Abstract
Late (L) domains are required for the efficient release of several groups of enveloped viruses. Three amino acid motifs have been shown to provide L-domain function, namely, PPXY, PT/SAP, or YPDL. The retrovirus Mason-Pfizer monkey virus (MPMV) carries closely spaced PPPY and PSAP motifs. Mutation of the PPPY motif results in a complete loss of virus release. Here, we show that the PSAP motif acts as an additional L domain and promotes the efficient release of MPMV but requires an intact PPPY motif to perform its function. Examination of HeLaP4 cells expressing PSAP mutant virus by electron microscopy revealed mostly late budding structures and chains of viruses accumulating at the cell surface with little free virus. In the case of the PPPY mutant virus, budding appeared to be mostly arrested at an earlier stage before induction of membrane curvature. The cellular protein TSG101, which interacts with the human immunodeficiency virus type 1 (HIV-1) PTAP L domain, was packaged into MPMV in a PSAP-dependent manner. Since TSG101 is crucial for HIV-1 release, this result suggests that the Gag-TSG101 interaction is responsible for the virus release function of the MPMV PSAP motif. Nedd4, which has been shown to interact with viral PPPY motifs, was also detected in MPMV particles, albeit at much lower levels. Consistent with a role of VPS4A in the budding of both PPPY and PTAP motif-containing viruses, the overexpression of ATPase-defective GFP-VPS4A fusion proteins blocked both wild-type and PSAP mutant virus release.
Retrovirus assembly and release are driven by the viral Gag polyprotein, which contains all determinants necessary for the formation of virus-like particles (5, 33). Within Gag, an interaction domain mediates Gag-Gag association, while a membrane binding domain directs Gag to the plasma membrane. Based on the sequence of Gag assembly and membrane association, two major pathways of virus assembly can be distinguished: betaretroviruses, such as Mason-Pfizer monkey virus (MPMV) and mouse mammary tumor virus (MMTV), assemble immature procapsids in the cytoplasm (B/D-type assembly). These procapsids are subsequently transported to the plasma membrane. Other retroviruses, e.g., the lentivirus human immunodeficiency virus (HIV), form electron-dense assembly complexes after targeting Gag to the plasma membrane (C-type assembly). Irrespective of the mode of Gag assembly, however, virus bud formation is generally initiated at the plasma membrane. The efficient separation of the bud from the cell membrane has been shown to be an active process which requires so-called late (L) domains (6). During or after budding, the virus undergoes maturation, i.e., the viral protease (PR) cleaves Gag into the mature matrix (MA), capsid (CA), and nucleocapsid (NC) proteins as well as other products, depending on the virus. This process leads to the condensation of the viral core and is essential for virus infectivity.
To date, three different sequence motifs have been shown to provide L-domain function: PT/SAP, PPXY, and YPDL. While L domains have first been described in retroviral Gag proteins (9, 12, 21, 25, 36, 37), they are also present in the M and matrix proteins of rhabdoviruses and filoviruses, respectively (1, 10, 11, 13). Most retroviruses except lentiviruses contain a PPXY motif, in some cases together with a PT/SAP motif. In contrast, most lentiviruses harbor only the PT/SAP motif. The lentivirus equine infectious anemia virus is the only virus that has been shown to use the YPDL motif as an L domain (25). The L-domain sequence is generally very conserved, and even subtle mutations result in the retention of virus buds at the plasma membrane (see, e.g., reference 3). The budding structures arrested at the cell surface stay immature in morphology.
Host proteins have been identified that directly bind to the different L-domain motifs, and it is believed that the recruitment of these proteins to the site of virus budding ultimately results in the membrane fission event required for virus detachment. The exact set of factors needed, however, and the mechanism of their action are not understood. TSG101, which functions in the vacuolar protein sorting (Vps) pathway (14), binds to the PT/SAP motif of HIV-1 and HIV-2 (8, 18, 34). Similarly to mutations in the PT/SAP motif, depletion of TSG101 from virus-producing cells causes the accumulation of virus buds tethered to the cell surface (8). PPXY motifs are potential binding sites for WW domains (32), and the WW domain containing Nedd-4 family E3 ubiquitin ligases has been shown to bind to retroviral PPXY L domains (7, 15, 39). A function of ubiquitin ligases in retrovirus budding is supported by the observations that a small percentage of Gag proteins within purified HIV and murine leukemia virus (MLV) is monoubiquitinated (19, 20) and that large amounts of free ubiquitin are present inside retroviruses (19, 26). In addition, the intracellular depletion of free ubiquitin by treatment with proteasome inhibitors has been shown to block release of HIV-1, HIV-2, and Rous sarcoma virus (RSV) at a very late stage (22, 29, 30). In the case of RSV, this block could be partially overcome by fusing ubiquitin to the C terminus of Gag (22).
It is presently not known if the machinery needed to separate viral and cellular membranes is identical or at least overlapping for viruses containing different L domains. The observation that the overexpression of ATPase-defective VPS4A proteins blocked release not only of HIV-1, carrying a PTAP motif, but also of MLV (8), which functionally depends on a PPPY motif (41), suggested that these two pathways may utilize the same downstream effectors. Like TSG101, VPS4 is a component of the cellular Vps pathway, normally involved in formation of multivesicular bodies (MVB) (14). A potential role of Nedd-4-like ubiquitin ligases in the MVB pathway and whether the PPXY and PT/SAP motifs act in a redundant or synergistic way in those viruses where both are found are unclear. Different L-domain motifs have been shown to be at least partially exchangeable between retroviruses (17, 21, 40). Some viruses (e.g., HIV-1) carry only one known L-domain motif, while several retroviruses (e.g., MPMV and human T-cell leukemia virus type 1 [HTLV-1]) as well as Ebola virus and vesicular stomatitis virus contain closely spaced or overlapping PPPY and PSAP motifs. In the case of MPMV, PPPY and PSAP motifs are found in the pp24/16 domain between the MA and CA regions of Gag. Yasuda et al. showed that the PPPY motif is essential for virus release (38). Thus, the MPMV PSAP motif alone is not sufficient to mediate virus release. Here, we report that the MPMV PSAP motif acts as an additional L domain and promotes MPMV release in a PPPY motif-dependent manner. Viruses that contain a mutated PSAP motif were defective at a late stage of release, since late budding structures and chained procapsids accumulated at the cell surface. Consistent with a function of TSG101 in MPMV release, we found endogenous TSG101 to be enriched in purified MPMV in a PSAP-dependent manner.
MATERIALS AND METHODS
MPMV expression constructs.
