Abstract
A major impediment to the use of adenovirus as a gene therapy vector and for vaccine applications is the host immune response to adenovirus hexon—the major protein component of the icosahedral capsid. A solution may lie in novel vectors with modified or chimeric hexons designed to evade the immune response. To facilitate this approach, we have distinguished the portion of hexon that all serotypes have in common from the hypervariable regions that are responsible for capsid diversity and type-specific immunogenicity. The common hexon core—conserved because it forms the viral capsid—sets boundaries to the regions where modifications can be made to produce nonnative hexons. The core has been defined from the large and diverse set of known hexon sequences by an accurate alignment based on the newly refined crystal structures of human adenovirus types 2 (Ad2) and Ad5 hexon. Comparison of the two hexon models, which are the most accurate so far, reveals that over 90% of the residues in each have three-dimensional positions that closely match. Structures for more distant hexons were predicted by building molecular models of human Ad4, chimpanzee adenovirus (AdC68), and fowl adenovirus 1 (FAV1 or CELO). The five structures were then used to guide the alignment of the 40 full-length (>900 residues) hexon sequences in public databases. Distance- and parsimony-based phylogenetic trees are consistent and reveal evolutionary relationships between adenovirus types that parallel those of their animal hosts. The combination of crystallography, molecular modeling, and phylogenetic analysis defines a conserved molecular core that can serve as the armature for the directed design of novel hexons.
The human adenovirus family has at least 49 distinct serotypes, which fall into six subgroups. These subgroups, A through F, are associated with a variety of pathologies and tissue tropisms (63). The subgroup C human adenoviruses, including types 2 (Ad2) and 5, have been widely used to develop vectors for gene therapy applications (77). Because of prior exposure, many patients have preexisting immunity to these common adenovirus types that limits their use (13). When introduced, they stimulate B cells to produce neutralizing antibodies that block vector uptake. Nevertheless, infection-induced humoral immunity to a particular serotype does not provide cross-immunity to serotypes in other subgroups (32). The main targets of type-specific antibodies are the adenovirus structural proteins hexon and fiber (49). As antifiber antibody neutralizes virus infectivity only in vitro (78) and not in vivo (25), the neutralizing antibody response relevant to vector applications is directed to hexon.
Hexon is the largest and most abundant of the structural proteins in the icosahedral adenovirus capsid (59, 62). The Ad2 virion has a particle mass of at least 150 × 106 Da (69), with hexon accounting for more than 83% of the capsid protein. The 240 copies of the homotrimeric hexon molecule are distributed symmetrically, with 12 copies forming each of the 20 capsid facets (12). With a length of 967 residues, the Ad2 hexon polypeptide chain is the longest of the known hexon sequences (109,064 Da [including the acetylated N terminus]). The other two major capsid proteins, the penton base and fiber, form the penton complex at each virion vertex. Two distinct viral attachment and cellular uptake events are required for infectivity, with each involving a separate site on the penton. Initially, a globular knob at the distal end of the fiber attaches to the host cell primary receptor. This is the coxsackie virus and adenovirus receptor (CAR) for Ad subgroups A, C, D, E, and F (7, 57, 67). For cells that lack CAR, the receptor is the major histocompatibility complex class I α2 subunit (for subgroup C) (17, 34) or sialic acid for Ad37 (subgroup D) (2). After binding, receptor-mediated endocytosis results from an interaction between an Arg-Gly-Asp sequence on the penton base and its αvβ integrin target (28, 45, 72).
Several different approaches have been taken to engineer the capsid of Ad5- or Ad2-based vectors to enhance infectivity and redirect native virus tropism. By exchanging fiber genes, vector uptake has been redirected to a non-CAR-mediated pathway (20, 56, 73, 75). Alternatively, retargeting can be achieved by attaching bispecific molecules. These include bifunctional peptides (19, 33) and chemically conjugated bispecific antibodies. The antibodies have been attached through an epitope incorporated into penton base (71, 74) or by specific recognition of the fiber knob domain (21, 27, 30, 47, 66, 70). These approaches show great promise for altering normal infection patterns and allowing specific targeting, but they do not overcome the problem of preexisting immunity.
One approach to circumventing preexisting host immunity is to mask the surface antigens with protective polymers such as polyethylene glycol (50), polylactic glycolic acid microspheres (44), or lipids (41, 46). Another approach is to derive vectors from different adenovirus serotypes (37, 42, 43, 48, 54). A recent example uses a nonhuman adenovirus isolated from chimpanzee (AdC68) (22). Detailed restriction endonuclease mapping (38, 76) and subsequent full-sequence analysis (22) have shown that AdC68 is closely related to Ad4. However, cross-reactivity of type-specific antisera between AdC68 and Ad4 is absent or diminished (22). Currently, efforts are under way to engineer the AdC68 vector for use as a vaccine carrier (79).
Another way to accomplish immunological distinction is to modify or replace the type-specific epitopes that are most likely located within seven hypervariable regions (HVRs) in the hexon sequence (15). The HVRs occur in loops at the top of the molecule that lie on the exterior of the virion and cover nearly its entire surface (60, 62). Genetic engineering approaches creating chimeric viruses have replaced the entire Ad5 hexon, or just the hexon FG1 loop, with that from Ad2 (26), replaced all four Ad5 (subgroup C) hexon loop domains with those of Ad12 (subgroup A) (58), and packaged the Ad5 genome into the capsids of Ad3 (subgroup B), Ad4 (subgroup E), or Ad9 (subgroup D) (51). Nevertheless, attempts to make a chimeric vector by replacing the Ad5 hexon with that of Ad7 (subgroup B) failed (26). The authors of that previous work suggested that the Ad7 hexon may be incompatible with the Ad5 100,000 folding protein, which is involved with hexon assembly and transport to the nucleus, or with other capsid proteins. The alteration of hexon epitopes need not be limited to switching serotypes. In fact, an 8-amino-acid sequence from the major antigenic site in the VP1 capsid protein of poliovirus type 3 has been introduced into two regions of the Ad2 hexon (16). These results show that the hexon structure is amenable to specific alterations but that these must be made with care, as some changes can have a profound effect on viral assembly and infectivity.
In this study, new high-resolution crystallographic refinements of the Ad2 and Ad5 hexon structures have been performed to take advantage of newer protocols and to resolve the observed differences between the previous Ad5 (Protein Data Bank [PDB] identifier, 1RUX) (60) and Ad2 (PDB identifier, 1HDX) (3) hexon models. To extend the diversity of the structural data, models for human Ad4, a chimpanzee adenovirus (AdC68), and an avian adenovirus (FAV1 or CELO) were built by molecular modeling. The five structures were then used in conjunction with sequence-based methods to define the common core of the hexon molecule that is required for capsid assembly. The resultant hexon armature describes the parts of the molecule that should not be violated when designing novel adenovirus-based vectors for efficacious human gene therapy or vaccine delivery.
MATERIALS AND METHODS
Crystallographic refinement. (i) Ad2 hexon.
A new crystallographic refinement of the Ad2 hexon structure was performed, based on a previous model for Ad5 hexon at 2.5-Å resolution (PDB identifier code, 1RUX) (60). Ad2 hexon crystals are isomorphous with those grown from the homologous Ad5 hexon (86% amino acid identity) (14, 24, 55). The methods for virus growth, hexon purification and crystallization (61), and X-ray data collection and processing (60) have been described previously. The Ad2 hexon diffraction data were collected at room temperature from a single crystal (dimensions, 0.6 × 0.5 × 0.5 mm) grown in 0.5 M sodium citrate buffer at pH 3.2. The Ad2 space group is P213 (a = b = c = 150.5 Å, α = β = γ = 90°), with one subunit of the hexon trimer in the asymmetric unit. The data include 48,199 of the 57,118 unique reflections in the resolution range from 10 to 2.2 Å (i.e., 84% complete). The overall agreement between related reflections, Rsymm, was 9.8%: Rsymm = Σh [Ih − average(Ih)]/Σh average(Ih), where Ih is the intensity of reflection h.
