Abstract
Limitin has sequence homology with alpha interferon (IFN-α) and IFN-β and utilizes the IFN-α/β receptor. However, it has no influence on the proliferation of normal myeloid and erythroid progenitors. In this study, we show that limitin has antiviral activity in vitro as well as in vivo. Limitin inhibited not only cytopathic effects in encephalomyocarditis virus- or herpes simplex virus (HSV) type 1-infected L929 cells, but also plaque formation in mouse hepatitis virus (MHV) type 2-infected DBT cells. In addition, administration of limitin to mice suppressed MHV-induced hepatitis and HSV-induced death. The antiviral activity may be mediated in part by 2′,5′-oligoadenylate synthetase, RNA-dependent protein kinase, and Mx protein, which inhibit viral replication or degrade viral components, because limitin induced their mRNA expression and enzyme activity. While limitin has antiviral activity as strong as that of IFN-α in vitro (the concentration that provided 50% inhibition of cytopathic effect is ∼30 pg/ml), IFN regulatory factor 1 (IRF-1) dependencies for induction of an antiviral state were different for limitin and IFN-α. In IRF-1-deficient fibroblasts, a higher concentration of limitin than of IFN-α was required for the induction of antiviral activity and the transcription of proteins from IFN-stimulated response element. The unique signals and the fewer properties of myelosuppression suggest that a human homolog of limitin may be used as a new antiviral drug.
Interferons (IFNs) are divided into two major groups of biologically active proteins. IFN-α, IFN-β, IFN-ω, and IFN-τ, which belong to the IFN-α/β family, have structural homology to each other and share a common receptor (36). IFN-γ is a product of activated T lymphocytes and natural killer cells (52). Although only small amounts of IFNs are produced under steady-state conditions, these can be markedly increased by virus infections or by exposure to double-stranded nucleic acids (50). IFNs are highly pleiotropic cytokines with potent immunoregulatory and antiproliferative properties in addition to their antiviral activities (9, 40, 41, 42, 65). IFNs regulate immune responses, such as major histocompatibility antigen expression, cytokine production, and T-lymphocyte activation (59). IFNs suppress the proliferation of many normal and transformed cells through prolongation of the G1 phase of the cell cycle and reduce the rate of entry into the S phase, as well as inducing apoptosis (7). The antiviral activity of IFNs results in part from induction of the enzymes 2′,5′-oligoadenylate synthetase (2′,5′-OAS) and RNA-dependent protein kinase (PKR) and of Mx protein, which inhibit viral replication or degrade viral components (59). These broad biological properties have suggested many clinical applications in cancer and virologic diseases (19, 66).
Limitin is an IFN-like cytokine that we recently cloned on the basis of its ability to arrest the growth of or kill a myelomonocytic leukemia cell line (37). The initial cDNA clone carried a 997-bp insert (EMBL-GenBank-DDBJ accession number AB024521) whose deduced protein is composed of 182 amino acid residues. It contains a signal peptide of 21 amino acids at the N-terminal end and one N-linked glycosylation site at amino acid residue 68 but lacks an internal transmembrane domain. In addition, limitin protein was detected in the supernatants of transfectants with an expression plasmid of limitin. Thus, limitin seems to be a secreted glycoprotein. Analysis with the translated protein sequence reveals that limitin has ∼30% identity with IFN-α, IFN-β, and IFN-ω. Computer modeling of limitin structure shows an IFN-like globular structure composed of five long α-helices and one short helix in the middle of the loop connecting helices B and C, with possible disulfide bonds between residues 52 and 157 and between residues 80 and 130 (36). In addition, receptor-binding assays and experiments using IFN-α/β receptor-deficient cells have indicated that limitin binds to and utilizes the IFN-α/βreceptor (37). Thus, limitin seems to be a new member of the IFN-α/β family. As has been reported for previously known IFNs, limitin inhibits not only B lymphopoiesis but also proliferation of a variety of lympho-hematopoietic cell lines (37). Limitin enhances major histocompatibility antigen expression on antigen-presenting cells, as well as the killer activity of cytotoxic T lymphocytes (60). However, limitin is unique among IFNs because it has no influence on the proliferation of normal myeloid and erythroid progenitors (37). Although limitin transduces signals via the IFN-α/β receptor, its signals could be different from those of IFN-α and IFN-β. In this study, we show that limitin has antivial activity, a major biological function of IFNs, and we clarify the difference between signals induced by limitin and IFN-α.
MATERIALS AND METHODS
Cells and mice.
A murine myeloid leukemia cell line (WEHI3), a murine fibroblast cell line (L929), and a murine brain tumor cell line (DBT) were maintained in Dulbecco's modified Eagle's medium (Nikken, Kyoto, Japan) supplemented with 10% fetal calf serum (Flow, Aurora, Ohio). Cultured fibroblasts were generated from murine lungs and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
IFN regulatory factor 1 (IRF-1)-deficient mice were kindly provided by T. Taniguchi (Tokyo University, Tokyo, Japan). C57BL/6 mice were purchased from Japan Clea (Tokyo, Japan) and used as control wild-type mice for IRF-1-deficient mice. BALB/c mice were purchased from Nippon SLC (Shizuoka, Japan). All mice were maintained under specific-pathogen-free conditions at the Institute for Experimental Animals, Osaka University. The mice were ∼6 to 10 weeks of age at the time of use.
Viruses and cytokines.
Encephalomyocarditis virus (EMCV) and L929 cells were kindly provided by M. Kohase (National Institute of Infectious Disease, Tokyo, Japan). EMCV was propagated in L929 cell cultures. Human herpes simplex virus type 1 (HSV-1) was isolated from the vesicle of a patient and propagated in L929 cell cultures. Mouse hepatitis virus type 2 (MHV-2) and DBT cells were kindly provided by F. Taguchi (National Institute of Neuroscience, Tokyo, Japan). MHV was propagated in DBT cell cultures (14). EMCV and HSV were titrated by the cytopathic effect (CPE) dye uptake method, and MHV was titrated by plaque assay.
Recombinant murine IFN-α was purchased from GIBCO BRL (Grand Island, N.Y.). The production and purification of recombinant limitin and anti-limitin polyclonal antibody were described in a previous article (38). The fusion proteins limitin-immunoglobulin (Ig) and CD44-Ig were prepared as described previously (60).
Gel filtration chromatography.
Analysis of purified recombinant limitin was performed by gel filtration chromatography using a Superdex 75 PC 3.2/30 column (Amersham Biosciences, Piscataway, N.J.). A mobile phase consisted of 20 mM Tris HCl, pH 8.0, and 150 mM sodium chloride. Eluted protein was detected by UV light at 280 nm. An LMW calibration kit (Amersham Biosciences) was used as a standard protein marker.
[3H]thymidine incorporation assay.
WEHI3 cells were incubated in flat-bottom 96-well microplates (Corning Costar, Tokyo, Japan) for 48 h. The cells were pulsed with 0.5 μCi of [3H]thymidine (Amersham, Tokyo, Japan)/well for the last 4 h of culture and then harvested onto glass filters (Wallac, Turku, Finland) with a semiautomatic cell harvester (Pharmacia), and incorporated radioactivity was measured with a liquid scintillation counter.
CPE dye uptake method.