The pp24/16 PPPY and PSAP motifs were mutated in the context of pSHRM15 (provided by E. Hunter [28]). All mutations were introduced by standard PCR mutagenesis, and amplified fragments were sequenced. A new MPMV expression plasmid (pMPMV) was constructed to achieve high levels of virus production after transient transfection. To this end, the MPMV proviral sequence (wild-type [wt] or mutant) was excised from the respective pSHRM15 plasmids and cloned into the EcoRI site of pcDNA3.1/Zeo(−) (Invitrogen). The region between the pcDNA3.1/Zeo(−) and MPMV transcription start sites was deleted by PCR. Detailed cloning procedures are available on request. The resulting constructs carrying the PPPY, PSAP, and double mutations were designated pMPMV/PY(−), pMPMV/PP(−), and pMPMV/2x(−), respectively. pSHRM15 derivatives were termed analogously.
Cell lines and transfections.
HeLaP4 and 293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum and antibiotics. For labeling experiments and analysis by electron microscopy (EM), HeLaP4 cells were transfected with FuGENE 6 (Roche) as instructed by the manufacturer. All other transfections were carried out by the calcium phosphate precipitation method.
Metabolic labeling.
For pulse-chase experiments, 4 × 106 HeLaP4 cells were transfected with 20 μg of the indicated plasmid. Twenty-four hours after transfection, cells were detached by using phosphate-buffered saline (PBS) containing 15 mM EDTA, washed, and starved for 20 min in methionine-free medium (ICN) containing 2% fetal calf serum. Then, cells were pulse-labeled for 30 min with [35S]methionine (2 mCi/ml; SJ-5050, Amersham). The pulse period was ended by adjusting the medium to 1 mM unlabeled methionine (Sigma), followed by removal of the labeling medium. Aliquots were chased in DMEM containing 0.4 mM methionine for the indicated period of time. For overnight labeling, 4 × 105 cells were transfected with 4 μg of plasmid. Thirty hours posttransfection, cells were rinsed with PBS and incubated with [35S]methionine (0.1 mCi/ml, diluted as above; SJ-5050, Amersham) for 12 h. Subsequent to pulse-chase or overnight labeling, cells were washed with PBS and lysed in RIPA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). Cell lysates were cleared by centrifugation (13,000 × g for 15 min) and preblocked by using a mixture of three preimmune sera and protein A Sepharose (Amersham) (1 h at 4°C). In the case of the pulse-chase experiments, virus-containing media were directly lysed by addition of 5× RIPA buffer. After overnight labeling, media were passaged through 450-nm-pore-size filters and virus was sedimented through sucrose cushions (20% [wt/vol] sucrose in PBS) by ultracentrifugation (90 min at 192,000 × g) and lysed in RIPA buffer. All samples were immunoprecipitated by using anti-MPMV CA antiserum. The immunoprecipitates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Gels were fixed in 30% methanol-10% acetic acid and dried. Labeled viral proteins were quantified by using a Bio-Rad Personal FX Phosphorimager and Bio-Rad Quantity One software.
Virus purification.
Virus was prepared from 293T cells transiently transfected with pMPMV or pMPMV/PP(−). Twelve hours after transfection, cells were rinsed with PBS, and the medium was replaced. Virus-containing medium was harvested 36 h after transfection and filtered through a 450-nm-pore-size filter. Virus was pelleted through sucrose cushions by ultracentrifugation (60 min at 53,000 × g), resuspended in 0.8 ml of PBS, and loaded onto a 6 to 18% Optiprep gradient as described previously (4). The visible virus band was isolated and diluted with PBS. Virus was recovered by ultracentrifugation (30 min at 164,000 × g). Purity of the preparation was monitored by silver staining and evaluated based on the loss of albumin. For external digestion of viral particles with trypsin, sucrose pellets were resuspended in 0.1 mg of trypsin/ml (ICN) in PBS and incubated at 30°C for 30 min. When virus was lysed with 0.1% Triton X-100 before trypsinization, a complete loss of the viral CA band was observed by Western blotting.
Western blotting and antibodies.
Western blotting was performed by using standard procedures. The following primary antibodies were used: rabbit anti-MPMV CA (provided by T. Ruml) (1:1,000); mouse anti-TSG101-4A10 from GeneTex (1:100); rabbit anti-Nedd-4 from Becton Dickinson Biosciences (1:5,000); rabbit anti-green fluorescent protein (GFP) that was raised against bacterially expressed GFP (1:1,000); and rabbit anti-14-3-3γ from Santa Cruz (1:1,000).
EM.
HeLaP4 cells (1 × 106) were transfected with 8 μg of wt or mutant pMPMV. At 48 h after transfection, cells were gently rinsed with PBS and fixed with ice cold 2.5% glutaraldehyde in 0.05 M sodium cacodylate (pH 7.2) for 20 min on ice. Fixed cells were scraped from the plates, collected by low-speed centrifugation (200 × g for 10 min at 4°C), successively stained with 2% osmium tretroxide and 0.5% uranyl acetate, and processed for ultrathin sectioning. Micrographs were taken with a Zeiss EM-10 electron microscope at 80 kV. The magnification indicator was routinely controlled by the use of a grating replica.
RESULTS
The PPPY and PSAP motifs both contribute to MPMV release.
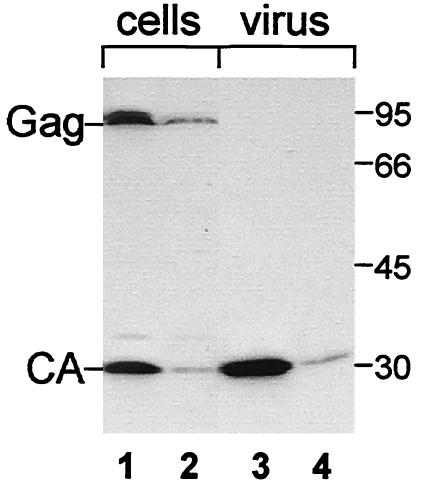
To facilitate the biochemical analysis of MPMV release, a novel MPMV expression plasmid was constructed. To this end, the MPMV transcription start site was placed under the control of a CMV promoter such that the resulting transcripts are identical to those derived from the LTR promoter. The new plasmid (pMPMV) and the parental construct (pSHRM15) were transfected into HeLaP4 cells, and cell extracts and virus released into the growth medium were analyzed by Western blotting 24 h after transfection. The same pattern of Gag and CA proteins was detected in both cases (Fig. 1). Transfection of pMPMV, however, reproducibly yielded significantly higher levels of Gag protein expression and virus release than transfection of pSHRM15 (Fig. 1, compare lanes 1 and 3 to lanes 2 and 4). In agreement with previously published results, intracellular processing of Gag was observed for both constructs (28, 35). The extent of intracellular processing did not correlate with the relative level of Gag expression.
FIG. 1.
Comparison of MPMV expression from pMPMV and pSHRM15. HeLaP4 cells were transfected with equal amounts of pMPMV (lanes 1 and 3) or pSHRM15 (lanes 2 and 4), and levels of intracellular (lanes 1 and 2) and virus-associated CA-reactive proteins (lanes 3 and 4) were analyzed by Western blotting with anti-CA antiserum. Molecular mass standards in kilodaltons are shown on the right; MPMV Gag and CA are identified on the left.