The starting model for Ad2 hexon refinement was the crystallographic model for Ad5 hexon (PDB identifier, 1RUX). This has seven segments (residues 5 to 135, 165 to 186, 193 to 250, 258 to 269, 278 to 430, 437 to 442, and 445 to 946) with intervening gaps where the polypeptide chain is disordered. The segments were renumbered to match the Ad2 hexon sequence (residues 5 to 135, 172 to 193, 204 to 261, 269 to 280, 290 to 442, 454 to 459, and 461 to 962). Of the 884 residues in the starting Ad2 model, 65 residues that differ in sequence from Ad5 were truncated at the β-carbon and modeled as Ala. To serve as a control for potential model bias, the remaining 110 residues with large aromatic side chains (i.e., Phe, Tyr, and Trp) were also modeled as Ala, and all water molecules were removed. All atomic temperature factors (B factors) were initialized to 15 Å2, a value typical of atomic positional disorder in macromolecular structures at room temperature. Initial refinement was performed as previously described for Ad5 (60) using TNT (68) modified to include the MLF1 maximum-likelihood target function, TNT-ML (53). Crystallographic R factors, which provide an estimate of model error, were monitored during refinement. The R factor is the sum of the absolute differences between experimentally observed (Fo) and calculated (Fc) amplitudes over all observed diffraction reflections (R = Σ |Fo − Fc|/Σ |Fo|). The working R (Rwork) is calculated with the reflections used for refinement, and an unbiased or free R (Rfree) is calculated with a subset of the data that has not been included in the refinement. Refinement, commencing with a rigid body, was performed using all data except for 5% used to monitor Rfree (10). The model was rebuilt between refinement rounds using program O (36).
To take advantage of the greater convergence potential of torsion angle dynamics, refinement was continued using the Crystallography and NMR System (CNS) software version 0.9a (11) with the protocol based on the supplied “refine.inp” procedure. For each round of CNS refinement, diffraction data from 6.0- to 2.2-Å resolution were used for initial B factor and bulk solvent corrections. Solvent molecules were harmonically restrained with a constant of 10. The model was first subjected to 200 steps of conjugate gradient minimization, and then torsion angle dynamics with slow-cooled annealing were performed from 5,000 to 300 K with 100-K temperature drops per cycle. Finally, another 200 steps of conjugate gradient minimization were applied, followed by 20 steps of restrained individual B-factor minimization. The model was rebuilt between refinement rounds as before, and water molecules were located with the waterpeaks script in the CCP4 program package (Collaborative Computational Project, 1994). The coordinates and structure factors have been deposited in the Protein Data Bank (identifier 1P2Z).
(ii) Ad5 hexon.
The same CNS methods that improved the Ad2 hexon model (above) were also applied to the previously published TNT-ML-refined 2.5-Å resolution Ad5 hexon structure (PDB identifier, 1RUX) (60). The Ad5 data set used was identical to that for 1RUX. The space group is P213 (a = b = c = 150.7 Å, α = β = γ = 90°), with one subunit of the Ad5 hexon trimer in the asymmetric unit. The diffraction data include 36,549 of 39,573 unique reflections (92% complete from 20- to 2.5-Å resolution) and have an Rsymm of 8.1%. The coordinates and structure factors have been deposited in the Protein Data Bank (identifier 1P30).
Molecular modeling.
The methods used to model the Ad4 and AdC68 hexons were described previously (22) and used to model the CELO hexon. Briefly, models of the homologous Ad4, AdC68, and CELO hexons were initially produced using the Swiss-PdbViewer protein modeling environment (29). Sequence alignments were used to guide the threading of model sequences onto the known molecular structures. These initial molecular models were then improved manually by moving gaps to exposed variable regions and optimizing side chain packing. Side chains or stretches of main chain that were missing from the crystal structures because of disorder were selected from libraries of side chain rotamers or known loop structures. The conformation of each model was further refined by energy minimization using the molecular mechanics program CHARMM (9). Graphical images prepared within the Swiss-PdbViewer program were exported and rendered with the Persistence of Vision Ray Tracer program (POV-Ray 2000; version 3.1g).
Structure- and sequence-based alignments.
The X-ray and homology model structures were each aligned with the Ad5 hexon model with transformation matrices calculated with the program LSQMAN (39) and applied with the program O (36). Secondary structure elements were assigned with PROMOTIF (35). A high-quality reference alignment was prepared by aligning the sequences corresponding with the five structurally aligned models. Forty unique full-length (>900 residues) hexon sequences were retrieved from a search of GenBank (6) with the Ad5 hexon sequence using BLASTP (1) (sequence accession numbers are listed below). The 35 unique and unmodeled full-length hexon sequences were then aligned with the Clustal_X program (64, 65) using the reference alignment as a profile. The multiple-alignment parameters used in this heuristic approach included the following: Gonnet 250 series matrices, sequence divergence delay of 30%, gap opening 10, gap extension 0.2, residue-specific and hydrophilic penalties included, gap separation distance of 4, and no end gap separation. Hexon sequence labels and GenBank accession numbers (in parentheses) are as follows: ad12_A (CAA51891), ad3_B (CAA54051), ad7_B (CAA88460), ad7a_B (AAD03667), ad16_B (CAA52726), ad2_C (AAA92215), ad5_C (AAD15293), ad21_B (BAB20066), ad34_B (BAB20014), ad35_B (BAB20015), unk (CAA09917), ad48_D (AAB17439), ad4_E (AAD03657), ad41_E (CAA36079), ad40_F (AAC13967), bovine3 (AAA42509), bovine4 (AAC41020), bovine5 (AAF20944), bovine6 (AAF20945), bovine7 (AAF63488), bovine8 (AAF63489), bovine10 (AAF82136), canine1 (AAB05443), canine2 (AAB38725), chimp68 or adC68 (AF394196), duck1 (CAA70809), equine1 (AAB88062), equine2 (AAB88060), fowl1 or CELO (AAC54912), fowl8 (AAD50344), fowl10 (AAA91647), frog1 (AAF86932), goat1 (AAF20946), murine1 (AAB48187), odocoileus (AAF13265), ovine7 (AAK56926), ovine287 (AAA84979), porcine3 (AAC99441), porcine5 (AAK26489), and turkey3 (AAC64532).
Phylogenetic analyses.
Phylogenetic analyses of the aligned hexon sequences were performed with PHYLIP (version 3.5.7c) (23). The following program protocols were used: seqboot to calculate 100 bootstrap trials, PROTPARS and CONSENSE for parsimony analysis, and a combination of PROTDIST using the Dayhoff PAM matrix (18), FITCH, and CONSENSE for distance-based analysis. The phylogenetic trees were displayed with TreeView 1.6.1 (52).
RESULTS
Crystallographic refinement.
Initial rigid body refinement of the Ad2 starting model reduced the Rwork and Rfree values from 38.3 to 33.2 and from 38.1 to 33.8%, respectively. The large aromatic side chains that were intentionally removed from the starting model as a control were immediately apparent in the resultant electron density. After four rounds of model building and TNT-ML refinement, the model contained 883 of 967 residues (91%), 296 water molecules, with an Rwork value of 19.0% and an Rfree value of 23.7%. Subsequent CNS refinement converged after five rounds to yield 887 of 967 Ad2 hexon residues (93%), five citrate molecules, 430 water molecules, with an Rwork value of 17.8% and an Rfree value of 22.1% at 2.2-Å resolution. For Ad5 hexon, the CNS protocol reduced the initial Rwork and Rfree values from 21.5 to 16.9 and 24.3 to 21.0%, respectively. The refinement converged after four more rounds to yield 891 of 951 Ad5 hexon residues (94%), 304 water molecules, with an Rwork value of 15.8% and an Rfree value of 20.7% at 2.5-Å resolution. The presence of citrate molecules reflects the composition of the crystallization buffer. The statistics for the final Ad2 and Ad5 hexon models are shown in Table 1.
TABLE 1.
Hexon refinement statistics
| Characteristic | Adenovirus
|
|
|---|---|---|
| Ad2 | Ad5 | |
| Resolution (Å) | 10.0-2.2 | 20.0-2.5 |
| No. of unique reflections | 48,199 | 36,549 |
| Rwork (%) | 17.8 | 15.8 |
| Rfree (%) | 22.1 | 20.7 |
| No. of residues | 887 | 891 |
| RMS bond deviation (Å) | 0.006 | 0.006 |
| RMS angle deviation (°) | 1.341 | 1.309 |
| No. of protein atoms Average B factor (Å2) | 6,914 | 7,000 |
| 18.0 | 26.4 | |
| No. of citrate molecules Average B factor (Å2) | 5 | 0 |
| 67.0 | ||
| No. of water molecules Average B factor (Å2) | 430 | 304 |
| 23.9 | 25.0 | |
Structure alignment.
While the fold of the newly refined Ad2 hexon model (Fig. 1) is generally similar to that of an earlier model (3), the alterations bring the new Ad2 model into close agreement with the Ad5 model. An alignment of the current two models with the program LSQMAN (39) shows an excellent match for 874 residues in the Ad2 (887 residues) and Ad5 (891 residues) models, with a root mean square (RMS) distance between Cα atoms of 0.3 Å. Of these 874 matching residues, 812 have the same side chains, whereas the remaining 62 residues differ. The matching residue positions for the two X-ray structures are as follows: for Ad5, positions 5 to 135, 165 to 186, 192 to 211, 213 to 250, 258 to 268, 279 to 305, 308 to 430, and 445 to 946; for Ad2, positions 5 to 135, 172 to 193, 203 to 222, 224 to 261, 269 to 279, 291 to 317, 320 to 442, and 461 to 962. As the hexon homology models for the AdC68, Ad4, and CELO hexons were based upon the Ad2 and Ad5 crystal structures, they are also closely related to one another.