Antiviral activity was measured by the CPE dye uptake method with minor modifications (2, 20, 70). Briefly, confluent monolayers of L929 cells were exposed to serial dilutions of limitin or IFN-α overnight, challenged with either EMCV or HSV until 75 to 95% CPE was evident, and stained with naphthol black solution (0.1% naphthol blue-black, 0.1% acetic bicarbonate, 9% acetic acid). The bound dye in the wells of the microtiter plates was eluted with 100 mM NaOH, and the optical density at 650 nm (OD650) was measured colorimetrically using a spectrophotometer (Emax precision microplate reader; Molecular Devices).
Plaque assay.
DBT cell monolayer cultures on six-well dishes were washed with phosphate-buffered saline (PBS) and inoculated with MHV for 1 h. The cultures were overlaid with minimal essential medium containing 1% Bacto Agar (Difco, Detroit, Mich.) and 10% tryptose phosphate broth. After 48 h of incubation, each well was stained by a second overlay containing 1:10,000 neutral red for 12 h, and then the plaques were counted (14).
Northern blot analysis.
Total RNAs were isolated using a TRIZOL reagent (Invitrogen, Carlsbad, Calif.). The RNAs (15 μg/lane) were electrophoresed through a formaldehyde-agarose gel and transferred onto a nylon membrane (Amersham, Arlington Heights, Ill.). The cDNA fragments were labeled with [32P]dCTP using a randomly primed DNA-labeling kit (Boehringer Mannheim, Indianapolis, Ind.) and hybridized to the membrane. The blots were then washed and autoradiographed. The cDNA fragment of Mx1 (12) was kindly provided by O. Haller (Freiburg University, Freiburg, Germany), and that of IRF-1 was a kind gift of Y. Kanakura (Osaka University, Osaka, Japan). The cDNA fragments of PKR and 2′,5′-OAS were amplified by PCR (16, 17) with the following primers: sense (5′-TTAGGTGGATTTGGTCAA-3′) (895 to 912) and antisense (5′-TCGGTCCTTGGGTTTCTCTG-3′) (1600 to 1617) for PKR and sense (5′-GTCTCCAAGGTGGTGAAGGG-3′) (201 to 220) and antisense (5′-TACTCAAGTTGTGTCGGGTC-3′) (781 to 800) for 2′,5′-OAS.
2′,5′-OAS activity.
Measurement of 2′,5′-OAS activity was performed using a 2′,5′-oligoadenyl-5′-triphosphate (2-5A) radioimmunoassay kit (Eiken Chemical Co., Tokyo, Japan) according to the manufacturer's instructions and with some modifications described elsewhere (48, 63). Briefly, L929 cells (105) were seeded in 24-well plates and stimulated with limitin for the appropriate time. The cells were then suspended in 1 ml of lysis buffer consisting of 10 mM HEPES (pH 7.5), 50 mM KCl, 3 mM Mg(CHOO)2, 0.3 mM EDTA, 1 mM 2-mercaptoethanol, 20% glycerol, and 0.5% NP-40 and centrifuged at 10,000 × g for 5 min at 4°C. Then, 50 μl of supernatant medium and cell lysates was mixed with 50 μl of poly(rI) · poly(rC) agarose beads and then incubated for 10 min at room temperature. After centrifugation at 400 × g for 5 min, the pellets were resuspended with 500 μl of ATP solution and incubated at 37°C for 1 h. To the mixture was added 100 μl of 125I-labeled 2-5A solution to quantify the 2-5A produced by enzymatic reaction of 2′,5′-OAS and ATP. The mixture was incubated at 37°C for 1 h and centrifuged at 800 × g for 30 min. After the beads were washed, their radioactivity was measured with a well-type gamma counter (WALLAC WIZARD 1470). The amounts of 2-5A synthesized by 2′,5′-OAS were calculated from the standard curve.
Infection.
Mice were infected intraperitoneally with an appropriate dose of virus and then treated intraperitoneally with limitin or PBS. The procedures were in compliance with the guidelines set by the institute for care and use of laboratory animals.
GPT enzyme activity.
The concentrations of glutamate pyruvate transaminase (GPT) in the sera of MHV-infected mice were measured using a Rikitech GPT IFCC assay kit (Roche Diagnostics, Tokyo, Japan).
Histopathological analysis.
Histopathological examinations were carried out using paraffin-embedded sections of livers from mice infected with MHV. The sections were deparaffinized and stained with hematoxylin-eosin.
Luciferase assay.
Luciferase assays were performed using the Dual-Luciferase Reporter System (Promega, Madison, Wis.), in which transfection efficiency was monitored by cotransfected pRL-SV40 (Promega), an expression vector of Renilla luciferase. The cultured cells were transfected with 500 ng of pCIS-ISRE (Stratagene, La Jolla, Calif.), together with 50 ng of pRL-SV40, by the Lipofectamine transfection method (Invitrogen). The transfected cells were lysed in buffer supplied by the manufacturer, followed by measurement of the firefly and Renilla luciferase activities on a luminometer (LB96P; Berthold Japan, Tokyo, Japan). The relative firefly luciferase activities were calculated by normalizing transfection efficiencies according to the Renilla luciferase activities.
RESULTS
Recombinant limitin protein.
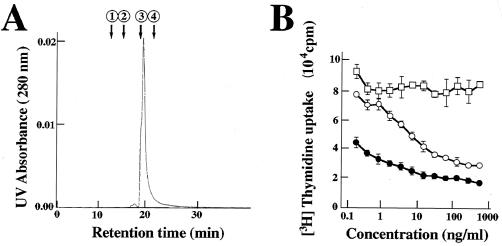
We made recombinant limitin protein from a stably limitin-producing Chinese hamster ovarian cancer cell line as previously described (38). When the purity of recombinant protein was evaluated by gel filtration chromatography, only one large peak whose molecular mass was ∼20 kDa was seen (Fig. 1A). The activity of recombinant limitin protein was also confirmed by the previously clarified limitin function (37). Limitin suppressed the proliferation of WEHI3 cells in a dose-dependent manner, and the activity of limitin was stronger than that of the fusion protein limitin-Ig at the same concentration (Fig. 1B). Therefore, our recombinant protein, whose purity is >98%, has higher specific activity than limitin-Ig. In this study, we used this lot of recombinant limitin protein to analyze the antiviral activity of limitin.
FIG. 1.
Characterization of recombinant limitin. (A) Gel filtration profile of limitin on a high-performance liquid chromatography Superdex 75 PC 3.2/30 column precalibrated with the standard protein markers. Recombinant limitin (1.5 μg) was applied, and the flow rate was 75 μl/min. The arrows indicate the elution peaks of the marker proteins (albumin, 67.0 kDa [1]; ovalbumin, 43.0 kDa [2]; chymotrypsinogen A, 25.0 kDa [3]; and RNase A, 13.7 kDa [4]). (B) WEHI3 cells (2.5 × 103) were seeded in each well and stimulated with the indicated dose of recombinant limitin (solid circles), limitin-Ig (open circles), and CD44-Ig (open squares) for 48 h. The results represent the means ± standard deviations of triplicate cultures. Similar results were obtained in three independent experiments.
Limitin has antiviral activity in vitro.