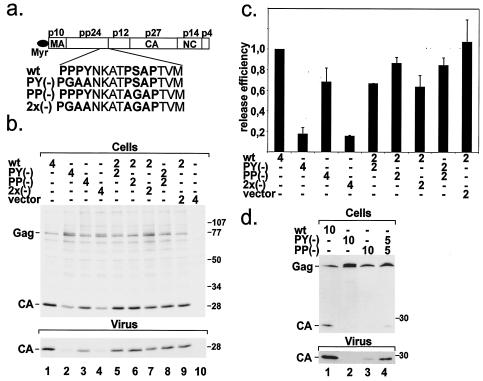
Within its pp24/16 domain, MPMV Gag carries a PSAP motif four amino acids downstream of a PPPY motif (Fig. 2a). To evaluate the relative contribution of the two motifs to MPMV release, we individually mutated the PPPY motif to PGAA and mutated the PSAP motif to AGAP. The mutations were introduced into plasmids pMPMV and pSHRM15. In addition, we constructed plasmids containing both mutations (Fig. 2a). The amino acid substitutions were chosen based on published data (12, 17, 34) and should result in complete inactivation of the respective motif. To determine the efficiency of wt and mutant virus release, HeLaP4 cells were transfected with the indicated plasmids and, 36 h after transfection, metabolically labeled with [35S]methionine for 12 h. After the labeling period, cells were lysed. Virus was collected by centrifugation through sucrose cushions. Cell extracts and virus samples were subjected to immunoprecipitation with an antiserum against the MPMV CA protein followed by SDS-PAGE and phosphorimage analysis. The result of a representative experiment is shown in Fig. 2b. Transfection of wt pMPMV yielded the typical pattern of cell-associated Gag and CA proteins and caused the release of virus into the growth medium (Fig. 2b, lane 1). It should be noted that all virus samples contained only completely processed CA and no precursor or intermediate processing products, independent of the processing defect observed in transfected cells (data not shown). Mutation of the PPPY motif resulted in a pronounced impairment of intracellular Gag processing and a defect in particle release (Fig. 2b, lane 2). Gag and a protein with slightly lower mobility than Gag (see below) accumulated in PY(−)-transfected cells, while the amount of processed CA protein was reduced. Analysis of the virus fraction (Fig. 2b, lower panel) revealed that the PPPY mutant displayed a strong defect in virus release. This result is in agreement with the report by Yasuda et al., who also observed a complete loss of virus release as well as a defect in Gag processing for a PPPY mutant (38). Analysis of PSAP mutant-transfected cells showed a slight impairment of processing and a significant reduction in virus release (Fig. 2b, lane 3). Thus, the PSAP motif cannot substitute for the PPPY motif, but it acts as an additional L domain and contributes to the efficient release of MPMV. The phenotype of PSAP inactivation was less pronounced than that of the PPPY mutation. Consistent with the absolute dependence of MPMV release on the intact PPPY motif, the PPPY and PSAP motif double mutant behaved like the PPPY mutant (Fig. 2b, lane 4).
FIG. 2.
(a) Schematic representation of MPMV Gag, showing the L-domain sequence within pp24 and the mutations introduced. (b) wt and L-domain mutant MPMV release. Cells were transfected with the indicated pMPMV constructs and metabolically labeled with [35S]methionine for 12 h. Cell-associated (upper panel) and virus-associated (lower panel) CA protein was immunoprecipitated with anti-CA antiserum, resolved by SDS-PAGE, and subjected to phosphorimage analysis. Note that no precursor or intermediate processing product was observed in the virus samples. (c) Quantification of wt and mutant release efficiencies. CA-containing bands were quantified. The release efficiency of wt and mutant virus as well as that of mixed virus populations was calculated by dividing the signal from virus-associated CA protein by the sum of the signals from cell- and virus-associated CA-containing proteins. The obtained numbers were normalized for wt release efficiency. n = 4 for columns 1, 2, 3, and 8; n = 2 for all other columns. (d) Steady-state levels of wt and mutant virus release. Cells were cotransfected with the indicated pSHRM15 plasmids, and cell- and virus-associated viral proteins (upper and lower panel, respectively) were analyzed by Western blotting with an anti-CA antiserum. Again, no precursor or intermediate processing product was observed in the virus samples.
It has been suggested that the magnitude of the defect in virus release caused by L-domain mutations may correlate with the level of Gag expression (31). The described experiments were performed with a CMV-driven high-level expression plasmid transfected by lipofection. To exclude the possibility that the intermediate phenotype of the PSAP mutant was a result of high levels of expression, we repeated the experiment by using transfection of pSHRM15 into HeLaP4 cells by calcium phosphate precipitation (Fig. 2d, lanes 1 to 3). As observed for transfection of the pMPMV plasmid, the PPPY mutant Gag protein remained largely unprocessed and exhibited a block to virus release (Fig. 2d, lane 2). Intracellular processing of Gag and virus release were also significantly reduced in the case of the PSAP mutant but were less impaired than for the PPPY mutant (Fig. 2d, compare lanes 1 to 3). Accordingly, the intermediate phenotype of the PSAP mutant is not due to overexpression.
To quantify the relative efficiencies of wt and mutant virus release from pMPMV-transfected cells, the virus-associated CA signal was related to the sum of the signals of the intracellular and virus-associated CA-containing proteins. Only the three major CA-containing bands (namely, Gag, the protein migrating slightly above Gag, and processed CA) detected in cell extracts were quantified. The obtained numbers were normalized for the methionine content of the respective proteins. The protein migrating above Gag was assumed to carry the same number of methionines as Gag. These quantifications indicated that virus release is reduced by approximately fivefold in the case of the PPPY and double mutants, while PSAP inactivation resulted in a reduction of virus release by roughly 30% (Fig. 2c).
Rescue of L-domain mutant Gag proteins.
It has been reported that RSV L-domain mutant Gag protein can be rescued into virus-like particles by coexpression of Gag protein carrying an intact L domain (36). These results suggest that only a minor fraction of Gag molecules of a budding virion needs to carry an L domain. To test the release efficiency of mixed populations of MPMV Gag proteins, we cotransfected equal amounts of mutant and wt plasmids and analyzed and quantified CA-reactive proteins in cell and virus extracts 24 h post transfection (Fig. 2b, lanes 5 to 7 and Fig. 2c). Cotransfection of the wt plasmid with either PPPY or double mutant pMPMV resulted in a 40% reduction of virus release compared to wt virus (Fig. 2b, compare lanes 1, 5, and 7; Fig. 2c), indicating that only a partial rescue occurred. A similar analysis for the PSAP motif mutant was not conclusive, because differences in virus release were too small (Fig. 2b, compare lanes 1 and 6; Fig. 2c).