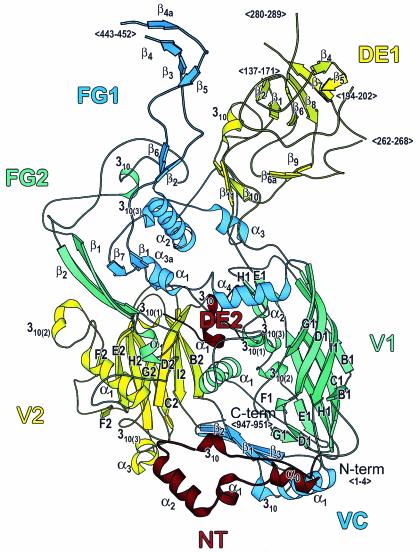
FIG. 1.
Hexon structure. Ribbon representation of the Ad2 hexon subunit. The view is perpendicular to the molecular threefold axis from the inside of the molecule. The top of the molecule, which contains the loops (DE1, FG1, and FG2), forms the outer surface of the viral capsid. The hexon base contains a small loop (DE2) and two eight-stranded “viral” jellyrolls (V1 and V2), which are separated by the connector, VC. The eight jellyroll β strands are labeled B to I. The N-terminal loop, NT, lies underneath the base. The secondary structural elements are labeled, as are the positions of chain segments that are not observed in the crystallographic model (<…>). The figure was made with MOLSCRIPT (40).
Hexon regions.
For simplicity, all hexon models will be discussed together in this section, although only two are based on crystallographic data. Each hexon structure can be divided into eight regions (Fig. 1; Table 2). The list of secondary structural elements (Table 3) shows a few that have been newly identified in the two X-ray structures (underlined) and some that are predicted in the homology models (in parentheses). The N-terminal region, 55 residues long in the Hominidae (human and chimpanzee) hexons and 60 for CELO, forms an extended loop on the basal surface of the trimer. It is important for stability, as it extends from one subunit, under a second, to touch the third. The additional five residues in CELO precede the α0 helix (Fig. 1; Table 3) and would be exposed at the edge of the base where they could be involved in intermolecular contacts in the viral capsid. Some flexibility is present at the N terminus, as the first four residues are disordered in both crystal structures. The NT-α0 helix secondary structural element is newly observed in the current Ad2 and Ad5 crystal structures (residues 5 to 12). The only significant difference between the two models in the N-terminal region is a different side chain conformation for His 52.
TABLE 2.
Hexon regions
| Region | Label | Ad2
|
Ad5
|
AdC68
|
Ad4
|
CELO
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Gaps | Total (undefined) | Sequence | Gaps | Total (undefined) | Sequencea | Total | Sequencea | Total | Sequencea | Total | ||
| N terminus | NT | 1-55 | 1-4 | 55 (4) | 1-55 | 1-4 | 55 (4) | 1-55 | 55 | 1-55 | 55 | 1-60 | 60 |
| First viral jellyroll | V1 | 56-114 | 198 | 56-114 | 198 | 56-114 | 198 | 56-114 | 198 | 61-119 | 197 | ||
| 335-405 | 323-393 | 305-375 | 306-376 | 288-357 | |||||||||
| 570-637 | 554-621 | 535-602 | 538-605 | 524-591 | |||||||||
| Extended loop | DE1 | 115-334 | 137-171 | 220 (61) | 115-322 | 136-164 | 208 (43) | 115-304 | 190 | 115-305 | 191 | 120-287 | 168 |
| 194-202 | 251-256 | ||||||||||||
| 262-268 | 271-278 | ||||||||||||
| 280-289 | |||||||||||||
| Extended loop | FG1 | 406-569 | 443-452 | 164 (10) | 394-553 | 431-436 | 160 (8) | 376-534 | 159 | 377-537 | 161 | 358-523 | 166 |
| 443-444 | |||||||||||||
| Viral jellyroll connector | VC | 638-660 | 963-967 | 53 (5) | 622-644 | 947-951 | 53 (5) | 603-625 | 53 | 606-628 | 53 | 592-614 | 52 |
| 938-967 | 922-951 | 903-932 | 906-935 | 913-941 | |||||||||
| Second viral jellyroll | V2 | 661-699 | 151 | 645-683 | 151 | 626-664 | 151 | 629-667 | 151 | 615-653 | 153 | ||
| 730-787 | 714-771 | 695-752 | 698-755 | 684-743 | |||||||||
| 884-937 | 868-921 | 849-902 | 852-905 | 859-912 | |||||||||
| Extended loop | DE2 | 700-729 | 30 | 684-713 | 30 | 665-694 | 30 | 668-697 | 30 | 654-683 | 30 | ||
| Extended loop | FG2 | 788-883 | 96 | 772-867 | 96 | 753-848 | 96 | 756-851 | 96 | 744-858 | 115 | ||
The initial Met residue predicted from the gene sequences of the homology models is not included, as it is not present in Ad2 and Ad5 hexons, where it is removed upon N-terminal acetylation.
TABLE 3.
Hexon secondary structure
| X-ray models
|
Homology modelsb
|
|||||
|---|---|---|---|---|---|---|
| Ad5 residues | Ad2 residues | Region | Elementa | C68 residues | Ad4 residues | CELO residues |
| NT | 2-5 (310) | 2-5 (310) | ||||
| 5-12 | 5-12 | NT | α0 | 6-11 | 6-11 | 12-18 |
| 18-23 | 18-23 | NT | α1 | 19-24 | 19-24 | |
| 24-35 | 24-35 | NT | α2 | 25-36 | 25-36 | 30-39 |
| 40-44 | 40-44 | NT | 310 | 40-45 | 40-45 | 46-50 |
| 64-67 | 64-67 | V1 | B1 | 65-68 | 65-68 | 70-73 |
| 71-74 | 71-74 | V1 | B1 | 72-75 | 72-75 | 77-80 |
| 78-87 | 78-87 | V1 | C1 | 79-88 | 79-88 | 85-92 |
| 93-94, 98-107 | 93-94, 98-107 | V1 | D1 | 93-108 | 93-108 | 99-100, 104-113 |
| 132-134 | 132-134 | DE1 | β1 | 134-135 | 138-139 | |
| 166-168 | 173-175 | DE1 | β2 | 140-141 | ||
| 173-174 | DE1 | β3 | 147-148 | 142-144 | ||
| 178-179 | 185-186 | DE1 | β4 | 151-152 | 152-156 | |
| 182-188 | 189-190 | DE1 | β5 | 158-163 | 162-166 | 162-166 |
| 191-194 | DE1 | β5a | 166-169 | 169-172 | ||
| 218-223 | 229-234 | DE1 | β6 | 193-194 | 193-195 | 197-198, 201-202 |
| DE1 | 204-206 (310) | |||||
| 236-237 | 247-248 | DE1 | β6a | |||
| 262-266 | 273-277 | DE1 | β7 | 232-234 | 233-238 | |
| 283-288 | 295-300 | DE1 | β8 | 259-263 | 248-253 | |
| 292-293 | 304-305 | DE1 | β9 | |||
| 297-301 | 309-313 | DE1 | β10 | 262-264 | ||
| 310-316 | 323-328 | DE1 | 310 | 293-297 | 294-299 | 275-280 |
| 317-320 | 329-332 | DE1 | β11 | 283-285 | ||
| 326-328 | 338-340 | V1 | E1 | 309-311 | 310-312 | 291-293 |
| 330-333 | 342-345 | V1 | 310(1) | |||
| 340-344 | 352-356 | V1 | 310(2) | |||
| 346-349 | 358-361 | V1 | E1 | 331-332 | 332-333 | 311-314 |
| V1 | 316-317 (310) | |||||
| Nonregular | Nonregular | V1 | F1 | Nonregular | Nonregular | Nonregular |
| 364-377 | 376-389 | V1 | α1 | 347-360 | 348-361 | 329-340 |
| 384-387 | 396-399 | V1 | 310(3) | |||
| 394-399 | 406-411 | FG1 | α1 | 359-364 | ||
| 400-401 | 412-413 | FG1 | β1 | 382-385 | 385-386 | 365-366 |
| 412-413 | 423-425 | FG1 | β2 | 395-396 | 397-397 | 377-378 |
| 423-425 | 435-437 | FG1 | β3 | 404-408 | 404-409 | |
| 428-429 | 440-441 | FG1 | β4 | 411-413 | ||