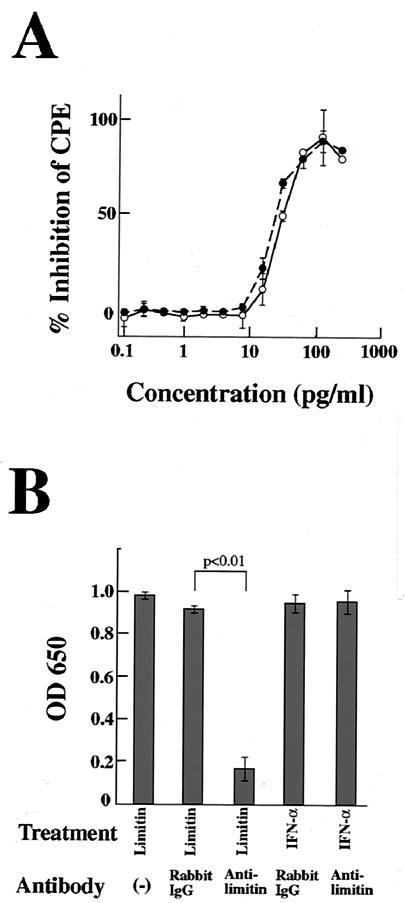
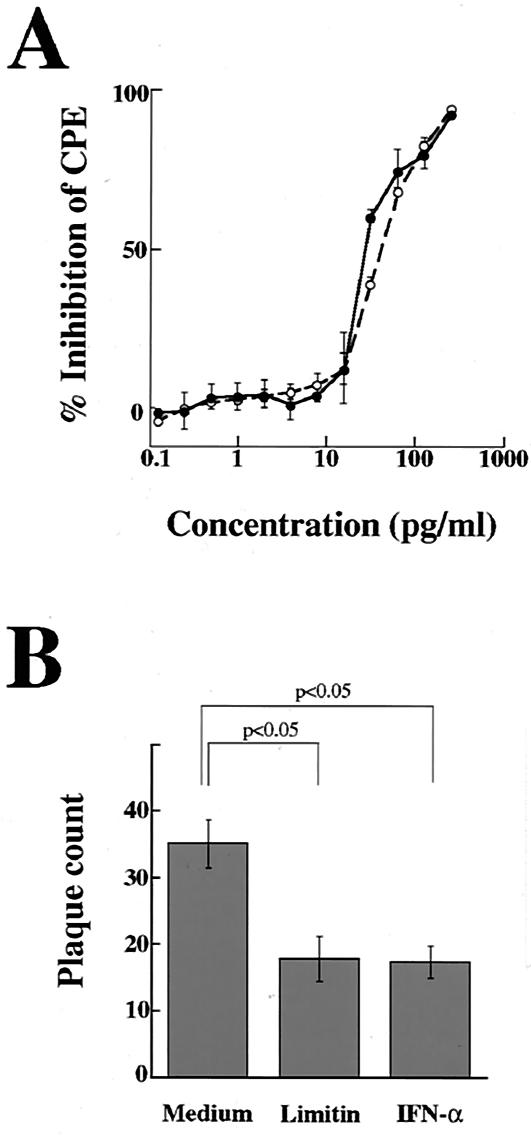
When L929 cells were infected with EMCV, CPE was detected within 24 h. As shown in Fig. 2A, CPE after EMCV infection was inhibited by limitin in a dose-dependent manner. The antiviral activity of limitin was evident at 15 pg/ml and maximal at 100 pg/ml, and 30 pg of limitin/ml provided 50% inhibition of CPE, which was similar to that of the control IFN-α. The inhibition of CPE by limitin was completely cancelled by the addition of anti-limitin polyclonal antibody (Fig. 2B). In contrast, the anti-limitin antibody could not cancel the inhibition of CPE by IFN-α. These facts indicate that the anti-limitin antibody is specific to limitin and that limitin truly induces an antiviral state in L929 cells. Although CPE of L929 cells was observed 72 h after HSV infection, limitin exhibited dose dependency for antiviral activity similar to that of IFN-α, and the dose of both limitin and IFN-α that provided 50% inhibition of CPE against HSV was also ∼30 pg/ml (Fig. 3A). Limitin, as well as IFN-α, also inhibited plaque formation by MHV in DBT cells (Fig. 3B). Therefore, limitin displays antiviral activity in vitro against three kinds of viruses: EMCV, HSV, and MHV. The concentration (30 pg/ml) of limitin and IFN-α that provided a 50% inhibition of CPE in L929 cells (with EMCV as the challenge virus) was defined as 1 experimental unit (EU)/ml (Fig. 2A) in the subsequent experiments (67).
FIG. 2.
Reduction of susceptibility of L929 cells to EMCV by limitin and IFN-α. (A) L929 cell monolayers on 96-well plates were pretreated with the indicated concentrations of limitin (solid circles) or IFN-α (open circles) overnight and challenged with EMCV (5 × 102 PFU/well). After 24 h of incubation, the cells protected from EMCV virulence were evaluated by the CPE dye uptake assay. The percent inhibition of CPE was calculated as 100 × [OD650 (each sample) − OD650 (virus control)/OD650 (cell control) − OD650 (virus control)]. The results represent the means ± standard deviations (SD) of triplicate cultures. Similar results were obtained in three independent experiments. (B) Limitin (1,000 pg/ml) or IFN-α (1,000 pg/ml) was incubated with the indicated antibodies (10 μg/ml) for 30 min on ice. L929 cells were cultured in the presence of the mixtures overnight and then challenged with EMCV (5 × 102 PFU/well). After 24 h of incubation, the cells protected from EMCV virulence were evaluated by the CPE dye uptake assay. The results represent the means ± SD of triplicate cultures. Similar results were obtained in three independent experiments.
FIG. 3.
Antiviral effects of limitin and IFN-α against HSV and MHV. (A) L929 cell monolayers on 96-well plates were pretreated with the indicated concentrations of limitin (solid circles) or IFN-α (open circles) overnight and challenged with HSV (50 PFU/well). After 72 h of incubation, the cells protected from EMCV virulence were evaluated by the CPE dye uptake assay. The results represent the means ± standard deviations (SD) of triplicate cultures. Similar results were obtained in three independent experiments. (B) DBT cells were seeded in a six-well dish, and then the subconfluent monolayer was treated with 30 EU of limitin/ml or 30 EU of IFN-α/ml for 24 h. The pretreated DBT cells were infected with MHV (103 PFU/well) for 1 h, followed by a plaque assay. The results represent the means ± SD of triplicate cultures. Similar results were obtained in three independent experiments.
In vivo antiviral activity of limitin.
