Abstract
Previous studies using wild-type Encephalomyocarditis virus (EMCV) and Mengo virus, which have long poly(C) tracts (61 to 146 C's) at the 5′ nontranslated region of the genome, and variants of these viruses genetically engineered to truncate or substitute the poly(C) tracts have produced conflicting data on the role of the poly(C) tract in the virulence of these viruses. Analysis of the nucleotide sequence of an EMCV strain isolated from an aborted swine fetus (EMCV 30/87) revealed that the virus had a poly(C) tract that was 7- to 10-fold shorter than the poly(C) tracts of other EMCV strains and 4-fold shorter than that of Mengo virus. Subsequently, we investigated the virulence and pathogenesis of this naturally occurring short-poly(C)-tract-containing virus in rodents, pigs, and nonhuman primates. Infection of C57BL/6 mice, pigs, and cynomolgus macaques resulted in similar EMCV 30/87 pathogenesis, with the heart and brain as the primary sites of infections in all three animals, but with different disease phenotypes. Sixteen percent of EMCV 30/87-infected pigs developed acute fatal cardiac failure, whereas the rest of the pigs were overtly asymptomatic for as long as 90 days postinfection (p.i.), despite extensive myocardial and central nervous system (CNS) pathological changes. In contrast, mice infected with ≥4 PFU of EMCV 30/87 developed acute encephalitis that resulted in the death of all animals (n = 25) between days 2 and 7 p.i. EMCV 30/87-infected macaques remained overtly asymptomatic for 45 days, despite extensive myocardial and CNS pathological changes and viral persistence in more than 50% of the animals. The short poly(C) tract in EMCV 30/87 (CUC5UC8) was comparable to that of strain 2887A/91 (C10UCUC3UC10), another recent porcine isolate.
Encephalomyocarditis virus (EMCV) is a picornavirus belonging to the Cardiovirus genus that infects many animal species including pigs (17), rodents (45), cattle (41), elephants (13), raccoons (47), marsupials (38), baboons, macaques, chimpanzees, and humans (2, 16, 21, 38, 44, 45). Rats and mice are the natural hosts of the virus, but pigs are the most commonly and severely infected domestic animals (5, 36). EMCV strains have been isolated from primates, pigs, and rodents (8, 15, 20, 22, 24, 29). EMCV Rueckert (EMCV R/45), the prototype strain, was isolated from a 5-year-old chimpanzee that suffered acute fatal myocarditis in 1945 (15), whereas EMC-M/58 virus was isolated from a naturally infected domestic pig suffering from severe myocarditis (29). A number of EMC-M/58 variants, including EMC-D/58, EMC-B/58, PV2/58, and PV21/58, were generated in laboratories and used to study the pathogenesis of viral diabetes (6, 7, 18, 25, 46, 49). Mengo virus, another cardiovirus that shares the same serotype as EMCV, was isolated from the cerebrospinal fluid of a paralyzed rhesus macaque in 1948 (8). EMCV 30/87 and 2887A/91 are more recent EMCV isolates obtained from aborted swine fetuses in the United States and Belgium, respectively, following natural EMCV outbreaks in domestic pigs (19, 20).