| 439-440 | 456-457 | FG1 | β4a | 421-423 | ||
| 449-451 | 465-467 | FG1 | β5 | 431-435 | 434-441 | |
| 459-460 | 475-477 | FG1 | β6 | 441-442 | 444-445 | 427-428 |
| 462-476 | 478-492 | FG1 | α2 | 444-462 | 447-465 | 430-444 |
| 477-480 | 493-496 | FG1 | 310(1) | |||
| 481-485 | 497-501 | FG1 | 310(2) | 463-467 | 466-470 | |
| 499-506 | 515-522 | FG1 | α3 | 484-489 | ||
| 509-514 | 525-530 | FG1 | 310(3) | 491-496 | 494-499 | |
| 522-523 | 538-539 | FG1 | β7 | 503-506 | 506-507 | 493-494 |
| 525-530 | 541-546 | FG1 | α3a | 507-513 | 510-516 | 496-501 (310) |
| 539-550 | 555-566 | FG1 | α4 | 523-530 | 526-533 | 510-519 |
| 554-561 | 570-577 | V1 | G1 | 536-541 | 539-544 | 525-527, 530-532 |
| V1 | 549-552 (310) | 552-555 (310) | 538-541 (310) | |||
| 572-573 | 588-589 | V1 | G1 | 554-555 | 557-558 | 543-544 |
| 576-585 | 592-601 | V1 | H1 | 558-567 | 561-570 | 548-552, 554-555 |
| 587-591 | 603-607 | V1 | α2 | 569-573 | 572-576 | 558-562 |
| 592-594 | 608-610 | V1 | H1 | 574-576 | 577-579 | 563-565 |
| V1 | 581-586 (α) | 584-589 (α) | ||||
| 606-617 | 622-633 | V1 | I1 | 588-601 | 591-604 | 577-588 |
| 622-634 | 638-650 | VC | α1 | 604-615 | 607-618 | 593-604 |
| 635-639 | 651-655 | VC | 310 | |||
| 640-643 | 656-659 | VC | β1 | 622-624 | 625-627 | 611-613 |
| 648-655 | 664-671 | V2 | B2 | 630-636 | 633-640 | 619-625 |
| 661-669 | 677-685 | V2 | C2 | 644-651 | 647-654 | 632-640 |
| 676-683 | 692-699 | V2 | D2 | 658-665 | 661-668 | 647-654 |
| 685-687 | 701-703 | DE2 | 310 | 667-669 | 670-672 | 656-658 |
| 704-709 | 720-725 | DE2 | α1 | 675-680 | ||
| 713-715 | 729-731 | V2 | 310(1) | |||
| 717-724 | 733-740 | V2 | E2 | 699-706 | 702-709 | 688-695 |
| 728-730 | 744-746 | V2 | F2 | 710-712 | 713-715 | 699-701 |
| 742-743 | 758-759 | V2 | F2 | 724-725 | 727-728 | 713-714 |
| 750-753 | 763-769 | V2 | 310(2) | 730-734 | 723-725 | |
| V2 | 736-737 (β) | 739-740 (β) | 727-728 (β) | |||
| V2 | 742-743 (β) | 745-746 (β) | 733-734 (β) | |||
| 761-774 | 777-790 | V2 | α1 | 744-756 | 747-759 | 735-746 |
| 784-788 | FG1 | 310 | ||||
| 792-796 | 808-813 | FG2 | α1 | 774-778 | 777-781 | |
| 797-805 | 814-821 | FG2 | β1 | 779-787 | 782-790 | 770-772, 775-778 |
| FG2 | 785-786 (β) | |||||
| FG2 | 791-792 (β) | |||||
| 817-821 | 833-837 | FG2 | 310 | 799-803 | 802-806 | |
| FG2 | 810-811 (β) | 813-814 (β) | ||||
| FG2 | 818-819 (β) | 821-822 (β) | ||||
| 857-865 | 873-880 | FG2 | β2 | 838-846 | 842-850 | 849-852, 855-857 |
| 870-873 | 886-889 | V2 | G2 | 851-854 | 855-858 | 862-865 |
| 886-889 | 902-906 | V2 | 310(3) | 866-870 | 870-874 | 877-881 |
| 890-895 | 906-910 | V2 | α3 | 871-887 | 875-881 | 882-888 |
| 898-906 | 914-922 | V2 | H2 | 879-887 | 883-891 | 890-898 |
| 913-920 | 929-936 | V2 | I2 | 894-901 | 898-905 | 905-912 |
| 922-928 | 938-944 | VC | β2 | 906-909 | 910-912 | 914-923 |
| 936-942 | 952-958 | VC | β3 | 917-919 | 926-934 | |
| VC | 928-929 (β) | 937-941 (310) | ||||
New secondary structure observed in the X-ray models is underlined.
Predicted secondary structure is shown in parentheses.
The N-terminal region is followed by the first of the two viral jellyrolls, V1 (Fig. 1; Table 2), with its eight strands labeled B to I. The size of V1 is essentially constant, with 198 residues in human hexons and 197 residues in CELO. The DE1 loop is an extended structure extending from strands D and E of V1 to form the largest domain in the Ad2 and Ad5 hexons (220 and 208 residues, respectively). In all other known hexon sequences, DE1 is smaller. DE1 is the most variable of the hexon regions and is flexible; the X-ray models have four gaps due to low electron density (Table 2). Ad5 hexon has two new very short strands in DE1 at residues 191 to 194 (β5a) and 236 to 237 (β6a). The FG1 loop, emerging between the F and G strands of V1, is smaller than the DE1 loop and relatively constant in size compared to DE1, varying from 159 residues (AdC68) to 166 residues (CELO). Two new secondary structural elements, FG1-β4a and FG1-α3a, are observed in both X-ray structures (Fig. 1; Table 3). The newly observed FG1-β4a strand hydrogen bonds with the previously observed FG1-β4 strand to form a second short antiparallel pair close to that formed by FG1-β3 and FG1-β5.
The viral jellyroll connector domain (VC) is located in the base of the molecule and serves, with V1-α1 and loop DE2, to separate the first (V1) and second (V2) viral jellyrolls and to clamp them together. VC is formed by stretches from the middle and end of the molecule (i.e., residues 638 to 660 and 938 to 967 in Ad2), but its size is almost invariant—53 residues for the human hexons and 52 residues for CELO. This conservation is particularly significant, as any alteration in VC size would alter the separation of V1 and V2 and so affect the hexon diameter (see Fig. 3). With 720 copies of hexon present in the capsid, any small change in the separation of V1 and V2 would be greatly magnified and have a significant effect on the overall size of the virion. VC extends to the C terminus, with the last five residues disordered in both X-ray structures.
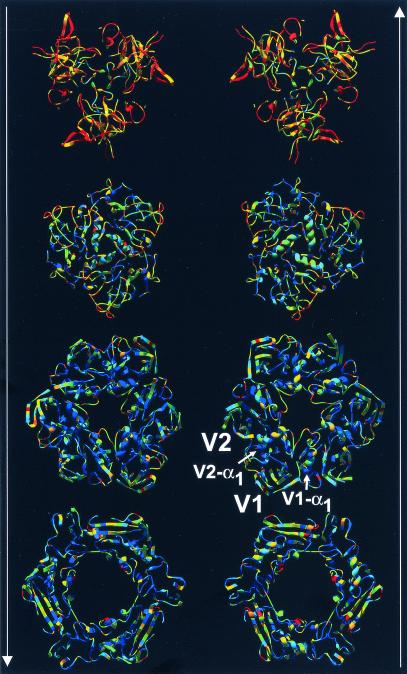
FIG. 3.
Hexon trimer colored by sequence diversity. Layers of the trimeric hexon molecule displayed as ribbons. The two columns show hexon slices viewed along the molecular threefold axis. The arrows indicate the direction of view from either the virion exterior top to bottom (left) or from the virion interior bottom to top (right). The structures are colored by sequence diversity from blue (conserved) to yellow (less conserved) to red (not conserved). The jellyrolls V1 and V2 and their α1 helices are labeled. Note that the size of the hexon's pseudohexameric base is dictated by structural elements, such as V1-α1 and V2-α1, which separate the jellyrolls. A PDB file of the hexon trimer with temperature factors (B factors) representing sequence diversity is available for download at http://bioinfo.wistar.upenn.edu/pub/RKB_hexon_var.pdb.