The experiments shown in the previous section suggested that both motifs contribute to MPMV release. To test whether the PPPY and PSAP motifs can complement each other in trans or need to be present on the same polyprotein, we performed cotransfection experiments. Transfecting equal amounts of pMPMV plasmids carrying either the PPPY or the PSAP mutation yielded higher levels of virus release than transfection of the same amount of either variant alone (Fig. 2b compare lanes 2, 3 and 8; Fig. 2c). A partial rescue of virus release was also observed by Western blot analysis of virus produced from HeLaP4 cells cotransfected with PPPY and PSAP mutant pSHRM15 (Fig. 2d). Thus, both motifs can complement each other in trans under low- and high-expression conditions.
Gag lacking the PPPY motif trans-dominant-negatively inhibits wt virus release.
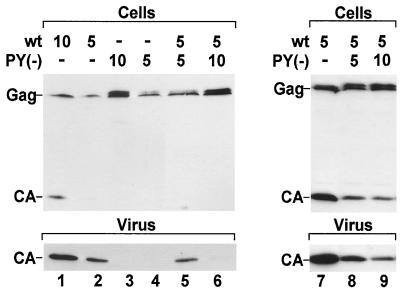
Since virus release is not completely rescued by expressing equal amounts of PPPY mutant and wt Gag polyproteins (Fig. 2b and c), we determined whether this mutant has a dominant-negative influence on wt virus release. wt and PPPY mutant pSHRM15 derivatives were cotransfected at a ratio of 1:1 and 1:2. In all cases, the total amount of transfected DNA was adjusted to 15 μg by using empty vector. As noted above (Fig. 2), cotransfection of equal amounts of PPPY mutant and wt proviral plasmids (5 μg each) did not yield more virus production than transfection of 5 μg of wt plasmid alone (Fig. 3, compare lanes 2 and 5). When wt and mutant plasmids were cotransfected at a ratio of 1:2, virus release was reduced to nearly undetectable levels, even though higher levels of Gag protein were observed in cell extracts. Cotransfection of wt and PPPY mutant plasmids also did not restore intracellular Gag processing, since only Gag and the protein slightly above Gag were detected in cell lysates (Fig. 3, lanes 5 and 6). The dominant-negative effect of the PPPY mutant Gag protein was also evident when high levels of expression were used (Fig. 3, lanes 7 to 9) and is therefore independent of the Gag expression level. In addition, we confirmed our finding by quantitative Western blotting (data not shown). Taken together, these results suggest that a molar excess of PPPY mutant Gag polyproteins over wt Gag has a dominant-negative influence on virus production. We also tested whether cotransfection of the MPMV PSAP mutant or of a HIV-1 L-domain mutant with the respective wt plasmid inhibited virus release. In both cases, no dominant-negative effect on virus release was observed (data not shown).
FIG. 3.
Effect of PPPY mutant Gag protein on wt virus release. HeLaP4 cells were cotransfected with the indicated pSHRM15 (lanes 1 to 6) or pMPMV (lanes 7 to 9) wt and PY(−) plasmids. In each case, the amount of total DNA was adjusted to 15 μg. Steady-state levels of cell- and virus-associated CA protein (upper and lower panel, respectively) were analyzed by Western blotting with anti-CA antiserum.
Analysis of wt and mutant Gag processing kinetics.
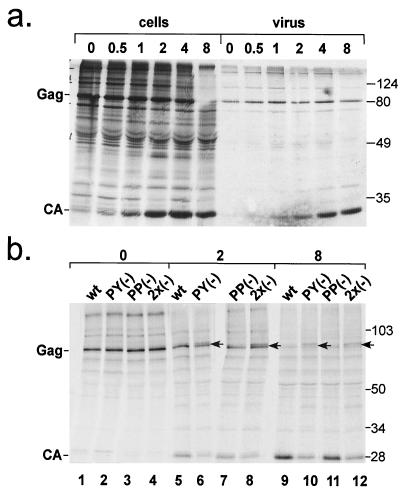
As shown above, intracellular processing of the PPPY and double-mutant Gag polyproteins was significantly impaired (Fig. 2). To analyze the processing kinetics of wt and mutant Gag proteins and to determine whether the protein migrating slightly above Gag represents a posttranslational modification of Gag, we performed pulse-chase experiments. For this, we transfected HeLaP4 cells with wt or mutant pMPMV constructs. Twenty-four hours after transfection, cells were pulse-labeled with [35S]methionine for 30 min and chased for the times indicated. Cell and virus extracts were immunoprecipitated with anti-CA antibodies. wt Gag processing and release kinetics are shown in Fig. 4a. Directly after the pulse, the Gag polyprotein was the predominant band detected (lane 1), and processing to CA became clearly detectable after a 1-h chase (lane 3). Most of the intracellular Gag was processed to CA after 8 h. Release of virus particles containing processed CA was detected after 1 h and increased over the entire chase period.
FIG. 4.
Kinetic analysis of wt and mutant Gag processing. (a) Kinetics of wt Gag processing and virus release. HeLaP4 cells were transfected with pMPMV and pulse-labeled with [35S]methionine for 30 min. Cells were chased in DMEM for the indicated time. Cell- and virus-associated CA-containing proteins (lanes 1 to 6 and 7 to 12, respectively) were immunoprecipitated with anti-CA antiserum, resolved by SDS-PAGE, and visualized by phosphorimage analysis. (b) wt and L-domain mutant processing kinetics. HeLaP4 cells were transfected with the indicated pMPMV constructs, labeled as described above, and chased for 0, 2, or 8 h. At the 2-h time point, a Gag-related band (arrow) that migrates slower than Gag was detected in cells expressing PPPY mutant and double mutant Gag.
A comparison of wt and mutant Gag processing kinetics is shown in Fig. 4b. Similar amounts of Gag polyproteins were observed in all cases after the pulse period, and the protein migrating above Gag was not detected in these samples (Fig. 4b, lanes 1 to 4). Processing kinetics of the PSAP variant were largely similar to the wt polyprotein (Fig. 4b, compare lanes 5 and 7 and lanes 9 and 11). A significant processing defect was observed for the PY(−) variant and for the double mutant, with both exhibiting reduced levels of CA after 2-h and 8-h chase periods (lanes 6 and 8 and lanes 10 and 12). Furthermore, the protein migrating slightly slower than Gag (marked by an arrow in lanes 6, 8, 10, and 12) became detectable in cells transfected with the PPPY or double mutant after a chase period of 2 h and remained present after 8 h. Therefore, this product corresponds to a posttranslationally modified version of Gag. We hypothesize that it represents Gag lacking the proline-rich C-terminal p4 domain (Fig. 2a). Cleavage of p4 by the viral PR may result in altered migration of the protein in SDS-PAGE. CA-containing cleavage products migrating slower than their precursors have been described for other retroviruses (23). Alternatively, the product migrating just above Gag could result from a posttranslational modification of Gag other than processing.
