBPα, and p21 proteins during the early lytic cycle (51; Wu et al., unpublished data). The mechanisms involved include both presumed piggyback targeting of RAP or ZTA to identified C/EBP binding sites in all four promoters and stabilization of the normally short-half-lived C/EBPα and p21 proteins by RAP and ZTA (48, 50, 51).
More recently, we found that interactions between KSHV RTA and C/EBPα also contribute to the autoregulatory loops acting on the RAP promoter (46). In addition to RTA, the cellular transcription factor C/EBPα is itself a potent activator of the DE class RAP promoter (46), acting through direct binding to an upstream C/EBP site present in the RAP promoter, which overlaps with and appears to constitute part of the previously identified type I RRE. Direct interactions between RTA and C/EBPα as well as between RTA and RAP occur in lytically infected cells as well as in in vitro assays (46). Presumed piggyback DNA binding by both RTA:C/EBPα and RAP:C/EBPα complexes mediated through the C/EBP binding site in the RRE evidently results in high-level cooperative activation of the RAP promoter in transient-cotransfection reporter gene assays, as well as in KSHV lytically infected cells.
The results described here now provide further evidence for a critical and important role of C/EBPα in KSHV lytic cycle regulation during both earlier and later events than just those associated with the DE RAP promoter. In particular, we have demonstrated here that C/EBPα on its own can positively regulate the KSHV IE class RTA promoter, as well as both the DE class PAN and MTA promoters, and that it contributes cooperatively to RTA autoregulation as well as to RTA mediated-enhancement of at least these two other downstream DE viral promoters, even where other direct and indirect mechanisms have already been described. Aspects of our present results also hint that, like both RAP and ZTA, the interaction with RTA might lead to stabilization of C/EBPα, and they also raise as yet untested questions about whether RTA alone might be capable of independently inducing cell cycle arrest and p21 levels.
RTA mRNA belongs to the IE class of transcripts whose expression is induced very early (even in the absence of new protein synthesis) upon primary infection or after reactivation from latency. However, the mechanism that initially triggers RTA promoter expression during the earliest stages of KSHV lytic reactivation is unknown. In cell culture, TPA and butyrate mimic this process, and RTA was originally reported to positively autoregulate its own promoter in cooperation with the cellular OCT1 protein (32). However, we have shown here that C/EBPα plays an even more important role than previously appreciated during the early stages of KSHV lytic reactivation. Not only does C/EBPα mediate G0/G1 host cell cycle arrest, but it also directly activates the expression of early viral transcriptional and posttranscriptional nuclear regulatory proteins (RTA and MTA), as well as that of a protein involved in initiating viral DNA lytic replication (RAP). We also demonstrated that RTA cooperates with C/EBPα to enhance activation of both the RTA and C/EBPα promoters, that RTA can induce endogenous C/EBPα proteins in uninfected cells, and that exogenously introduced C/EBPα can induce RTA protein expression in latently infected PEL cells.
We have previously suggested that the ability of C/EBPα and RAP to cooperatively transactivate both the cellular C/EBPα and viral RAP promoters provides a mutually self-reinforcing loop to boost the levels of both proteins at early times in KSHV-infected cells (51). However, as shown here by Northern and Western blotting, the increase of C/EBPα mRNA and protein levels in latently infected PEL cells began to occur within 3 h after TPA treatment, whereas the appearance of RAP mRNA and protein did not occur until 12 h. Therefore, the presence of a much earlier trigger of the C/EBPα promoter (other than RAP) seems logical. Furthermore, in the DG75 KSHV-negative control cells, we know that TPA treatment itself does not induce endogenous C/EBPα expression in B cells, although the introduction of RTA alone does. Obviously, that model must now be modified to argue that C/EBPα and RTA also produce a self-reinforcing loop that reciprocally activates both promoters and protein levels at an even earlier stage and that subsequent RAP-mediated induction of C/EBPα expression occurs predominantly at later stages and functions to further enhance and maintain the levels of both C/EBPα and p21 proteins to promote cell cycle arrest (48). In fact, the highest levels of C/EBPα mRNA and protein were indeed observed between 12 and 24 h, a period when RAP protein is abundantly expressed.
However, there is still an interesting dilemma. Despite the fact that C/EBPα can activate the RTA promoter and that either KSHV RAP or RTA alone strongly induces C/EBPα expression in PEL cells, RAP alone (unlike RTA) is unable to induce RTA, vIL6, or other markers of the lytic cycle when introduced into latently infected PEL cells (9, 29, 39). It is probable that the promoters for lytic cycle viral genes, especially RTA, are transcriptionally silenced or repressed during latency and that even the high levels of C/EBPα protein induced by the exogenously introduced RAP alone is not sufficient to reverse the latent repression of RTA. In fact, the introduction of exogenous C/EBPα into latent BCBL1 cells was much more effective in inducing endogenous RAP expression (82%) than endogenous RTA expression (37%), suggesting that C/EBPα itself probably cannot fully overcome the latency-associated repression of the RTA promoter. Similarly, C/EBPα and RTA together boosted the level of cells coexpressing RAP to 98% (46) but that of RTA as shown here to only 51%. Presumably, this differential effect is caused by the presence of negative repressor elements within the KSHV RTA promoter similar to those in the EBV ZTA promoter (1), which may only be relieved by lytic cycle-inducing agents such as the protein kinase C agonist TPA, histone acetylation by butyrate, or perhaps B-cell receptor signaling. Alternatively, maybe only RTA but not RAP can access or displace chromatin appropriately, or the unknown relative contributions of C/EBPα protein stabilization compared to transcriptional effects in the PELs may also influence these results.