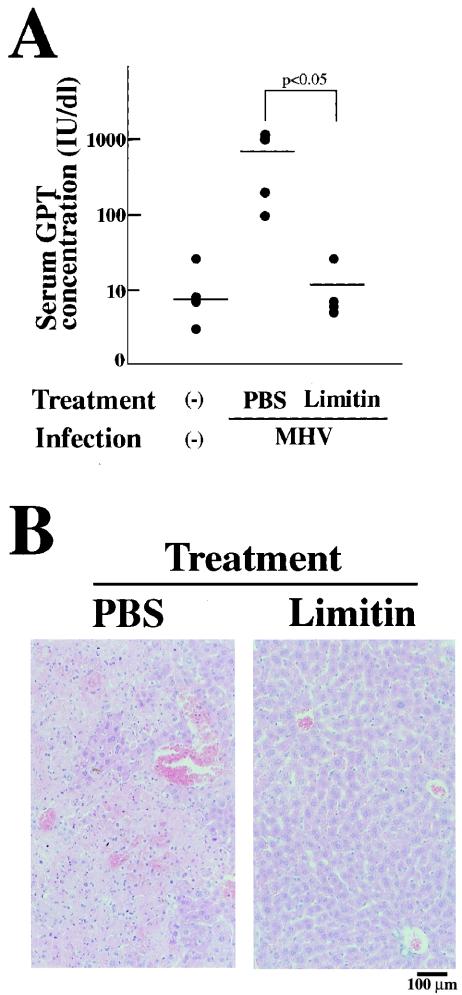
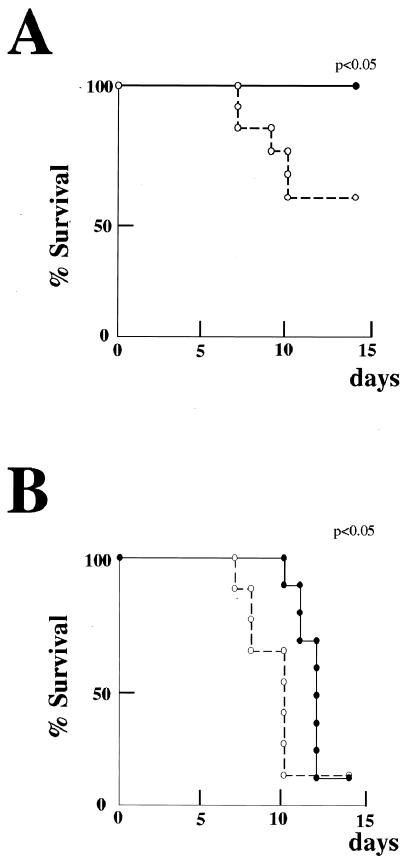
To investigate the antiviral activity of limitin in vivo, limitin was injected intraperitoneally every day into MHV-infected mice (31). The mice injected with PBS, but not those injected with limitin, started to die within 5 days after MHV infection (data not shown). We evaluated the GPT concentrations in serum with the GPT IFCC assay and analyzed hepatitis histologically 3 days after MHV infection. In mice injected with PBS, the GPT concentrations in serum were markedly elevated (Fig. 4A), and their liver sections revealed massive infiltration of lymphocytes, massive necrosis of hepatocytes, and intrahepatic hemorrhage (Fig. 4B). In mice injected with limitin, neither elevation of the GPT concentration in serum nor the hepatitis effects in liver sections were observed. When BALB/c mice were infected with HSV (43, 68), they started to die within 7 days after infection, and ∼50% of the mice in the PBS-treated group died during our observation period (Fig. 5A). In the limitin-treated group, all mice survived throughout the period. HSV was then inoculated into athymic nude mice (8, 35). Although limitin could not rescue the HSV-infected mice, administration of limitin apparently prolonged their survival compared with that with PBS injection (Fig. 5B). Therefore, limitin injection produces antiviral status against MHV, as well as HSV, in mice.
FIG. 4.
Repression of MHV infection in vivo by limitin. (A) Ten BALB/c mice were divided into two groups. One group was injected with 3 × 104 EU of limitin/day. The other group was treated with PBS as controls. The mice were treated intraperitoneally with PBS or limitin on days −1, 0, 1, and 2 relative to MHV exposure (103 PFU/mouse). After 72 h of virus inoculation, the mice were anesthetized with pentobarbital and bled to measure the activity of GPT in serum. Each dot represents the GPT concentration in the indicated mouse. Similar results were obtained in two independent experiments. (B) Two mice from each group were anatomized, and their livers were investigated histologically. Magnification, ×100.
FIG. 5.
Prolongation of survival of HSV-challenged mice by limitin. (A) Twenty-four BALB/c mice were separated into two groups and challenged with HSV (5 × 104 PFU/mouse). The mice were treated with 3 × 104 EU of limitin (solid circle) or PBS (open circles)/mouse on days −1, 0, 1, 2, 3, 4, and 5 relative to HSV exposure. The duration of observation for survival was 14 days. Similar results were obtained in two independent experiments. (B) Eighteen athymic nude mice were separated into two groups and challenged with HSV (104 PFU/mouse). The mice were treated with 3 × 104 EU of limitin (solid circles) or PBS (open circles)/mouse on days −1, 0, 1, 2, 3, 4, and 5 relative to HSV exposure. The duration of observation for survival was 14 days. Similar results were obtained in two independent experiments. Statistical analysis was performed by the Kaplan-Meier method, and the P value was calculated by the generalized Wilcoxon test.
Induction of antivirus-related genes.
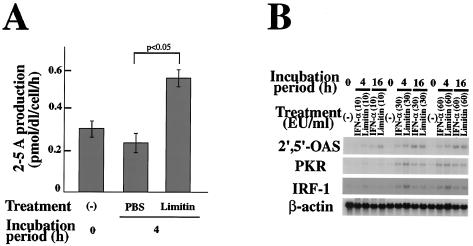
PKR activates eIF-2α, which inhibits mRNA translation (26, 30, 45, 72). 2′,5′-OAS activates RNase L, leading to RNA degradation, by producing 2-5A (5, 21, 53, 71). Mx1 is a GTPase that affects the intracellular transport of viral particles (3, 39). Whole-cell lysates of L929 cells stimulated with limitin contained approximately twice the 2′,5′-OAS enzyme activity of controls (Fig. 6A) (0.24 ± 0.05 pmol/dl/cell/h with PBS versus 0.55 ± 0.04 pmol/dl/cell/h with limitin at 4 h). In addition, the enhanced enzyme activity was also detected in the supernatants of limitin-stimulated L929 cells (data not shown). Message of 2′,5′-OAS was also induced after limitin stimulation, and it was enhanced till 16 h after the stimulation (Fig. 6B) (69). Although PKR mRNA consists of three transcripts of 6.0, 4.0, and 2.5 kbp (16), only 6-kbp mRNA was induced by limitin and IFN-α in L929 cells. Only weak message of Mx protein was induced within 8 h after limitin stimulation (data not shown), because L929 cells were derived from C3H/He mice, which belong to the phenotypically Mx− strain (54, 55). When L929 cells were stimulated with limitin, messages of 2′,5′-OAS and PKR were induced as strongly as with IFN-α (Fig. 6B). Therefore, limitin induces 2′,5′-OAS, PKR, and Mx proteins, which mainly mediate an antiviral state via IFNs (11, 56, 57, 58, 62).
FIG. 6.
Induction of antiviral molecules by limitin. (A) Whole-cell lysates of L929 cells stimulated with limitin (30 EU/ml) for 4 h were prepared and subjected to measurement of 2′,5′-OAS activity using a 125I radioimmunoassay kit. The results were represented as 2-5A production by each sample within 1 h. The assay range of this method is 10 to 810 pmol/dl. The results represent the means ± standard deviations of triplicate cultures. Similar results were obtained in three independent experiments. (B) Total RNAs of L929 cells stimulated with the indicated doses of limitin or IFN-α for 4 or 16 h were prepared and subjected to Northern blot analysis and probed with the indicated materials. β-Actin was used as an internal control. Similar results were obtained in three independent experiments.
Different requirements for IRF-1 by limitin and IFN-α for induction of antiviral activity.