Nucleotide sequencing of most EMCV strains and Mengo virus has identified a poly(C) tract, consisting of 61 to 146 cytosine residues (C61 to C146), occasionally interrupted by 1 to 3 uridine residues, located at the 5′ nontranslated region (NTR) of the positive-sense RNA genome (1, 9, 10, 19, 31, 49). Studies to determine the role of the poly(C) tract in virus replication, virulence, and host range have produced conflicting findings. For instance, in earlier studies, truncation of the poly(C) tract of Mengo virus M/48 (from C44UC10 to C8- or C13UC10) attenuated the virus in mice, resulting in lower virus titers in the brain and development of milder meningoencephalitis (11, 33). However, later, more-extensive experiments using a number of inbred strains of mice demonstrated that both short- and long-poly(C)-tract variants of Mengo virus M/48 were highly virulent in newborn mice of any strain (34). Studies using both EMCV R/45 and EMCV PV2/58 demonstrated no correlation between the length of the poly(C) tract and virulence. Truncation of the poly(C) tract in EMCV R/45 from C115UC3UC10 to C4, C9, or C20 produced only slight decreases in virulence among 12-week-old SJL mice (14). Strain PV2/58 (containing 118 C's) was cloned, and three viruses with different lengths of the poly(C) tract were generated; rPV2/dT had the poly(C) tract replaced by a poly(U) tract, rPV2/C20 had the poly(C) tract replaced with 20 C's, and rPV2/ran had the poly(C) tract replaced with a random sequence of 238 nucleotides (48). Infection of mice with either wild-type PV2/58, rPV2/dT, rPV2/C20, or rPV2/ran resulted in insignificantly higher virus loads and degrees of pathological lesions in mice infected with wild-type PV2/58 than in mice infected with recombinant viruses, suggesting that the poly(C) tract was not essential for virulence in mice (48).
Another interesting aspect of EMCV pathogenesis is the ability of these viruses to cause interspecies infections (16, 22, 38, 39). EMCV outbreaks in zoos in Australia and the United States have involved multiple animal species including lemurs, squirrels, macaques, mandrills, chimpanzees, hippopotami, kangaroos, and possibly humans (38, 45). The few documented cases of EMCV infection in humans have been associated with fever, neck stiffness, lethargy, delirium, headaches, or vomiting (12, 28). In Germany, strains of the virus have been isolated from children suffering from meningitis and encephalitis, but no causal relationship between EMCV and the symptoms has been demonstrated (12). There is renewed interest in pig-to-human zoonotic viruses because of advances in xenotransplantation as a means of overcoming the acute shortage of transplantation tissues and organs for humans. Porcine cells, tissues, and organs are the primary animal tissues being considered for human transplantation because of the similarities in anatomical and physiological features between humans and pigs, the availability of the species, and the relative ease of breeding pigs (27, 35, 37, 40, 43).
It has recently been demonstrated that infection of 5-week-old pigs with EMCV 30/87 resulted in the deaths of 16% of the pigs 2 to 3 days after infection from acute myocarditis, characterized by extensive lysis of sarcoplasm, cellular degeneration, and mineralization in the myocardium (4). The rest of the infected pigs (84%) did not develop clinical illness but showed virus persistence for as long as 90 days in cardiomyocytes and central nervous system (CNS) cells, which was associated with extensive apoptosis and viral antigen expression. More importantly, EMCV 30/87 productively infected primary human cardiomyocytes, resulting in production of 100 to 1,000 PFU of infectious virus per cell within 6 h. In the present study, we demonstrate from the nucleotide sequence that EMCV 30/87 has the shortest poly(C) tract of any naturally occurring EMCV strain, and we determine its virulence and pathogenesis in rodents, pigs, and nonhuman primates.
MATERIALS AND METHODS
Viruses.
An EMCV strain isolated from naturally infected pigs in Minnesota in 1987 (EMCV 30/87) was used in all our experiments (20). The virus was amplified by infection of HeLa cells, which were maintained in RPMI 1640 medium supplemented with 5% fetal bovine serum, l-glutamine, antibiotics, and antimycotics. The nucleotide sequences of EMCV 30/87 were compared to the sequences of Mengo virus M/48 and EMCV strains R/45, EMC-B/58, EMC-D/58, PV2/58, PV21/58, and 2887A/91. The year of the parent isolate has been added to the designation of each of these strains, and the same year is given to laboratory variants of a particular isolate (e.g., EMC-B/58, EMC-D/58, PV2/58, PV21/58).
Nucleotide sequencing of EMCV 30/87.