EM analysis of virus assembly and release.
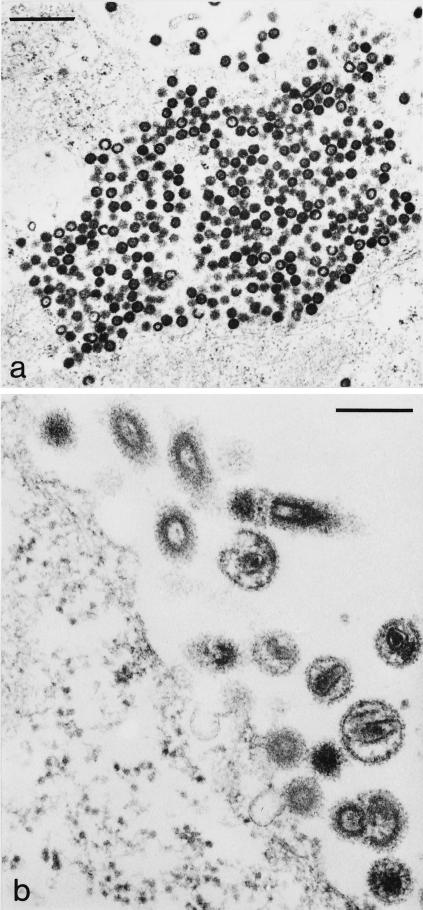
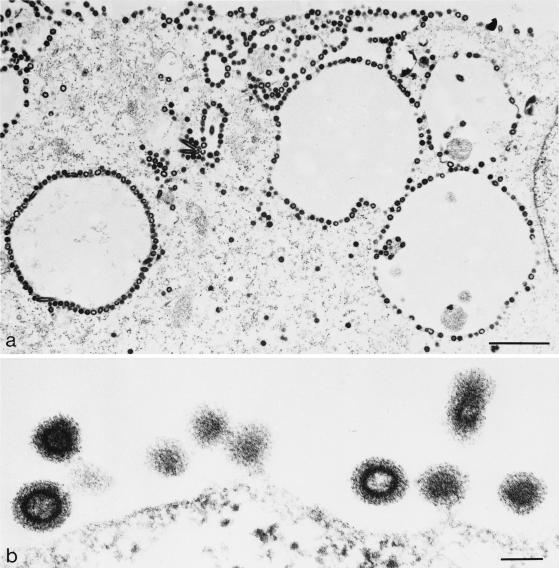
Deletion of either the PPPY or the PSAP motif impaired MPMV release, but the effect of the PSAP mutation was much less pronounced, and virus production was still observed in this case (Fig. 2). Analysis of particle formation by EM is a more direct way to determine virus release at the morphological level. We therefore performed thin-section EM analysis of HeLaP4 cells transfected with wt, PPPY mutant, PSAP mutant, or double-mutant pMPMV. In all cases, the typical sites of intracellular assembly of immature procapsids were observed (shown representatively for cells transfected with wt pMPMV in Fig. 5a). Furthermore, thin sections of cells transfected with the wt plasmid contained many procapsids budding from the plasma membrane as well as numerous mature and immature virions (Fig. 5b). In contrast, no mature virions were detected in thin sections of cells transfected with the PPPY mutant MPMV (Fig. 6). Instead, many immature procapsids accumulated underneath the plasma membrane and appeared to be arrested at an early stage of bud formation (Fig. 6a). Typical late budding structures connected to the plasma membrane by a thin membrane stalk as well as chained budding structures containing more than one procapsid were also present, indicating that the early budding arrest was overcome by some procapsids (Fig. 6b). Interestingly, in several cells, procapsids were found to decorate intracellular vesicles (Fig. 6a). Again, the majority of these procapsids failed to initiate bud formation, and no bulging of the membrane was observed.
FIG. 5.
Thin-section EM analysis of HeLaP4 cells transfected with pMPMV (wt). Bars, 500 nm (a) and 200 nm (b).
FIG. 6.
Thin-section EM analysis of HeLaP4 cells transfected with PPPY mutant pMPMV. Bars, 1 μm (a) and 100 nm (b).
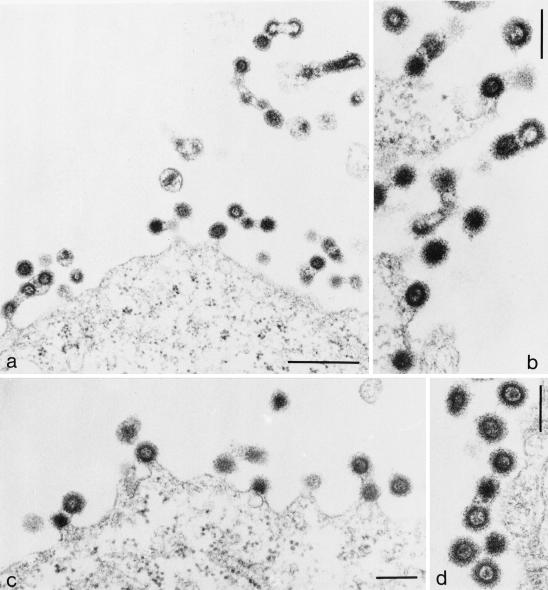
Thin-section EM analysis of cells transfected with PSAP mutant pMPMV revealed a different phenotype. In this case, we observed predominantly late immature budding structures, many of which were tethered to each other (Fig. 7a to d). Some of these budding chains containing multiple procapsids were apparently released from the cell (Fig. 7a, upper right). Less extracellular particles were observed than in the case of wt pMPMV, but some mature virions containing a condensed core were detected (e.g., Fig. 7a, middle). For a more quantitative determination of the late defect of the PSAP variant, the relative abundance of different budding structures and free mature and immature particles was counted for wt and PSAP mutant pMPMV-transfected cells (Table 1). This analysis confirmed that much less mature virus was observed in the case of the PSAP variant, and the amount of total cell-free virus detected accounted for only 16% of all procapsids, compared to 60% for the wt constructs. On the other hand, the relative abundance of budding structures and virus chains was increased in the case of the PSAP variant. Only 7% of wt procapsids were found in virus chains attached to the plasma membrane, while 26% of PSAP mutant procapsids were found in such chains. Nearly one-half of the chains containing multiple procapsids were apparently released from the cell, i.e., no connection to the plasma membrane was visible. The relative abundance of immature procapsids was also significantly higher for the PSAP variant, indicating that maturation is impaired in this case. Taken together, these results showed that the PSAP mutant is indeed defective at the stage of virus release. Interestingly, the budding arrest in the case of the PSAP variant appeared to be at a later stage than in the PPPY variant, with more advanced budding structures extending further away from the plane of the plasma membrane. As in the biochemical experiments described above, the double mutant was indistinguishable from the PPPY mutant at the morphological level as well.