The V2 domain is the smaller of the two viral jellyrolls, with 151 residues for all Hominidae hexons and 153 for CELO (Table 2). V2 also has loop extensions between the DE and FG strands, but they are much smaller than their counterparts in V1. Their structural similarities indicate that the jellyrolls arose by gene duplication (3). The double jellyroll is a key feature that distinguishes viruses such as adenovirus and bacteriophage PRD1 from viruses with single-jellyroll capsid proteins like the picornaviruses (4). Trimeric double-jellyroll coat proteins have pseudohexagonal shapes that make them well suited to accurate assembly of large viral capsids (12). The DE2 loop is the smallest of the four hexon loops, and its length is remarkably conserved at 30 residues in all the hexon structures. This conservation probably arises because DE2 plays the same role as VC in separating and stabilizing V1 and V2—but at the upper half of the jellyrolls. The path of the Ad2 residues—706 Leu→707 Gly→708 Ser—differs from that of the corresponding residues in Ad5 (residues 690 to 692), even though the residues are the same. The flexibility at this point explains how the frog and turkey hexons can accommodate a Trp residue in place of the Gly that is present in the other 38 hexon sequences. The Ad2 Gly 707 residue lies on the basal surface, so the Trp in frog and turkey may be involved in hexon-hexon interactions in the capsid.
The FG2 loop, which extends between the F and G strands of V2, is much larger than the DE2 loop. FG2 has 96 residues in the Hominidae hexon models and 115 in CELO. In contrast to the other upper loops, DE1 and FG1, FG2 has no disordered residues in the X-ray structures. It is also much smaller and lower in height. FG2 forms an intriguing solvent-accessible pocket underneath the tower that accommodates hydrophobic residues of quite different sizes in different hexons. As they are normally buried, such residues are usually conserved. The pocket explains the puzzling variability in these residues, which lie within a generally conserved region in the sequence, but the feature has no obvious function. In Ad2, the pocket is occupied by Val 861, with Phe 845 in Ad5, and Ala, Tyr, or Trp in other hexons.
Sequence alignment.
The structural superposition of the hexon crystallographic and homology models aligned their five sequences and produced a profile to guide the alignment of the remaining 35 sequences. The resultant sequence alignment (Fig. 2) is colored to reveal conserved chemical properties using default Clustal_X definitions. Sequence diversity is indicated by the Clustal_X quality scores, which are shown as a histogram (Fig. 2) and as a colored molecule (Fig. 3). The conservation pattern reflects hexon's function as an architectural building block. Its pseudohexagonal shape (Fig. 3) enables hexons to closely pack and form a closed spherical capsid. The inside of the molecule's central cavity and its upper surface both vary, particularly in the tower regions at the top, while the inside and lower surfaces are conserved. Thus, the regions that make up the base of the molecule, NT (77% identity and 83% similarity), V1 (76% identity and 84% similarity), VC (76% identity and 81% similarity), V2 (79% identity and 87% similarity), and DE2 (81% identity and 86% similarity) have highly conserved sequences (Fig. 2 and 3). In contrast, the regions forming the top of the molecule, DE1 (37% identity and 46% similarity), FG1 (62% identity and 71% similarity), and FG2 (64% identity and 72% similarity) are less highly conserved (Fig. 2 and 3).
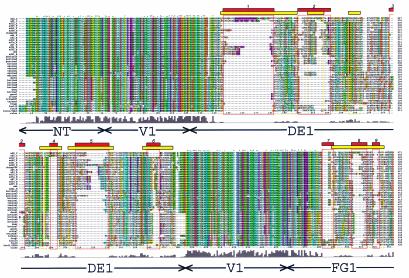
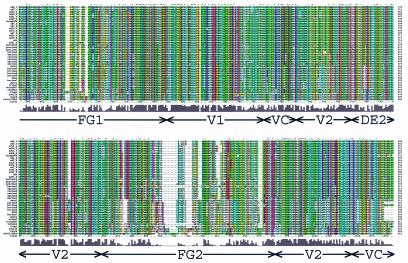
FIG. 2.
Hexon sequence alignment. Multiple-sequence alignment of 40 hexon sequences calculated with Clustal_X (64, 65). An alignment profile based on the structural alignment of two hexon crystal structures and three homology models was used to guide the multiple-sequence alignment. The default Clustal_X color scheme indicates significant features. All G (orange) and P (yellow) are colored. Frequent occurrences of a property at a sequence position are colored: hydrophobic, blue; hydrophobic tendency, light blue; basic, red; acidic, purple; hydrophilic, green; unconserved, white. Conserved positions are marked as complete (*), strong (:), or weak (.). A histogram indicates the conservation level, and the eight molecular regions are labeled. Previously assigned (15) HVRs (HVR1 to -9) are boxed in yellow, and the new assignments (HVR1 to -9) are boxed in red. A Clustal_X alignment file is available for download at http://bioinfo.wistar.upenn.edu/pub/RKB_hexon.aln.
The most divergent of the 1,064 positions in the hexon are the 208 (20%) with Clustal_X quality scores in the bottom 10th percentile. These unconserved residues are distributed throughout the hexon structure (colored red in Fig. 3), but all are located on the molecular surface. Many of these residues fall at the top of the molecule in the HVRs defined by Crawford-Miksza and Schnurr (15). The new alignment made it possible to assign the HVRs quantitatively as stretches of more than five residues with quality scores in the bottom 10th percentile (with at most one residue allowed to have a higher score). This procedure defines the HVR boundaries (Fig. 2) more precisely and yields nine regions: HVR1 (residues 146 to 181), HVR2 (199 to 221), HVR3 (264 to 270), HVR4 (288 to 293), HVR5 (307 to 330), HVR6 (359 to 366), HVR7 (482 to 489), HVR8 (503 to 513), and HVR9 (900 to 911).
Phylogenetic analysis.
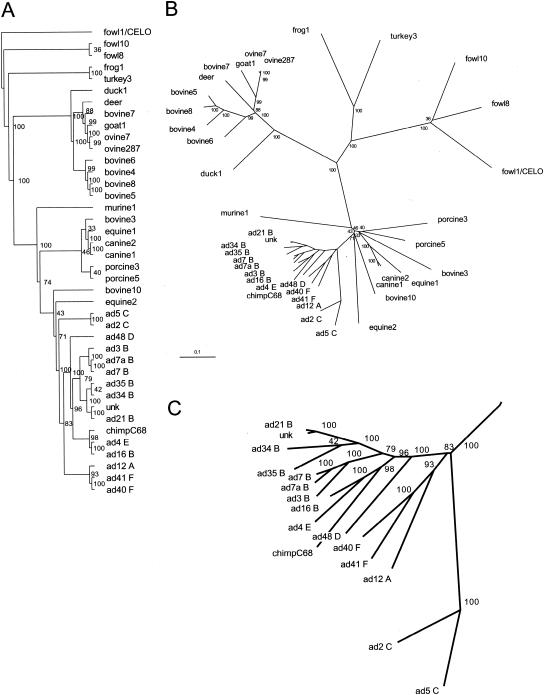
The results of the phylogenetic analyses of the 40 aligned full-length hexon sequences are summarized in Fig. 4. Parsimony analysis builds an evolutionary tree depicting the order requiring the least number of residue alterations to change each sequence to another. Sequences that group together are more likely to have evolved together. The parsimony analysis for the hexon (Fig. 4A) shows that the Hominidae hexons clearly segregate from all others and that the human hexons form groups consistent with their serotypes. The hexon of unknown origin (unk) is likely to be in subgroup B. That Ad16 (subgroup B) seems more similar to Ad4 (group E) than to others in its subgroup illustrates the limitations of an analysis based solely on sequence.
FIG. 4.
Phylogenetic analysis of adenovirus hexon. Phylogenetic trees of 40 full-length hexon sequences calculated with the Phylip program package (23). (A) An unrooted tree generated by parsimony analysis with PROTPARS and CONSENSE. (B) An unrooted tree generated by distance matrix analysis with PROTDIST (Dayhoff's PAM 001 scoring matrix), FITCH (global rearrangements option), and CONSENSE. Branch lengths are proportional to the number of substitutions, and the scale bar represents 10 mutations per 100 sequence positions. Bootstrap values for the 100 trials are indicated for each branch. (C) A close-up view of the Hominidae adenovirus hexon branches.
Phylogenetic distance analysis estimates the divergence time between each pair of sequences. The smaller the distance, the more likely it is that the pair has evolved from a common ancestor. The results of the distance matrix analysis are shown as trees (Fig. 4B and C). The chimpanzee hexon (AdC68) is clearly grouped with the human serotypes and is most similar to Ad4 and Ad16. It is clear from the data that the adenoviruses share a common origin. For the human, equine, porcine, bovine, canine, ovine, and fowl hosts, the different hexons largely group together. As noted previously (5, 31), there are at least two distinct groups of bovine hexons. Bovine3 and -10 hexons group close to human hexons, whereas bovine4, -5, -6, -7, and -8 hexons lie elsewhere. The phylogenetic distances presented here are shorter than earlier measurements (31), as the current calculation includes all positions in the full-length hexon sequences. It is curious that turkey and duck hexons are less closely related to the fowl hexon group than to the frog hexon and the main bovine hexon group, respectively.