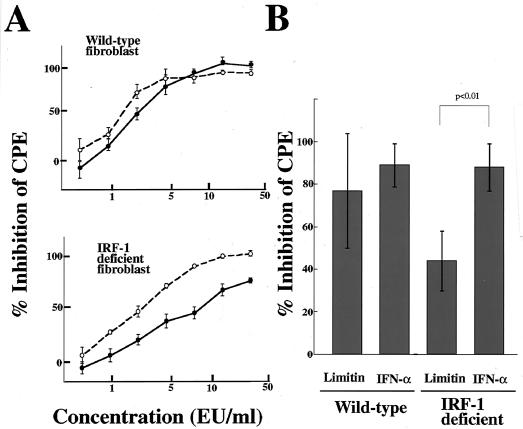
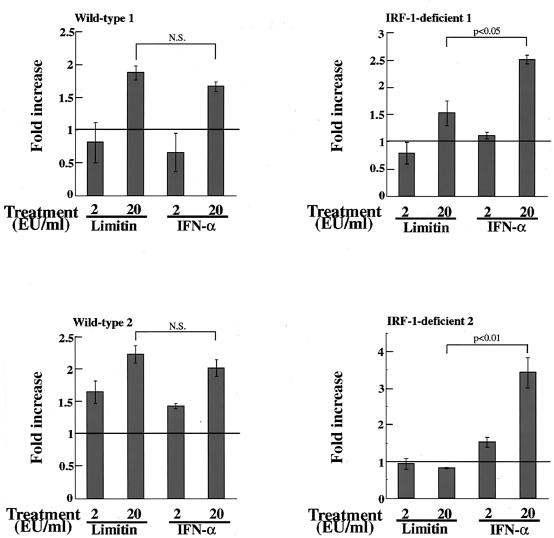
Activation of signal transducer and activator of transcription 1 (STAT1) is essential for the antiviral activity of IFNs (10). A heterotrimer composed of STAT1, STAT2, and IRF-9 binds to the interferon-stimulated response element (ISRE) (22, 27) that exists in the promoters of 2′,5′-OAS, PKR, and Mx1 genes (6, 15, 61). The STAT1 homodimer is another mediator, and it transduces IRF-1 (49), which recognizes consensus promoter sequences in part overlapping the ISRE (32). Our Northern blotting revealed that limitin enhanced IRF-1 gene expression within 2 h in L929 cells (data not shown), and the message was induced more strongly than by IFN-α 4 h after stimulation (Fig. 6B). To further examine the contribution of the IRF-1-dependent pathway, we performed a CPE dye uptake assay using IRF-1-deficient fibroblasts. As shown in Fig. 7A, the antiviral activity of limitin was impaired more severely than that of IFN-α in IRF-1-deficient fibroblasts (the doses that provided 50% inhibition of CPE were 3 EU/ml with limitin versus 3 EU/ml with IFN-α in wild-type fibroblasts and 7 to 8 EU/ml with limitin versus 3 to 4 EU/ml with IFN-α in IRF-1-deficient fibroblasts). The reduced antiviral activity of limitin was observed in fibroblasts established from six independent IRF-1-deficient mice (Fig. 7B). The relatively higher requirement for IRF-1 for the limitin-induced antiviral state was also confirmed by ISRE-luciferase reporter assays. IRF-1-deficient fibroblasts stimulated by limitin produced less luciferase protein from the ISRE sequence than those stimulated by IFN-α, while similar luciferase production was observed in wild-type fibroblasts (Fig. 8). Therefore, IRF-1 is more critical for the limitin-mediated antiviral state than for the IFN-α-mediated antiviral state.
FIG. 7.
Antiviral activities of limitin and IFN-α in fibroblasts established from wild-type and IRF-1-deficient mice. (A) Fibroblast monolayers on 96-well plates were pretreated with the indicated concentrations of limitin (solid circles) or IFN-α (open circles) overnight and challenged with EMCV (5 × 102 PFU/well). After 24 h of incubation, the cells protected from EMCV virulence were evaluated by the CPE dye uptake assay. The results represent the means ± standard deviations (SD) of triplicate cultures. Similar results were obtained in three independent experiments. (B) Fibroblast monolayers on 96-well plates were pretreated with limitin (10 EU/ml) or IFN-α (10 EU/ml) overnight and challenged with EMCV (5 × 102 PFU/well). After 24 h of incubation, the cells protected from EMCV virulence were evaluated by the CPE dye uptake assay. The results represent the means ± SD of the results obtained from six sets of fibroblasts independently established from wild-type and IRF-1-deficient mice.
FIG. 8.
ISRE-dependent luciferase activity after stimulation of limitin and IFN-α. The pCIS-ISRE plasmid was transfected into fibroblasts derived from wild-type or IRF-1-deficient mice. The transfectants were stimulated with the indicated concentrations of limitin or IFN-α for 6 h. The cells were lysed and subjected to measurement of luciferase activity. The data are expressed as the fold increase over the average for controls and represent the means ± standard deviations of triplicate assays. Similar results were obtained with four sets of fibroblasts independently established from wild-type and IRF-1-deficient mice. N.S., not significant.
DISCUSSION
A novel IFN-like cytokine, limitin, has in vitro and in vivo antiviral activity in part via induction of PKR, Mx protein, and 2′,5′-OAS, which inhibit viral replication and degrade viral components. Although limitin has antiviral activity as strong as that of IFN-α, IRF-1 dependencies for induction of an antiviral state were different in limitin and IFN-α. These findings support the notion that a human homolog of limitin may be applicable as a new therapeutic drug.
Mice lacking the IFN-α/β receptor are abnormally sensitive to certain kinds of viruses, suggesting a critical role for IFN-α/β cytokines in this type of immune defense (33). As described here, limitin has antiviral activity like those of IFN-α and IFN-β. Limitin inhibited not only CPE in EMCV- or HSV-infected L929 cells, but also plaque formation in MHV-infected DBT cells, indicating that limitin displayed antiviral activity against at least three types of viruses: EMCV (RNA nonenveloped), HSV-1 (DNA enveloped), and MHV-2 (RNA enveloped) (29, 46). We also showed that administration of limitin to mice suppressed MHV-induced hepatitis and HSV-induced death. These facts suggest that limitin may be applicable clinically as an antiviral drug. Interestingly, the antiviral effects of limitin were higher in BALB/c mice than in athymic nude mice. Thus, in addition to direct effects of antiviral activity, limitin seems to have indirect antiviral activity, such as induction of IFNs, enhancement of apoptosis of virus-infected cells, activation of natural killer cells, and induction of nitric oxide, which eliminate invading viruses in vivo. Viruses are intracellular pathogens, and IFNs are thought to constitute the first line of defense in host resistance to virus infections before immune mechanisms come into play (59). It is noteworthy that the antiviral effects of IFNs and limitin in vivo may be different. IFNs seem to inhibit the expansion and/or extension of virus infections, because IFN-α and IFN-β are induced by double-stranded nucleic acids that are frequently produced as a result of viral replication (50). Previous data from immunohistological staining showing that limitin is produced constitutively in healthy mice (38) may suggest that limitin mainly inhibits invasion of the body by viruses. This possibility may also be supported by the fact that the main limitin-producing cells are T lymphocytes and bronchial epithelial cells, a major entrance for viruses into the body.