In summary, our model suggests that C/EBPα plays a central and highly complex role in KSHV lytic cycle reactivation in a process that involves multiple steps. The initial lytic switch at the RTA promoter is likely to be caused by a cellular activation or differentiation signal that is mimicked by chemical agents and/or cellular stress signals (such as TPA or butyrate). This trigger probably involves removal of cellular repressors from the RTA promoter that may in turn have been elevated or sequestered by latency-associated viral proteins. Once derepression is achieved, initial low levels of RTA may in turn cooperate with already existing low levels of C/EBPα to upregulate endogenous C/EBPα protein expression. After heightened C/EBPα protein levels are achieved, these could cooperate with RTA to further upregulate the RTA IE promoter. Both proteins then cooperate to activate the downstream DE class RAP promoter, and as levels of RAP build up, a second reinforcing loop between C/EBPα and RAP is established, which also involves stabilization of the C/EBPα (and p21) proteins, including their relocalization into PML oncogenic domains, and all leading to cell cycle arrest. Independently of C/EBPα, there are also more powerful direct effects of RTA or interactions of RTA with the cellular factor CBF1 that act on certain DE promoters such as PAN and MTA, and presumably also those for the core viral replication proteins, thus pushing the cells further into the lytic cycle and stimulating viral DNA replication and the formation of DNA replication compartments (RC).
The concepts presented here also raise new questions that have yet to be addressed experimentally about (i) whether RTA (like RAP) can independently stabilize the usually short half-lived C/EBPα protein, followed by inducing p21 expression and G1 cell cycle arrest; (ii) whether C/EBPα is itself one of the normal cellular triggers of RTA expression during KSHV reactivation in PELs and DMVECs, and perhaps any differentiation or cell cycle arrest events that may activate C/EBPα expression can themselves lead directly to the triggering of KSHV reactivation; and (iii) whether in addition antagonists of C/EBPα, perhaps including C/EBPβ, C/EBPδ, and CHOP10, may also contribute toward establishment, maintenance, or reactivation from latency.
Acknowledgments
These studies were funded by National Cancer Institute Research Grants (R01 CA73585 and RO1 CA81400) to G.S.H. from the National Institutes of Health. S.E.W. was supported by a postdoctoral fellowship in the Viral Oncology Program at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins School of Medicine. F.Y.W. was partially supported by the Anti-Cancer Drug Development Training Program (T32 CA09243).
We thank M. Daniel Lane for the generous gift of rabbit anti-C/EBPα PAb and all G.S.H. laboratory members for valuable discussions.
REFERENCES
- 1.Bryant, H., and P. J. Farrell. 2002. Signal transduction and transcription factor modification during reactivation of Epstein-Barr virus from latency. J. Virol. 76:10290-10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 4.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Chang, P.-J., D. Shedd, L. Gradoville, M.-S. Cho, L.-W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., J. Lee, Y. Wang, D. Huang, R. Ambinder, and S. Hayward. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and ZTA interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc. Natl. Acad. Sci. USA 96:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou, C.-J., L. J. Poole, P. Kim, D. M. Ciufo, J. S. Cannon, C. M. apRhys, D. J. Alcendor, J. C. Zong, R. F. Ambinder, and G. S. Hayward. 2002. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3421-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Y. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 12.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the RTA gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 12a.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the cooperative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, T. E., J. H. Albrecht, M. Nakanishi, and G. J. Darlington. 2001. CCAAT/enhancer-binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J. Biol. Chem. 276:29200-29209. [DOI] [PubMed] [Google Scholar]
- 17.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansen, L. M., I. Iwama, T. A. Lodie, K. Sasaki, D. W. Felsher, T. R. Golub, and D. G. Tenen. 2001. c-Myc is a critical target for C/EBPa in granulopoiesis. Mol. Cell. Biol. 21:3789-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 21.Lane, M. D., Q. Q. Tang, and M. S. Jiang. 1999. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem. Biophys. Res. Commun. 266:677-683. [DOI] [PubMed] [Google Scholar]
- 22.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jk (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 27.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas, J., J. C. Zong, D. J. Alcendor, D. M. Ciufo, L. J. Poole, R. T. Sarisky, C. J. Chiou, X. Zhang, X. Wan, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 29.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 31.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expressions. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saveliev, A. K., F. X. Zhu, and Y. Yuan. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301-304. [DOI] [PubMed] [Google Scholar]
- 34.Schulz, T. F. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 79:1573-1591. [DOI] [PubMed] [Google Scholar]
- 35.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 36.Slomiany, B. A., K. L. D'Arigo, M. M. Kelly, and D. T. Kurtz. 2000. C/EBPα inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol. 20:5986-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 39.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 43.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roesler, and N. A. Timchenko. 2001. C/EBPα arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 44.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 45.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, S. E., F. Y. Wu, M. Fujimuro, J. C. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of the CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, X., E. Scott, C. L. Sawyers, and A. D. Friedman. 1999. C/EBPα bypasses granulocyte colony-stimulating factor signals to rapidly induce PU1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood 94:560-571. [PubMed] [Google Scholar]
- 48.Wu, F. Y., Q.-Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. D. Lane, and G. S. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi's sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, F. Y., J.-H. Ahn, D. J. Alcendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, F. Y., H. Chen, S. E. Wang, C. apRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein α interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, F. Y., S. E. Wang, Q. Q. Tang, M. Fujimuro, C.-J. Chiou, Q. Cheng, H. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein (RAP) is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21CIP-1. J. Virol. 77:8893-8914. [DOI] [PMC free article] [PubMed]
- 52.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, D. E., P. Zhang, N. D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., R. Wang, A. Sweat, R. Goldstein, R. Horvat, and B. Chandran. 1999. Activation of human herpesvirus 8 (KSHV) thymidine kinase (TK) TATAA-less promoter by KSHV ORF50 gene product is SP1 dependent. DNA Cell Biol. 17:735-742. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]