FIG. 7.
Thin-section EM analysis of HeLaP4 cells transfected with PSAP mutant pMPMV. Bars, 500 nm (a) and 200 nm (b to d).
TABLE 1.
Relative abundance of wt and PSAP mutant budding structuresa
| Procapsid type | Relative abundance (%) of the indicated structure containing mature capsids
|
Relative abundance (%) of the indicated structure containing immature procapsids
|
||||||
|---|---|---|---|---|---|---|---|---|
| Free virus | Single buds | Chain buds | Free chains | Free virus | Single buds | Chain buds | Free chains | |
| wt | 37.7 | 0.8 | 1.1 | 0 | 22.6 | 31.5 | 6.3 | 0 |
| PSAP(−) | 7.5 | 0.4 | 0 | 0 | 8.6 | 57.6 | 14.9 | 11 |
Quantification of wt and PSAP mutant budding structures as detected by EM. HeLa P4 cells were transfected with the respective pMPMV plasmids, and 523 wt and 491 PSAP mutant procapsids or virus cores at or beyond the stage of budding were counted. Immature procapsids and mature cores were counted separately. Budding structures were classified as either free virus, buds containing one procapsid, buds containing more than one procapsid in a chain, or procapsid chains that were apparently released. In the case of chains, the number of procapsids present in all chains was counted rather than the number of chains.
TSG101 and Nedd-4 are incorporated into MPMV particles.
The interaction of the HIV-1 PT/SAP motif with TSG101 has been shown to be essential for HIV-1 release (8), and the binding of Nedd-4-like proteins to the PPPY motif is presumed to be necessary for the function of this late motif. Accordingly, the cellular binding partners should be recruited to the budding site and may be incorporated into virions. The overexpression of a segment of TSG101 by transfection led to incorporation of this protein into HIV-1 particles (2). To test whether endogenous cellular TSG101 or Nedd-4 are incorporated into MPMV virions, we analyzed highly purified particle preparations.
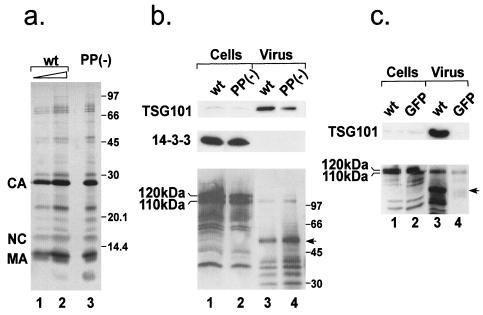
To this end, 293T cells were transfected with either wt or a PSAP mutant pMPMV. Media were gathered 36 h after transfection and filtered, and virus was centrifuged through cushions of 20% sucrose. Virus samples were further purified by velocity gradient centrifugation as described previously (4). The visible virus band was collected and diluted, and virus was recovered by ultracentrifugation. Purity of samples was evaluated by silver staining. Judging from the silver-stained gel, the general protein content and the degree of processing were very similar for wt and PSAP mutant virus (Fig. 8a). The viral Gag-derived proteins represented the major constituents of the virion. To examine the presence of TSG101 in purified MPMV, similar amounts of wt and PSAP mutant virus (the same ratio used in Fig. 8a, lanes 1 and 3) were probed for incorporation of endogenous TSG101 by Western blotting. TSG101 was readily detected in wt virus preparations, while the amount of TSG101 was significantly reduced in the PSAP mutant virus preparation (Fig. 8b, lanes 3 and 4), and no Tsg101 was detected in parallel preparations from the medium of mock-transfected cells (Fig. 8c, lanes 3 and 4). The data in Fig. 8b slightly exaggerate the amount of TSG101 present in PSAP mutant virus preparations, because more PSAP mutant virus than wt virus was loaded (compare Fig. 8a, lanes 1 and 3 or Fig. 8c, lanes 3 and 4). Quite clearly, however, TSG101 was present in PSAP mutant virus preparations, albeit at a reduced level. To determine whether other cytosolic proteins were nonspecifically incorporated into MPMV particles, we probed for the protein 14-3-3γ, which localizes mostly to the cytosol (data not shown). While the 14-3-3 protein was highly abundant in cells, it was completely absent from virus preparations (Fig. 8b). Thus, TSG101 is clearly enriched in particle preparations compared to cell extracts, while other cytosolic proteins are largely excluded. The incorporation of TSG101 into MPMV was confirmed by analysis of virions that had been treated with trypsin, which degrades proteins outside of the virion, but did not reduce the amount of TSG101 in the preparation (data not shown).
FIG. 8.
Incorporation of cellular proteins into MPMV. (a) wt and PSAP mutant virus was purified by centrifugation through sucrose cushions followed by sedimentation in an Optiprep velocity gradient. wt (lanes 1 and 2) and PSAP mutant (lane 3) virus protein content was analyzed by silver staining. (b) Comparable amounts of lysates from wt and mutant virus-expressing cells (lanes 1 and 2) as well as highly purified virus (the same amounts as in panel a, lanes 1 and 3) were probed for the cellular proteins TSG101 (upper panel), 14-3-3γ (middle panel), and Nedd-4 (lower panel) by Western blotting. In the lower panel, the major Nedd-4-related bands migrating at 120 and 110 kDa are indicated. The arrow marks a band reacting with Nedd-4 antiserum which does not comigrate with one of the major viral proteins detected in the Ponceau S stain (not shown). (c) To analyze whether TSG101 and Nedd-4 are released from 293T cells in the absence of MPMV expression, we probed sucrose pellets from culture media of cells transfected with pMPMV or EGFP expression vectors (lanes 3 and 4) for TSG101 (upper panel) and Nedd-4 (lower panel). The arrow marks the same product as in panel b.
The PPPY motif binding protein Nedd-4 was readily detected in cell lysates (Fig. 8b, lanes 1 and 2), but a protein migrating at the position of intact Nedd-4 was detected only in small amounts in wt and PSAP mutant virus preparations (Fig. 8b, lanes 3 and 4). The stronger signal in lane 4 than in lane 3 probably reflects the higher amount of virus loaded for the PSAP mutant (compare Fig. 8a, lanes 1 and 3). Besides intact Nedd-4, several other reactive proteins were detected with Nedd-4 antiserum in virus preparations. Some of these proteins may correspond to cross-reaction with (overloaded) viral proteins or have also been detected in cell lysates of untransfected cells (not shown), but this does not account for the protein with an apparent molecular mass of 50 kDa (Fig. 8b, arrow), which was not present in parallel preparations from mock-transfected cells (Fig. 8c, arrow). The presence of Nedd-4 and the smaller immunoreactive products inside MPMV virions was also confirmed by analysis of trypsin-treated virus (data not shown).