IFNs can act directly at various stages of the virus infectious cycle, including penetration, transcription, translation, and budding from the cell surface (59). Some IFN-inducible proteins, such as PKR and 2′,5′-OAS, inhibit translation of viral mRNA, and 2′,5′-OAS also leads to RNA degradation (5, 21, 26, 30, 45, 53, 71, 72). Another IFN-inducible protein, Mx protein, inhibits virus replication (3, 39). PKR, Mx protein, and 2′,5′-OAS may participate in the limitin-induced antiviral state, because limitin can enhance these proteins in fibroblasts. The available information suggests that there are two major pathways for induction of gene expression of PKR, Mx protein, and 2′,5′-OAS. In general, ligation of IFN-α and IFN-β to the IFN-α/β receptor causes phosphorylation of two Janus kinases, Jak1 and Tyk2, and the activated Jak1 then phosphorylates STAT1 and STAT2 (49, 59). The phosphorylated Stat1-Stat2 complexes combine with IRF-9 and migrate to the nucleus, where they bind to DNA (the ISRE sequence) (22, 27) and initiate the transcription of PKR, Mx1, and 2′,5′-OAS (6, 15, 61). Another important pathway is mediated by the Stat1-Stat1 homodimer, which binds to IFN-γ-activated sequence, resulting in transcription of IRF-1 (59). IRF-1 recognizes consensus promoter sequences in part overlapping the ISRE (13, 34). In addition, at least nine IRF members have been identified (47), and some interact with each other, resulting in modification of their transcriptional activities. Although signals from the IFN-α/β receptor are complex, the important roles of Stat1, IRF-1, and IRF-9 in IFN-induced antiviral activity have been clarified by experiments using knockout mice (10, 23, 24). Our data clearly indicate that signals for induction of an antiviral state are different for limitin and IFN-α, while limitin has antiviral activity as strong as that of IFN-α. Transcriptional activity from ISRE sequence, as well as the antiviral activity of limitin, was impaired more severely than that of IFN-α in IRF-1-deficient fibroblasts. Thus, the IRF-1-dependent signaling pathway is more critical for the limitin-mediated antiviral state than for the IFN-α-mediated antiviral state. Although direct proof is lacking, the phosphorylation rates of Stat1 and Stat2 or the association rates of Stat1-Stat1 and Stat1-Stat2-IRF9 might be different for limitin and IFN-α. Alternatively, limitin might induce more or fewer signals that modify the activities of the IRF family than IFN-α.
This is the first report showing that signals are different for limitin and IFN-α. While limitin transduces signals via the IFN-α/β receptor (36), there are some precedents for the unique signals of limitin. Differences between IFN-α and IFN-β affinities for the IFN-α/β receptor have been described, as well as between subtypes of IFN-α. The β-R1 gene is induced by IFN-β but not by IFN-α (44). Association of a 95- to 100-kDa tyrosine-phosphorylated protein with the IFN-α/β receptor was found in an IFN-β- but not an IFN-α-treated myeloma cell line (1). On the other hand, recent advances in the structural and functional analysis of IFN-α/β cytokines allow us to discuss the interactions between IFNs and the IFN-α/β receptor. The three-dimensional crystal structures of murine IFN-α and IFN-β revealed that they have a globular structure with five α-helices (A to E) and four loops (51). Experiments using monoclonal antibodies against IFNs (4, 25) and site-directed mutagenesis (64) revealed the functional importance of the N-terminal half of the loop AB and the loop DE, together with the nearest segments of the helices D and E. The sequence identity between limitin and IFNs is still ∼60%, even in the functionally important portions. The diversity of signals among IFN-α/β cytokines, including limitin, might be explained by differences in their interactions with the IFN-α/β receptor or by differences in their sites that bind to the IFN-α/β receptor.
IFN-α and IFN-β are used for the treatment of patients with AIDS-related Kaposi's sarcoma, as well as chronic hepatitis B and C (19, 66). IFN-α-treated patients have a significantly higher rate of HBe antigen seroconversion and suppression of hepatitis B virus DNA. IFN-α decreases the hepatitis C virus load and reduces the risk for development of hepatocellular carcinoma (18). In spite of wide acceptance of IFNs as therapeutic drugs, they produce some adverse reactions. Some patients chronically treated with IFNs experience weight loss, bone marrow suppression, alopecia, thyroid dysfunction, and depression and other psychiatric disorders (28). These severe problems sometimes require dose reduction or discontinuation of treatment. Another clinical problem is that several virus-derived proteins enable viruses themselves to evade the antiviral activities of IFN-α and IFN-β. The conventional IFN monotherapy is effective in only a small fraction of patients with chronic hepatitis C. Furthermore, the hepatitis C virus titer often rebounds after cessation of IFN treatment. To increase the sustained response rate, increasing IFN dosage, prolonging the duration of IFN treatment, and/or combination therapy with other antiviral drugs, such as ribavirin, is often required, although adverse reactions to IFNs also increase. Limitin has some important and unique features described here and elsewhere. Limitin has fewer myelosuppressive properties than IFN-α, indicating that high-dose- and long-duration therapy with limitin may be done more safely than with IFN-α and IFN-β. Limitin used different signals for induction of antiviral activity than IFN-α and IFN-β, suggesting that limitin might be effective for IFN-α-resistant patients. A human homolog of limitin or an engineered cytokine with useful features of the limitin structure could be superior to IFN-α and IFN-β in clinical utility.
Acknowledgments
This work was supported in part by grants from the Japan Research Foundation for Clinical Pharmacology, the Foundation for Research on Leukemia, the Yamanouchi Foundation for Research on Metabolic Disorders, and the Mochida Memorial Foundation for Medical and Pharmaceutical Research, as well as the Ministry of Education, Science and Culture of Japan and the Japan Society for Promotion of Science.
REFERENCES
- 1.Abramovich, C., L. M. Shulman, E. Ratovitski, S. Harroch, M. Tovey, P. Eid, and M. Revel. 1994. Differential tyrosine phosphorylation of the IFNAR chain of the type I interferon receptor and of an associated surface protein in response to IFN-alpha and IFN-beta. EMBO J. 13:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. A. 1971. Semi-micro, dye-binding assay for rabbit interferon. Appl. Microbiol. 21:723-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnheiter, H., M. Frese, R. Kambadur, E. Meier, and O. Haller. 1996. Mx transgenic mice—animal models of health. Curr. Top. Microbiol. Immunol. 206:119-147. [DOI] [PubMed] [Google Scholar]
- 4.Cebrian, M., E. Yague, M. O. de Landazuri, M. Rodiguez-Moya, M. Fresno, N. Pezzi, S. Llamazares, and F. Sanchez-Madrid. 1987. Different functional sites on rIFN-alpha 2 and their relation to the cellular receptor binding site. J. Immunol. 138:484-490. [PubMed] [Google Scholar]