While predictions of the origins of the adenoviruses should become clearer as more sequences are examined, at present the hexon data suggest that they form at least three and possibly four distinct groups that are evolving independently. It is clear from Fig. 4 that the Hominidae adenoviruses evolved from a common ancestor, with the closest separate group including adenoviruses infecting horses, pigs, cows, dogs, and mice. More distant is the group infecting ducks, cows, deer, goats, and sheep. The turkey and frog adenoviruses appear to be members of a third group, with the fowl adenoviruses as the most distant group. These conclusions are strengthened by the fact that the results of the distance-based analyses (Fig. 4B) are consistent with those of the parsimony analysis (Fig. 4A), as the two methods rely on different assumptions. Although the phylogenetic tree shown in Fig. 4B is not based on extensive data, it indicates that the evolution of the animal adenoviruses parallels that of their hosts. For example, adenoviruses infecting Hominidae (humans, chimpanzees) or Artiodactyla (cloven-hooved animals, such as cows, deer, goats, and sheep) cluster together. It further suggests that horizontal transfer is not common, despite the intimate contact between humans and animals such as chickens. The extent to which adenovirus transfer can occur between closely related hosts, such as chimpanzees and humans, is still unclear.
DISCUSSION
HVRs and epitopes.
To make reproducible assignments for the HVRs, we introduced a quantitative approach. Seven HVRs in the hexon sequence were described by Crawford-Miksza and Schnurr (15). The present analysis of the positions of the most divergent residues for 40 full-length hexon sequences reveals nine HVRs. In general, the positions of the newly defined HVRs correlate with those defined previously, but the greater number of sequences and the use of hexon structural data to guide their alignment has allowed a more precise definition of these regions. We previously showed that the HVRs lie in the extended loops at the top of the molecule (60). This is in agreement with the present analysis, in which the DE1 loop contains HVR1 to -6 and the FG1 loop contains HVR7 to -9. The alignment positions (Fig. 2) of the current (previous) HVR assignments are as follows: HVR1, 146 to 181 (144 to 197); HVR2, 199 to 221 (206 to 216); HVR3, 264 to 270 (235 to 242); HVR4, 288 to 293 (282 to 296); HVR5, 307 to 330 (302 to 333); HVR6, 358 to 366 (355 to 375); HVR7, 482 to 489, HVR8, 503 to 513, and HVR9, 518 to 522 (HVR7, 489 to 525).
The criteria used for the new assignments are more stringent than those used earlier and define the HVRs more tightly. In consequence, they focus on “hot spots” where sequence modifications are likely to be possible without structural disruption. The new assignments for HVR1, -4, -5, and -6 are subsets of those defined earlier. Similarly, it is now clear that the earlier HVR7 should be defined more precisely as HVR7, -8, and -9. The one exception to the general pattern of agreement with the previous assignments is HVR3. Its original definition places it in a well-conserved segment in the current sequence alignment (Fig. 2).
In the absence of evidence to physically map virus-specific neutralizing epitopes, it is reasonable to assume that they lie within one or more of the HVRs. The HVRs lie within the loops at the top of the hexon molecule that form the virion surface, and so it is not unexpected that the most significant differences between the hexon sequences are at these locations. Recently, Moffat et al. (48) showed that rabbit and mouse anti-Ad5, antibovine (bovine3), and antiporcine (porcine3) sera do not cross-react. This finding shows that their neutralizing epitopes differ significantly and is consistent with the idea that the epitopes must lie in positions of sequence variation—particularly as bovine3 and porcine3 are quite closely related (Fig. 4).
In an early effort to engineer the adenovirus, Crompton et al. (16) inserted an 8-amino-acid sequence from the major antigenic site in the poliovirus type 3 VP1 capsid protein into two regions of Ad2 hexon. Antisera specific for the poliovirus sequence efficiently neutralized the modified adenovirus, and antisera raised against the modified adenovirus recognized the VP1 capsid protein of poliovirus type 3. Substitution into residues 284 to 291 of the DE1 loop (corresponding to alignment positions 309 to 330 in HVR5; Fig. 2) resulted in a virus with wild-type growth characteristics, whereas substitution into residues 442 to 449 of the FG1 loop for Ad2 hexon (alignment positions 499 to 506) yielded viruses that grew poorly. This result was difficult to understand earlier, as the FG1 loop insertion site is within the region previously assigned to HVR7 (15). In contrast, the insertion extends beyond the range now assigned to HVR8. This suggests that the modification lay outside the most malleable region of hexon and so altered parts of its structure involved in virion formation. It is very encouraging that the more precisely defined HVRs presented here explain the earlier results. The explanation suggests that the new definitions will also be more accurate in predicting which hexon sites are amenable to future modification.
The topology of the hexon monomer (Fig. 1), with two axially oriented jellyrolls and long loop extensions, suggests a flexible structure capable of supporting a wide variety of sequence lengths. However, in the native trimeric form (Fig. 3), the jellyrolls are closely packed and the long loops that emerge between jellyroll strands are tightly wrapped around one another to form compact tower domains. In addition, the hexon base interacts closely with other protein partners in the virion. Due to these severe structural constraints, hexon sequences longer than that for Ad2 are likely to arise from insertions into one or more of the HVRs.
Crystallography.
In our earlier crystallographic analysis of the Ad5 hexon (60), we applied new crystallographic methods, including maximum-likelihood refinement and the modeling of bulk solvent, that were not available at the time that the Ad2 hexon structure was first determined (3). The results suggested that a reevaluation of Ad2 would be rewarding (60). The present study shows that this was not only worthwhile but that the Ad5 structure could be improved still further.
Initially, the methods used in the Ad5 hexon structure determination (60) improved the Ad2 model so it was of similar quality to that for Ad5 (PDB identifier, 1RUX). Subsequently, still further improvements were made in crystallographic refinement procedures. The program CNS (11) can now combine simulated-annealing and torsion angle dynamics with refinement against the maximum-likelihood target function developed for TNT-ML (53). Application of the latest methods has now produced excellent models for both the Ad2 and Ad5 hexons.
This study shows that determining crystal structures for closely related molecules can be rewarding. Only by determining the Ad5 hexon structure was the necessity to revisit the Ad2 structure made apparent. Likewise, this process suggested further improvements for Ad5. The quality of the current models is such that no further improvements are anticipated with the available diffraction data.
Hexon stabilization.
While monitoring the quality and stability of recombinant adenovirus preparations by anion exchange high performance liquid chromatography, Blanche et al. (8) noted that the retention time of the hexon peak increased with storage time. They showed that the shift corresponded to an increase in isoaspartate residue formation and thus resolved the long-standing puzzle of why chromatograms exhibit a trailing shoulder in the hexon peak (61). To reduce the rate of Asn deamidation, Blanche et al. (8) changed all four accessible Asn residues engaged in Asn-Gly pairs in the Ad5 hexon to Leu (Asn 243, Asn 254, and Asn 436) or Ala (Asn 275). These modifications reduced the microheterogeneity in the recombinant adenovirus without adversely affecting virus infectivity. In the Ad5 hexon X-ray model, only Asn 243 in loop DE1 has interpretable density, while the other three asparagines are located in disordered—or flexible—segments of loops DE1 and FG1. The other 39 full-length hexon sequences do not have Asn residues at these positions, but all have other Asn-Gly pairs. For example, the chimpanzee AdC68 hexon has Asn’s forming Asn-Gly pairs at positions 414, 486, and 533 in loop FG1. The homology model indicates that each of these is surface accessible and so would be subject to isoaspartate formation. For optimum stability, appropriate modifications should be made to all such Asn-Gly pairs when designing adenovirus vectors for clinical use.
Adenovirus-based vector design.
Adenoviruses are maintained episomally in infected cells and do not integrate with the host genome. Consequently, the transgene is lost as cells infected with a recombinant adenovirus vector turn over so that repeated administrations of vector are necessary. Problems then arise with preexisting or acquired antiadenoviral immunity. A conservative approach to overcoming this obstacle is to design and use adenovirus-based vectors with modified hexons.
By far the most common serotype of adenovirus-based vectors for human gene therapy is Ad5. A recent study by Youil et al. (80) lists several reasons why this is so. First, Ad5 is the most extensively studied serotype. Vectors based on another serotype would require additional studies to determine safety and tropism profiles before human use. Second, sequencing and annotation would be required before new replication-incompetent adenovirus vectors with deleted early genes could be designed. Third, alternative serotypes have an uncharacterized potential for targeting different cell types, due to variability in the cellular receptors and binding strategies used for attachment. Fourth, serotypes outside human subgroup C are likely to require the generation of new complementing cell lines to support their propagation.