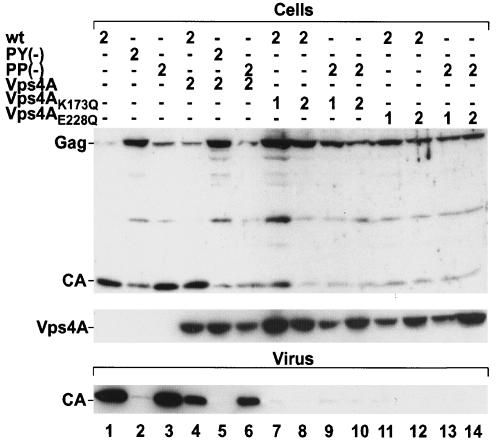
VPS4A is required for wt and PSAP mutant MPMV release.
The cellular AAA type ATPase VPS4A, which is a component of the Vps pathway involved in MVB sorting (14), has been shown to be required for the release of both HIV-1 and MLV. We therefore wanted to test whether VPS4A is also required for MPMV budding and whether the residual budding observed for the MPMV PSAP mutant depends on VPS4A function as well. For this, we cotransfected 293T cells with wt or PSAP mutant pMPMV and with either empty vector, wt GFP-VPS4A, or dominant-negative GFP-VPS4A mutants, which are defective for ATP binding (VPS4AK173Q) or ATP hydrolysis (VPS4AE228Q) (Fig. 9). As a control, we also transfected PPPY mutant pMPMV. As observed for HeLa cells, 293T cells transfected with PPPY mutant pMPMV exhibited no virus release (Fig. 9, lanes 1 and 2). In contrast, there was no significant phenotype for PSAP mutant MPMV in 293T cells (Fig. 9, lanes 1 and 3), indicating that the requirement for the PSAP motif depends on the cell type used. Upon cotransfection of wt GFP-VPS4A with pMPMV or the L-domain mutant plasmids, less virus was released than from cells cotransfected with empty vector (Fig. 9, compare lanes 4 to 6 and lanes 1 to 3). A similar finding was reported by Garrus et al., who observed a reduction of virus release upon cotransfection of wt GFP-VPS4A and HIV-1 (8). Strikingly, cotransfection of either VPS4A mutant construct reduced virus release for both wt pMPMV and the PSAP variant to levels as low as that observed for the PPPY mutant construct (Fig. 9, lanes 7 to 14). Furthermore, in the presence of ATPase-defective VPS4A variants, wt and PSAP mutant Gag exhibited a defect in processing similar to that of PPPY mutant Gag proteins. This result indicates that VPS4A is essential for MPMV release and acts downstream of both the PSAP and the PPPY motif.
FIG. 9.
Effect of coexpression of wt or ATPase-defective GFP-VPS4A fusion proteins on MPMV release. 293T cells were cotransfected with the indicated pMPMV plasmids. In each case, the amount of total DNA was adjusted to 4 μg. Steady-state levels of cell- and virus-associated CA protein (upper and lower panel, respectively) were analyzed by Western blotting with anti-CA antiserum, and the expression of the VPS4A constructs was confirmed by Western blotting with an anti-GFP antiserum (middle panel).
DISCUSSION
Here, we present a detailed analysis of the relative contributions of the closely spaced MPMV PPPY and PSAP motifs to MPMV particle production. In agreement with previous reports (38), a block to viral release was observed for MPMV lacking the PPPY motif. Procapsids accumulated underneath the plasma membrane or on intracellular vesicles, displaying an apparent defect mostly in the early stages of budding. Surprisingly, release of particles from cells cotransfected with equal amounts of wt and PPPY mutant constructs was inefficient as well, and a twofold excess of PPPY mutant Gag protein over wt Gag completely abolished virus release in a trans-dominant-negative way. Thus, at least one-half of the MPMV Gag molecules needs to carry an intact PPPY motif to allow efficient virus release. This is in contrast to findings which showed that one-fifth of HIV-1 Gag molecules carrying a PTAP L domain were sufficient to allow efficient particle release (17). Conceivably, there is a certain threshold for factor recruitment which may be different depending on the virus and/or the host cell.
While the MPMV PSAP motif cannot replace a functional PPPY motif (38; this study), our results clearly showed that the PSAP motif functions as a second L domain and promotes virus release in the context of an intact PPPY motif. The phenotype of the PSAP mutation was less pronounced than that of the PPPY mutation, since virus release was reduced by only a factor of two when PSAP was changed. The release defect was much more evident on thin-section EM analysis, however, which revealed a late budding arrest, with most procapsids in immature budding structures or budding chains containing multiple procapsids. These budding chains were apparently released from transfected cells, thus contributing to the extracellular virus detected. The phenotype of a construct carrying both the PPPY and PSAP mutations was indistinguishable from that of the PPPY mutant. Thus, in the context of MPMV Gag, PPPY and PSAP motifs are not redundant. This is different from results observed in substitution experiments, where the PPPY L domains of RSV and MLV were artificially exchanged for a HIV-1-derived PTAP motif. In these studies, the first 12 or 18 residues of HIV-1 p6 were used to replace the respective PPPY L-domain peptide, thereby reconstituting particle release (21, 40).
Like MPMV, HTLV-1, vesicular stomatitis virus, and Ebola virus contain closely spaced or overlapping L-domain sequences. The Ebola virus VP40 protein harbors the peptide P7TAPPEY13. Martin-Serrano et al. showed that substitution of P7, which disrupts the PTAP but not the PPXY motif, severely reduced the release of VP40 (17). This result indicated that the Ebola virus PTAP motif contributes to virus release as well, similarly to the result observed for MPMV in this study. One may speculate that the combined function of two different L-domain motifs is also required for efficient release of the other viruses that contain more than one L domain. Furthermore, the Ebola virus VP40 L-domain peptide has been shown to functionally replace the HIV-1 PSAP motif of the HIV-1 Gag polyprotein (17). Like in the context of VP40, the P7 mutation, which leaves the PPPY motif intact, resulted in a complete loss of HIV-like particle release (17). Similarly, Strack et al. replaced the entire p6 domain of HIV-1 Gag with a VP40-derived peptide (31). Consistent with the observation by Martin-Serrano et al., substituting Y13 induced no phenotype. When the same Ebola virus-derived peptide was used in the context of a minimal Gag protein, which lacked the globular domain of MA and the N-terminal part of CA and in which the NC-sp2 region was replaced by a leucine zipper, the PT/SAP motif did not suffice to promote particle release. Instead, as was observed in this study for MPMV, a PPPY motif was required to achieve release of particles. Strack et al. suggested that in the context of HIV-1, the PT/SAP motif cooperates with NC-p1 or the remainder of p6 (31). Thus, it seems likely that whether or not a PT/SAP motif can function as an L domain depends on the context of the Gag protein.