- 5.Clemens, M. J., and B. R. Williams. 1978. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell 13:565-572. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, B., D. Peretz, D. Vaiman, P. Benech, and J. Chebath. 1988. Enhancer-like interferon responsive sequences of the human and murine (2′-5′) oligoadenylate synthetase gene promoters. EMBO J. 7:1411-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creasey, A. A., J. C. Bartholomew, and T. C. Merigan. 1980. Role of G0-G1 arrest in the inhibition of tumor cell growth by interferon. Proc. Natl. Acad. Sci. USA 77:1471-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Clercq, E., Z. Zhen-Xi, J. Descamps, and K. Huygen. 1981. E-5-(2-bromovinyl)-2′-deoxyuridine vs. interferon in the systemic treatment of infection with herpes simplex virus of athymic nude mice. J. Infect. Dis. 143:846-852. [DOI] [PubMed] [Google Scholar]
- 9.De Maeyer, E., and J. De Maeyer-Guignard. 1998. Type I interferons. Int. Rev. Immunol. 17:53-73. [DOI] [PubMed] [Google Scholar]
- 10.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443-450. [DOI] [PubMed] [Google Scholar]
- 11.Floyd-Smith, G., Q. Wang, G. C. Sen, I. M. Kerr, and R. E. Brown. 1999. Transcriptional induction of the p69 isoform of 2′,5′-oligoadenylate synthetase by interferon-beta and interferon-gamma involves three regulatory elements and interferon-stimulated gene factor 3. Exp. Cell Res. 246:138-147. [DOI] [PubMed] [Google Scholar]
- 12.Frese, M., M. Prins, A. Ponten, R. W. Goldbach, O. Haller, and P. Zeltz. 2000. Constitutive expression of interferon-induced human MxA protein in transgenic tobacco plants does not confer resistance to a variety of RNA viruses. Transgenic Res. 9:429-438. [DOI] [PubMed] [Google Scholar]
- 13.Harada, H., T. Fujita, M. Miyamoto, Y. Kimura, M. Maruyama, A. Furia, T. Miyata, and T. Taniguchi. 1989. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58:729-739. [DOI] [PubMed] [Google Scholar]
- 14.Hirano, N., K. Fujiwara, and M. Matumoto. 1976. Mouse hepatitis virus (MHV-2). Plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20:219-225. [PubMed] [Google Scholar]
- 15.Hug, H., M. Costas, P. Staeheli, M. Aebi, and C. Weissmann. 1988. Organization of the murine Mx gene and characterization of its interferon- and virus-inducible promoter. Mol. Cell. Biol. 8:3065-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Icely, P. L., P. Gros, J. J. Bergeron, A. Devault, D. E. Afar, and J. C. Bell. 1991. TIK, a novel serine/threonine kinase, is recognized by antibodies directed against phosphotyrosine. J. Biol. Chem. 266:16073-16077. [PubMed] [Google Scholar]
- 17.Ichii, Y., R. Fukunaga, S. Shiojiri, and Y. Sokawa. 1986. Mouse 2-5A synthetase cDNA: nucleotide sequence and comparison to human 2-5A synthetase. Nucleic Acids Res. 14:10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai, Y., S. Kawata, S. Tamura, I. Yabuuchi, S. Noda, M. Inada, Y. Maeda, Y. Shirai, T. Fukuzaki, I. Kaji, H. Ishikawa, Y. Matsuda, M. Nishikawa, K. Seki, Y. Matsuzawa, et al. 1998. Relation of interferon therapy and hepatocellular carcinoma in patients with chronic hepatitis C. Ann. Intern. Med. 129:94-99. [DOI] [PubMed] [Google Scholar]
- 19.Itri, L. M. 1992. The interferons. Cancer 70:940-945. [PubMed] [Google Scholar]
- 20.Johnston, M. D., N. B. Finter, and P. A. Young. 1981. Dye uptake method for assay of interferon activity. Methods Enzymol. 78:394-399. [DOI] [PubMed] [Google Scholar]
- 21.Kerr, I. M., and R. E. Brown. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. USA 75:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kessler, D. S., S. A. Veals, X. Y. Fu, and D. E. Levy. 1990. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 4:1753-1765. [DOI] [PubMed] [Google Scholar]
- 23.Kimura, T., Y. Kadokawa, H. Harada, M. Matsumoto, M. Sato, Y. Kashiwazaki, M. Tarutani, R. S. Tan, T. Takasugi, T. Matsuyama, T. W. Mak, S. Noguchi, and T. Taniguchi. 1996. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells 1:115-124. [DOI] [PubMed] [Google Scholar]
- 24.Kimura, T., K. Nakayama, J. Penninger, M. Kitagawa, H. Harada, M. Toshifumi, N. Tanaka, R. Kamijo, J. Vilcek, T. W. Mak, and T. Taniguchi. 1994. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264:1921-1924. [DOI] [PubMed] [Google Scholar]
- 25.Kontsek, P., L. Borecky, E. Kontsekova, I. Macikova, A. Kolcunova, M. Novak, and V. Krchnak. 1991. Mapping of two immunodominant structures on human interferon alpha 2c and their role in binding to cells. Mol. Immunol. 28:1289-1297. [DOI] [PubMed] [Google Scholar]
- 26.Lebleu, B., G. C. Sen, S. Shaila, B. Cabrer, and P. Lengyel. 1976. Interferon, double-stranded RNA, and protein phosphorylation. Proc. Natl. Acad. Sci. USA 73:3107-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy, D. E., D. S. Kessler, R. Pine, and J. E. Darnell, Jr. 1989. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 3:1362-1371. [DOI] [PubMed] [Google Scholar]
- 28.Malik, U. R., D. F. Makower, and S. Wadler. 2001. Interferon-mediated fatigue. Cancer 92:1664-1668. [DOI] [PubMed] [Google Scholar]
- 29.Meager, A. 2002. Biological assay for interferons. J. Immunol. Methods 261:21-36. [DOI] [PubMed] [Google Scholar]
- 30.Meurs, E., K. Chong, J. Galabru, N. S. Thomas, I. M. Kerr, B. R. Williams, and A. G. Hovanessian. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379-390. [DOI] [PubMed] [Google Scholar]
- 31.Minnagawa, H., A. Takenaka, S. Mohri, and R. Mori. 1987. Protective effect of recombinant murine interferon beta against mouse hepatitis virus infection. Antivir. Res. 8:85-95. [DOI] [PubMed] [Google Scholar]
- 32.Miyamoto, M., T. Fujita, Y. Kimura, M. Maruyama, H. Harada, Y. Sudo, T. Miyata, and T. Taniguchi. 1988. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 54:903-913. [DOI] [PubMed] [Google Scholar]
- 33.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 34.Naf, D., S. E. Hardin, and C. Weissmann. 1991. Multimerization of AAGTGA and GAAAGT generates sequences that mediate virus inducibility by mimicking an interferon promoter element. Proc. Natl. Acad. Sci. USA 88:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagafuchi, S., H. Oda, R. Mori, and T. Taniguchi. 1979. Mechanism of acquired resistance to herpes simplex virus infection as studied in nude mice. J. Gen. Virol. 44:715-723. [DOI] [PubMed] [Google Scholar]
- 36.Oritani, K., P. W. Kincade, C. Zhang, Y. Tomiyama, and Y. Matsuzawa. 2001. Type I interferons and limitin: a comparison of structures, receptors, and functions. Cytokine Growth Factor Rev. 12:337-348. [DOI] [PubMed] [Google Scholar]
- 37.Oritani, K., K. L. Medina, Y. Tomiyama, J. Ishikawa, Y. Okajima, M. Ogawa, T. Yokota, K. Aoyama, I. Takahashi, P. W. Kincade, and Y. Matsuzawa. 2000. Limitin: an interferon-like cytokine that preferentially influences B-lymphocyte precursors. Nat. Med. 6:659-666. [DOI] [PubMed] [Google Scholar]
- 38.Oritani, K., S. Hirota, T. Nakagawa, I. Takahashi, S. Kawamoto, M. Yamada, N. Ishida, T. Kadoya, Y. Tomiyama, P. W. Kincade, and Y. Matsuzawa. 2003. T lymphocytes constitutively produce limitin, an interferon-like cytokine characterized as a heat- and acid-stable and heparin-binding glycoprotein. Blood 101:178-185. [DOI] [PubMed] [Google Scholar]
- 39.Pavlovic, J., and P. Staeheli. 1991. The antiviral potentials of Mx proteins. J. Interferon Res. 11:215-219. [DOI] [PubMed] [Google Scholar]