As described above, the insertion of an 8-amino-acid peptide corresponding to a poliovirus epitope into the Ad2 hexon by Crompton et al. (16) shows that the hexon molecule is amenable to careful modification. The most comprehensive hexon modification made to an Ad5-based vector was to replace the entire hexon with that of another serotype, Ad2 (26). Expression levels were monitored after intravenous injections of an Ad5-based vector containing the gene for liver chloramphenicol acetyltransferase. Expression was reduced only 100-fold in animals preimmunized with an Ad2 hexon-bearing Ad5 chimera, as opposed to 10,000-fold when an Ad5 vector was used for preimmunization. Similar results were observed when the FG1 loop of Ad5 was replaced with that of Ad2 (26). Attempts to create an Ad7 hexon-bearing Ad5 chimera were unsuccessful, which indicated that some residues cannot be readily substituted. In another study, Roy et al. (58) created a viable, albeit lower-titer, Ad12-Ad5 chimera by replacing all four Ad5 hexon loops (i.e., DE1, FG1, DE2, and FG2) with those of Ad12 (subgroup A).
More recently, Youil et al. (80) have expanded these studies by creating chimeric recombinant Ad5 constructs in which the hexon gene was replaced with that from several serotypes in subgroups A through E. Of the 18 constructs created, only those with the Ad1, Ad2, Ad6, and Ad12 hexon genes formed viable viruses. Ad1, Ad2, and Ad6 (like Ad5) are all in subgroup C. Only one non-subgroup-C hexon (i.e., Ad12, subgroup A) was able to substitute for Ad5 hexon. Viable vectors were not formed by a second subgroup A hexon (Ad18), or by four from subgroup B (Ad7, -11, -16, and -35), eight from subgroup D (Ad9, -10, -13, -15, -17, -19, -27, and -37), and one from subgroup E (Ad4). These authors echo the suggestion by Gall et al. (26) that the failure to produce viable virions is most likely due to structural incompatibilities between the alternate hexon proteins and the 100-kDa scaffolding protein, which is required for assembly, or with the other capsid proteins (i.e., penton, fiber, and protein IX) (80).
Summary.
We have applied crystallography, molecular modeling, and phylogenetic techniques to determine the conserved core of the hexon molecule and to locate sites that are most likely amenable to modification. The approach used here shows how excellent results can be achieved by combining both experimental and theoretical structural data with a heuristic alignment algorithm. The resultant sequence alignment uses all currently available data and produces phylogenies that are consistent with experimental groupings of adenovirus serotypes. The current alignment includes a wider range of hexon sequences than examined previously and allows a more precise definition of the HVRs. The new definitions now explain experimental results that were published previously. They show that the hexon molecule has a conserved core that forms the capsid and dictates its size and so should not be altered. The nine newly defined HVRs on the molecular surface are primary candidates for hexon modification.
Acknowledgments
We thank Donatella Pascolini for her early contributions to protein purification and diffraction data collection. We also thank the instructors and staff of the Nucleic Acid and Protein Sequence Analysis Workshop at the Pittsburgh Supercomputing Center—in particular David W. Deerfield II and Hugh B. Nicholas. We thank Carmen San Martín and Stacy D. Benson for helpful discussions regarding virus structure and crystallographic refinement techniques.
This work was supported by the National Institute of Allergy and Infectious Diseases (grant number AI-17270) and by the National Cancer Institute through the Wistar Institute Cancer Center (grant number CA 09171).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 3.Athappilly, F. K., R. Murali, J. J. Rux, Z. Cai, and R. M. Burnett. 1994. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 Å resolution. J. Mol. Biol. 242:430-455. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, D. H., R. M. Burnett, and D. I. Stuart. 2002. Evolution of viral structure. Theor. Pop. Biol. 61:461-470. [DOI] [PubMed] [Google Scholar]
- 5.Benkö, M., B. Harrach, and J. C. D'Halluin. 1990. Molecular cloning and physical mapping of the DNA of bovine adenovirus serotype 4; study of the DNA homology among bovine, human, and porcine adenoviruses. J. Gen. Virol. 71:465-469. [DOI] [PubMed] [Google Scholar]
- 6.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2000. GenBank. Nucleic Acids Res. 28:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 8.Blanche, F., B. Cameron, S. Somarriba, L. Maton, A. Barbot, and T. Guillemin. 2001. Stabilization of recombinant adenovirus: site-directed mutagenesis of key asparagine residues in the hexon protein. Anal. Biochem. 297:1-9. [DOI] [PubMed] [Google Scholar]
- 9.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187-217. [Google Scholar]
- 10.Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. [DOI] [PubMed] [Google Scholar]
- 11.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 12.Burnett, R. M. 1997. The structure of adenovirus, p. 209-238. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 13.Chirmule, N., K. Propert, S. Magosin, Y. Qian, R. Qian, and J. M. Wilson. 1999. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 6:1574-1583. [DOI] [PubMed] [Google Scholar]
- 14.Cornick, G., P. B. Sigler, and H. S. Ginsberg. 1971. Characterization of crystals of type 5 adenovirus hexon. J. Mol. Biol. 57:397-401. [DOI] [PubMed] [Google Scholar]
- 15.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crompton, J., C. I. A. Toogood, N. Wallis, and R. T. Hay. 1994. Expression of a foreign epitope on the surface of the adenovirus hexon. J. Gen. Virol. 75:133-139. [DOI] [PubMed] [Google Scholar]
- 17.Davison, E., I. Kirby, T. Elliott, and G. Santis. 1999. The human HLA-A*0201 allele, expressed in hamster cells, is not a high-affinity receptor for adenovirus type 5 fiber. J. Virol. 73:4513-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins, p. 345-352. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5, suppl. 3. National Biomedical Research Foundation, Washington, D.C.
- 19.Dmitriev, I., E. Kashentseva, B. E. Rogers, V. Krasnykh, and D. T. Curiel. 2000. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J. Virol. 74:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas, J. T., B. E. Rogers, M. E. Rosenfeld, S. I. Michael, M. Feng, and D. T. Curiel. 1996. Targeted gene delivery by tropism-modified adenoviral vectors. Nat. Biotechnol. 14:1574-1578. [DOI] [PubMed] [Google Scholar]
- 22.Farina, S. F., G.-P. Gao, Z. Q. Xiang, J. J. Rux, R. M. Burnett, M. R. Alvira, J. Marsh, H. C. J. Ertl, and J. M. Wilson. 2001. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 75:11603-11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 24.Franklin, R. M., U. Pettersson, K. Åkervall, B. Strandberg, and L. Philipson. 1971. Structural proteins of adenovirus. V. Size and structure of the adenovirus type 2 hexon. J. Mol. Biol. 57:383-395. [DOI] [PubMed] [Google Scholar]
- 25.Gall, J., A. Kass-Eisler, L. Leinwand, and E. Falck-Pedersen. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gall, J. G. D., R. G. Crystal, and E. Falck-Pedersen. 1998. Construction and characterization of hexon-chimeric adenoviruses: specification of adenovirus serotype. J. Virol. 72:10260-10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman, C. K., B. E. Rogers, J. T. Douglas, B. A. Sosnowski, W. Ying, G. P. Siegal, A. Baird, J. A. Campain, and D. T. Curiel. 1997. Targeted gene delivery to Kaposi's sarcoma cells via the fibroblast growth factor receptor. Cancer Res. 57:1447-1451. [PubMed] [Google Scholar]
- 28.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 29.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 30.Haisma, H. J., J. Grill, D. T. Curiel, S. Hoogeland, V. W. van Beusechem, H. M. Pinedo, and W. R. Gerritsen. 2000. Targeting of adenoviral vectors through a bispecific single-chain antibody. Cancer Gene Ther. 7:901-904. [DOI] [PubMed] [Google Scholar]
- 31.Harrach, B., and M. Benkö. 1999. Phylogenetic analysis of adenovirus sequences, p. 309-339. In W. S. M. Wold (ed.), Adenovirus methods and protocols—methods in molecular medicine, vol. 21. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 32.Hierholzer, J. C., Y. O. Stone, and J. R. Broderson. 1991. Antigenic relationships among the 47 human adenoviruses determined in reference horse antisera. Arch. Virol. 121:179-197. [DOI] [PubMed] [Google Scholar]