The EM results obtained in this study suggest that the defects of the PPPY and the PSAP variants of MPMV are both at the stage of virus release but are not identical. Most PPPY variant budding structures appeared to be much closer to the plane of the plasma membrane than was observed for the PSAP variant, indicating an earlier budding defect for the PPPY variant. Some late budding structures connected to the plasma membrane or to each other by only a thin membrane stalk were observed for the PPPY variant as well, but this was not common. In most cases, procapsids completely failed to initiate budding and did not induce membrane curvature at all. Le Blanc et al. reported a similar finding for HTLV-1, indicating that the intact PPPY motif may be required at a relatively early stage for the initiation of budding (16). If the PPPY motif acts upstream of the PSAP motif, this could explain why an intact PPPY motif is needed for the action of PSAP as an L domain (see also below).
In several cells, we observed intracellular vesicles decorated with PPPY mutant viral procapsids that did not initiate budding. Conceivably, some wt MPMV procapsids may also be transported to intracellular vesicles, but this may be normally overlooked, because it constitutes a minor pathway which becomes visible only under conditions of budding arrest. It is interesting in this regard that budding of HIV-1 into intracellular major histocompatibility complex class II vesicles has recently been reported, and this may be an important route of release at least in some cell types (27). Alternatively, these decorated vesicles may be the result of a partial defect in viral assembly even earlier than budding, i.e., during procapsid trafficking, or may be due to the expression of large amounts of viral Gag proteins.
TSG101 has been shown to bind to the HIV-1 PTAP L domain, and this interaction is essential for virus release (8, 34). We find readily detectable amounts of endogenous TSG101 present within MPMV particles, suggesting that the protein is recruited to the budding site and incorporated via its interaction with the PSAP motif. This incorporation is likely to be specific, because Tsg101 was not detected in preparations from mock-transfected cells, there was no incorporation of a highly expressed cytosolic protein into particles, and the virion levels of TSG101 were strongly reduced in the case of the PSAP variant. The finding that TSG101 incorporation is not abolished in particles lacking the PSAP motif raises the question whether TSG101 packaging is also specific in this case. Furthermore, it is unclear if the amount of TSG101 still detected accounts for the intermediate phenotype caused by PSAP mutation. Based on published data, TSG101 should not be able to bind to the inactivated PSAP motif (12, 17, 34). Since TSG101 shows weak binding to ubiquitin alone (8), it could be recruited by binding to ubiquitinated Gag protein. The fact that an L-domain phenotype was observed, even though some TSG101 was present in virus preparations, may indicate that a relatively high level of TSG101 is required at the budding site.
Besides TSG101, we also detected the E3 ubiquitin ligase Nedd-4 in MPMV particles, albeit at low levels. Nedd-4 incorporation was unaltered by mutation of the PSAP motif, as expected. Recently, a Nedd-4-like protein named Bul-1 has been reported to interact with MPMV Gag in a PPPY motif-dependent way, and Bul-1 overexpression was shown to stimulate MPMV release (39). Accordingly, Nedd-4 itself may be only weakly recruited by the MPMV Gag polyprotein, explaining its low concentration inside viral particles. It should be noted, however, that the major Nedd-4 immunoreactive protein in viral preparations migrated with an apparent mobility of ca. 50 kDa, which suggests that significantly more Nedd-4 may be incorporated into viral particles and cleaved by the viral PR. The 50-kDa protein was not detectable in cytoplasmic extracts, and the signal could not be assigned to any of the major viral proteins. It seems unlikely that a cellular protein is incorporated into virions at levels high enough to be detectable by unspecific binding of the antibody, and the possibility of PR-mediated cleavage of Nedd-4 is currently under investigation.
As observed for the release of HIV-1 and MLV (8), MPMV particle production was completely abolished by overexpression of dominant-negative variants of the cellular ATPase VPS4A as well. Virus release was also blocked in the case of the PSAP variant of MPMV, indicating that VPS4A promotes virus release independently of the PT/SAP motif and that the two L-domain motifs target the same pathway. This supports and extends previous findings for MLV, which also contains an essential PPPY motif but carries an additional PSAP motif in its MA domain, which may also contribute to virus release.
In summary, our results indicate that both L-domain motifs of MPMV contribute to virus release, with the PPPY motif being essential and a PPPY mutation causing budding arrest at an earlier stage than a PSAP mutation. In the case of the VPS machinery-dependent sorting of endosomal proteins into MVBs, initial monoubiquitination of cargo and adapter proteins is followed by recruitment of the TSG101-containing ESCRT-1 complex and further complexes of the Vps pathway (14, 24). By analogy, one may speculate that PPPY-dependent recruitment of Nedd-4 (or a Nedd-4 like protein) serves to monoubiquitinate Gag or a Gag-associated protein with subsequent binding of TSG101, both through the PSAP motif within Gag and through the ubiquitin moiety. Since TSG101 was incorporated into virions even in the absence of a PSAP motif, this motif in the case of MPMV may mainly serve to recruit sufficient amounts of TSG101 for efficient virus release, e.g., in cell types where TSG101 concentrations are limiting. This hypothesis predicts that TSG101 would also be required for the release of an MPMV variant lacking the PSAP motif, and this is under investigation. In the case of MLV, however, virus release was not severely altered following depletion of cellular TSG101 (8), indicating that the PPPY motif can function independently of TSG101 at least in this case.
Acknowledgments
We thank T. Ruml for providing the MPMV anti-CA antiserum, W. Sundquist for providing expression plasmids for wt and mutant VPS4A, and E. Hunter for providing pSHRM15.
E.G. was supported by the Boehringer Ingelheim Fonds.
REFERENCES
- 1.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dettenhofer, M., and X. F. Yu. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 6.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature 381:744-745. [DOI] [PubMed] [Google Scholar]
- 8.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 9.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 15.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 18.Myers, E. L., and J. F. Allen. 2002. Tsg101, an inactive homologue of ubiquitin ligase e2, interacts specifically with human immunodeficiency virus type 2 gag polyprotein and results in increased levels of ubiquitinated gag. J. Virol. 76:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 20.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pepinsky, R. B., I. A. Papayannopoulos, E. P. Chow, N. K. Krishna, R. C. Craven, and V. M. Vogt. 1995. Differential proteolytic processing leads to multiple forms of the CA protein in avian sarcoma and leukemia viruses. J. Virol. 69:6430-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 25.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 27.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 28.Rhee, S. S., H. X. Hui, and E. Hunter. 1990. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J. Virol. 64:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudol, M. 1996. Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 65:113-132. [DOI] [PubMed] [Google Scholar]
- 33.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 34.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weldon, R. A., Jr., W. B. Parker, M. Sakalian, and E. Hunter. 1998. Type D retrovirus capsid assembly and release are active events requiring ATP. J. Virol. 72:3098-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]