- 40.Pestka, S., J. A. Langer, K. C. Zoon, and C. E. Samuel. 1987. Interferons and their actions. Annu. Rev. Biochem. 56:727-777. [DOI] [PubMed] [Google Scholar]
- 41.Pestka, S. 1986. Interferon from 1981 to 1986. Methods Enzymol. 119:3-14. [DOI] [PubMed] [Google Scholar]
- 42.Pfeffer, L. M., C. A. Dinarello, R. B. Herberman, B. R. Williams, E. C. Borden, R. Bordens, M. R. Walter, T. L. Nagabhushan, P. P. Trotta, and S. Pestka. 1998. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 58:2489-2499. [PubMed] [Google Scholar]
- 43.Pinto, A. J., P. S. Morahan, M. Brinton, D. Stewart, and E. Gavin. 1990. Comparative therapeutic efficacy of recombinant interferons-α, -β, and -γ against alphatogavirus, bunyavirus, flavivirus, and herpesvirus infections. J. Interferon Res. 10:293-298. [DOI] [PubMed] [Google Scholar]
- 44.Rani, M. R., G. R. Foster, S. Leung, D. Leaman, G. R. Stark, and R. M. Ransohoff. 1996. Characterization of beta-R1, a gene that is selectively induced by interferon beta (IFN-beta) compared with IFN-alpha. J. Biol. Chem. 271:22878-22884. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, W. K., A. Hovanessian, R. E. Brown, M. J. Clemens, and I. M. Kerr. 1976. Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature 264:477-480. [DOI] [PubMed] [Google Scholar]
- 46.Sainz, B., Jr., and W. P. Halford. 2002. Alpha/beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J. Virol. 76:11541-11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato, M., T. Taniguchi, and N. Tanaka. 2001. The interferon system and interferon regulatory factor transcription factors—studies from gene knockout mice. Cytokine Growth Factor Rev. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 48.Sawai, H., H. Taira, K. Ishibashi, M. Itoh, H. Tanaka, and K. Shigenobu. 1988. Detection of 2′,5′-oligoadenylate synthetase activity in serum and in supernatant of cultivated lymphocytes: response to interferons or interferon inducers. Biomed. Res. 9:59-66. [Google Scholar]
- 49.Schindler, C., and J. E. Darnell, Jr. 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64:621-651. [DOI] [PubMed] [Google Scholar]
- 50.Sekellick, M. J., and P. I. Marcus. 1982. Interferon induction by viruses. VIII. Vesicular stomatitis virus: [+/−]DI-011 particles induce interferon in the absence of standard virions. Virology 117:280-285. [DOI] [PubMed] [Google Scholar]
- 51.Senda, T., T. Shimazu, S. Matsuda, G. Kawano, H. Shimizu, K. T. Nakamura, and Y. Mitsui. 1992. Three-dimensional crystal structure of recombinant murine interferon-beta. EMBO J. 11:3193-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shtrichman, R., and C. E. Samuel. 2001. The role of gamma interferon in antimicrobial immunity. Curr. Opin. Microbiol. 4:251-259. [DOI] [PubMed] [Google Scholar]
- 53.Slattery, E., N. Ghosh, H. Samanta, and P. Lengyel. 1979. Interferon, double-stranded RNA, and RNA degradation: activation of an endonuclease by (2′-5′)An. Proc. Natl. Acad. Sci. USA 76:4778-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staeheli, P., and J. G. Sutcliffe. 1988. Identification of a second interferon-regulated murine Mx gene. Mol. Cell. Biol. 8:4524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staeheli, P., R. Grob, E. Meier, J. G. Sutcliffe, and O. Haller. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8:4518-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staeheli, P., P. Danielson, O. Haller, and J. G. Sutcliffe. 1986. Transcriptional activation of the mouse Mx gene by type I interferon. Mol. Cell. Biol. 6:4770-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staeheli, P., O. Haller, W. Boll, J. Lindenmann, and C. Weissmann. 1986. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44:147-158. [DOI] [PubMed] [Google Scholar]
- 58.Staeheli, P., M. A. Horisberger, and O. Haller. 1984. Mx-dependent resistance to influenza viruses is induced by mouse interferons alpha and beta but not gamma. Virology 132:456-461. [DOI] [PubMed] [Google Scholar]
- 59.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi, I., H. Kosaka, K. Oritani, W. R. Heath, J. Ishikawa, Y. Okajima, M. Ogawa, S. Kawamoto, M. Yamada, H. Azukizawa, S. Itami, K. Yoshikawa, Y. Tomiyama, and Y. Matsuzawa. 2001. A new IFN-like cytokine, limitin, modulates the immune response without influencing thymocyte development. J. Immunol. 167:3156-3163. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka, H., and C. E. Samuel. 1994. Mechanism of interferon action: structure of the mouse PKR gene encoding the interferon-inducible RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 91:7995-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomis, D. C., J. P. Doohan, and C. E. Samuel. 1992. Mechanism of interferon action: cDNA structure, expression, and regulation of the interferon-induced, RNA-dependent P1/eIF-2 alpha protein kinase from human cells. Virology 188:33-46. [DOI] [PubMed] [Google Scholar]
- 63.Uno, K., T. Sato, Y. Takada, K. Fujioka, Y. Suginoshita, K. Kakimi, F. Moriyasu, and T. Kishida. 1998. A bioassay for serum interferon based on induction of 2′,5′-oligoadenylate synthetase activity. J. Interferon Cytokine Res. 18:1011-1018. [DOI] [PubMed] [Google Scholar]
- 64.Uze, G., G. Lutfalla, and K. E. Mogensen. 1995. Alpha and beta interferons and their receptor and their friends and relations. J. Interferon Cytokine Res. 15:3-26. [DOI] [PubMed] [Google Scholar]
- 65.Walter, M. R., R. Bordens, T. L. Nagabhushan, B. R. Williams, R. B. Herberman, C. A. Dinarello, E. C. Borden, P. P. Trotta, S. Pestka, and L. M. Pfeffer. 1998. Review of recent developments in the molecular characterization of recombinant alfa interferons on the 40th anniversary of the discovery of interferon. Cancer Biother. Radiopharm. 13:143-154. [DOI] [PubMed] [Google Scholar]
- 66.Weiss, K. 1998. Safety profile of interferon-alpha therapy. Semin. Oncol. 25:9-13. [PubMed] [Google Scholar]
- 67.Winkelman, G. L., R. M. Robert, A. J. Peterson, A. P. Alexenko, and A. D. Ealy. 1999. Identification of the expressed forms of ovine interferon-tau in the periimplantation conceptus: sequence relationships and comparative biological activities. Biol. Reprod. 61:1592-1600. [DOI] [PubMed] [Google Scholar]
- 68.Yamada, M., Y. Arao, A. Hatano, F. Uno, and S. Nii. 1988. Effect of recombinant mouse interferon-β on acute and latent herpes simplex infection in mice. Arch. Virol. 99:101-109. [DOI] [PubMed] [Google Scholar]
- 69.Yan, C., P. B. Sehgal, and I. Tamm. 1989. Signal transduction pathways in the induction of 2′,5′-oligoadenylate synthetase gene expression by interferon alpha/beta. Proc. Natl. Acad. Sci. USA 86:2243-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yousefi, S., M. R. Escobar, and C. W. Gouldin. 1985. A practical cytopathic effect/dye-uptake interferon assay for routine use in the clinical laboratory. Am. J. Clin. Pathol. 83:735-740. [DOI] [PubMed] [Google Scholar]
- 71.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]
- 72.Zilberstein, A., P. Federman, L. Shulman, and M. Revel. 1976. Specific phosphorylation in vitro of a protein associated with ribosomes of interferon-treated mouse L cells. FEBS Lett. 68:119-124. [DOI] [PubMed] [Google Scholar]