- 33.Hong, S. S., A. Galaup, R. Peytavi, N. Chazal, and P. Boulanger. 1999. Enhancement of adenovirus-mediated gene delivery by use of an oligopeptide with dual binding specificity. Hum. Gene Ther. 10:2577-2586. [DOI] [PubMed] [Google Scholar]
- 34.Hong, S. S., L. Karayan, J. Tournier, D. T. Curiel, and P. A. Boulanger. 1997. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 16:2294-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchinson, E. G., and J. M. Thornton. 1996. PROMOTIF—a program to identify and analyze structural motifs in proteins. Protein Sci. 5:212-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 47:110-119. [DOI] [PubMed] [Google Scholar]
- 37.Kass-Eisler, A., L. Leinwand, J. Gall, B. Bloom, and E. Falck-Pedersen. 1996. Circumventing the immune response to adenovirus-mediated gene therapy. Gene Ther. 3:154-162. [PubMed] [Google Scholar]
- 38.Kitchingman, G. R. 1982. Restriction mapping and molecular cloning of adenovirus type 4 (subgroup E) DNA. Gene 20:205-210. [DOI] [PubMed] [Google Scholar]
- 39.Kleywegt, G. J. 1996. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 52:842-857. [DOI] [PubMed] [Google Scholar]
- 40.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 41.Lee, S. G., S. J. Yoon, C. D. Kim, K. Kim, D. S. Lim, Y. I. Yeom, M.-W. Sung, D. S. Heo, and N. K. Kim. 2000. Enhancement of adenoviral transduction with polycationic liposomes in vivo. Cancer Gene Ther. 7:1329-1335. [DOI] [PubMed] [Google Scholar]
- 42.Mack, C. A., W.-R. Song, H. Carpenter, T. J. Wickham, I. Kovesdi, B.-G. Harvey, C. J. Magovern, O. W. Isom, T. Rosengart, E. Falck-Pedersen, N. R. Hackett, R. G. Crystal, and A. Mastrangeli. 1997. Circumvention of anti-adenovirus neutralizing immunity by administration of an adenoviral vector of an alternate serotype. Hum. Gene Ther. 8:99-109. [DOI] [PubMed] [Google Scholar]
- 43.Mastrangeli, A., B.-G. Harvey, J. Yao, G. Wolff, I. Kovesdi, R. G. Crystal, and E. Falck-Pedersen. 1996. “Sero-switch” adenovirus-mediated in vivo gene transfer: circumvention of anti-adenovirus humoral immune defenses against repeat adenovirus vector administration by changing the adenovirus serotype. Hum. Gene Ther. 7:79-87. [DOI] [PubMed] [Google Scholar]
- 44.Mathews, C. B., G. Jenkins, J. M. Hilfinger, and B. L. Davidson. 1999. Poly-l-lysine improves gene transfer with adenovirus formulated in PLGA microspheres. Gene Ther. 6:1558-1564. [DOI] [PubMed] [Google Scholar]
- 45.Mathias, P., T. Wickham, M. Moore, and G. Nemerow. 1994. Multiple adenovirus serotypes use αv integrins for infection. J. Virol. 68:6811-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meunier-Durmort, C., R. Picart, T. Ragot, M. Perricaudet, B. Hainque, and C. Forest. 1997. Mechanism of adenovirus improvement of cationic liposome-mediated gene transfer. Biochim. Biophys. Acta 1330:8-16. [DOI] [PubMed] [Google Scholar]
- 47.Miller, C. R., D. J. Buchsbaum, P. N. Reynolds, J. T. Douglas, G. Y. Gillespie, M. S. Mayo, D. Raben, and D. T. Curiel. 1998. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res. 58:5738-5748. [PubMed] [Google Scholar]
- 48.Moffatt, S., J. Hays, H. HogenEsch, and S. K. Mittal. 2000. Circumvention of vector-specific neutralizing antibody response by alternating use of human and non-human adenoviruses: implications in gene therapy. Virology 272:159-167. [DOI] [PubMed] [Google Scholar]
- 49.Norby, E. 1969. The structural and functional diversity of adenovirus capsid components. J. Gen. Virol. 5:221-236. [DOI] [PubMed] [Google Scholar]
- 50.O'Riordan, C. R., A. Lachapelle, C. Delgado, V. Parkes, S. C. Wadsworth, A. E. Smith, and G. E. Francis. 1999. PEGylation of adenovirus with retention of infectivity and protection from neutralizing antibody in vitro and in vivo. Hum. Gene Ther. 10:1349-1358. [DOI] [PubMed] [Google Scholar]
- 51.Ostapchuk, P., and P. Hearing. 2001. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J. Virol. 75:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 53.Pannu, N. S., and R. J. Read. 1996. Improved structure refinement through maximum likelihood. Acta Crystallogr. Sect. A Found. Crystallogr. 52:659-668. [Google Scholar]
- 54.Parks, R. J., C. M. Evelegh, and F. L. Graham. 1999. Use of helper-dependent adenoviral vectors of alternative serotypes permits repeat vector administration. Gene Ther. 6:1565-1573. [DOI] [PubMed] [Google Scholar]
- 55.Pereira, H. G., R. C. Valentine, and W. C. Russell. 1968. Crystallization of an adenovirus protein (the hexon). Nature 219:946-947. [DOI] [PubMed] [Google Scholar]
- 56.Roelvink, P. W., G. M. Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science 286:1568-1571. [DOI] [PubMed] [Google Scholar]
- 57.Roelvink, P. W., A. Lizonova, J. G. M. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy, S., P. S. Shirley, A. McClelland, and M. Kaleko. 1998. Circumvention of immunity to the adenovirus major coat protein hexon. J. Virol. 72:6875-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rux, J. J., and R. M. Burnett. 1999. Adenovirus capsid proteins, p. 5-16. In P. Seth (ed.), Adenoviruses: from basic research to gene therapy applications, Medical Intelligence Unit 15. R. G. Landes, Austin, Texas.
- 60.Rux, J. J., and R. M. Burnett. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1:18-30. [DOI] [PubMed] [Google Scholar]
- 61.Rux, J. J., D. Pascolini, and R. M. Burnett. 1999. Large-scale purification and crystallization of adenovirus hexon, p. 259-275. In W. S. M. Wold (ed.), Adenovirus methods and protocols—methods in molecular medicine, vol. 21. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 62.San Martín, C., and R. M. Burnett. 2003. Structural studies on adenoviruses, p. 57-94. In W. Doerfler and P. Böhm (ed.), Adenoviruses: model and vectors in virus host interactions—current topics in microbiology and immunology, vol. 272. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 63.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 64.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tillman, B. W., T. D. de Gruijl, S. A. Luykx-de Bakker, R. J. Scheper, H. M. Pinedo, T. J. Curiel, W. R. Gerritsen, and D. T. Curiel. 1999. Maturation of dendritic cells accompanies high-efficiency gene transfer by a CD40-targeted adenoviral vector. J. Immunol. 162:6378-6383. [PubMed] [Google Scholar]
- 67.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tronrud, D. E., L. F. Ten Eyck, and B. W. Matthews. 1987. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallogr. Sect. A Found. Crystallogr. 43:489-501. [Google Scholar]
- 69.van Oostrum, J., and R. M. Burnett. 1985. Molecular composition of the adenovirus type 2 virion. J. Virol. 56:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watkins, S. J., V. V. Mesyanzhinov, L. P. Kurochkina, and R. E. Hawkins. 1997. The “adenobody” approach to viral targeting: specific and enhanced adenoviral gene delivery. Gene Ther. 4:1004-1012. [DOI] [PubMed] [Google Scholar]
- 71.Wickham, T. J., G. M. Lee, J. A. Titus, G. Sconocchia, T. Bakács, I. Kovesdi, and D. M. Segal. 1997. Targeted adenovirus-mediated gene delivery to T cells via CD3. J. Virol. 71:7663-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 73.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 74.Wickham, T. J., D. M. Segal, P. W. Roelvink, M. E. Carrion, A. Lizonova, G. M. Lee, and I. Kovesdi. 1996. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J. Virol. 70:6831-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wigand, R., M. Mauss, and T. Adrian. 1989. Chimpanzee adenoviruses are related to four subgenera of human adenoviruses. Intervirology 30:1-9. [DOI] [PubMed] [Google Scholar]
- 77.Wivel, N. A., G.-P. Gao, and J. M. Wilson. 1999. Adenovirus vectors: the development of human gene therapy, p. 87-110. In T. Friedman (ed.), The development of human gene therapy. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 78.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiang, Z., G.-P. Gao, A. Reyes-Sandoval, C. J. Cohen, Y. Li, J. M. Bergelson, J. M. Wilson, and H. C. J. Ertl. 2002. Novel, chimpanzee serotype 68-based adenoviral vaccine carrier for induction of antibodies to a transgene product. J. Virol. 76:2667-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Youil, R., T. J. Toner, Q. Su, M. Chen, A. Tang, A. J. Bett, and D. Casimiro. 2002. Hexon gene switch strategy for the generation of chimeric recombinant adenovirus. Hum. Gene Ther. 13:311-320. [DOI] [PubMed] [Google Scholar]