EMCV 30/87 RNA from virus-infected HeLa cells was extracted by using a guanidinium thiocyanate-phenol-chloroform reagent (TRIzol; Invitrogen, San Diego, Calif.). The RNA, derived both from a mixed population of low-passage-number EMCV 30 and from a plaque-purified clone, was reverse transcribed using oligo(dT) primers with Superscript II reverse transcriptase (Invitrogen) at 42°C for 50 min. Initial primers for amplification of 800 bp at the 5′ untranslated end of the EMCV 30/87 sequence were designed by using EMCV strain R/45 (GenBank accession number M81861). Subsequent primers designed from EMCV 30/87 sequences were used to complete the rest of the genome. After reverse transcription, PCR was performed using 10 μl of the cDNA. For PCR, the cDNA was denatured at 94°C for 5 min, followed by 30 cycles of denaturation (at 94°C for 1 min), annealing (at 60°C for 1 min), and elongation (at 72°C for 1 min). The PCR products were gel purified (Qiagen, Valencia, Calif.) and either cloned into the PCR2.1 plasmid vector (Invitrogen) or directly sequenced. The subcloned EMCV 30/87 gene segments were sequenced with T7 forward primers and M13 reverse primers. Three independent clones of each segment, and more than five clones for the poly (C) tract, were sequenced. For the 5′ and 3′ ends of EMCV 30/87, the GeneRace kit (Invitrogen) was used as described previously (26).
Nucleotide sequence analysis.
Nucleotide sequence editing and prediction of amino acid sequences were performed with DNAStar (Madison, Wis.) software. Alignments were performed by the CLUSTALW method (42) by using the following previously sequenced EMCV strains: Mengo virus M/48 (GenBank accession number L22089), EMCV R/45 (accession number M81861), EMC-B/58 (accession number M22457), EMC-D/58 (accession number M22458), PV2/58 (accession number X87335), PV21/58 (accession number X74312), and 2887A/91 (accession number AF356822). Nucleotide and predicted amino acid sequences were compared for each of the 12 genes and proteins (leader, VP1, VP2, VP3, VP4, 2A, 2B, 2C, 3A, 3B, 3C, and 3D) and also for the long 5′ and short 3′ NTRs. To determine relationships among the EMCV strains, phylogenetic analysis of the predicted amino acid sequences for the translated region was performed with PAUP (phylogenetic analysis using parsimony) (40) software utilizing both parsimony and neighbor-joining analyses. The two analyses resulted in duplicate phylogenetic relationships and were each evaluated with 2,000 bootstrap replicates.
Infection of mice.
Four- to 6-week-old C57BL/6 mice were purchased from Jackson Laboratory and housed in the University of Minnesota biosafety level-2 mouse facilities at the St. Paul campus. Handling of animals, including feeding and euthanasia, was in conformity with the National Institutes of Health and University of Minnesota institutional animal care guidelines. Mice were intraperitoneally inoculated with 104, 100, or 2 PFU of EMCV 30/87 in a 1-ml volume and were monitored for clinical signs of EMCV disease. Terminally sick mice were euthanatized, whereas mice that did not develop clinical disease (mice inoculated with 2 PFU) were sacrificed at day 14, 21, or 45 postinfection (p.i.).
Infection of pigs.
Thirty-seven 5-week-old pigs were obtained from an EMCV-free swine herd (Midwest Research Swine, Gibbon, Minn.) and placed in negative-pressure isolation units at the University of Minnesota animal facilities. An enzyme-linked immunosorbent assay (ELISA) was used to confirm that the pigs were negative for EMCV antibodies before infection. Animals were intraperitoneally inoculated with 2.9 × 108 PFU of EMCV 30/87 in a 1-ml volume. Handling of animals, including feeding and euthanasia, was in conformity with National Institutes of Health and University of Minnesota institutional animal care guidelines. Four to six pigs were euthanized on days 7, 21, 45, and 90 p.i. by using pentobarbital sodium. Tissues from the brain, heart, kidney, liver, spleen, skeletal muscle, and pancreas were collected for isolation of infectious virus and viral RNA, histopathology, and immunohistochemistry.
Infection of cynomolgus macaques.
Fourteen 2- to 4-year-old specific-pathogen-free cynomolgus macaques (Macaca fascicularis) were purchased from Charles River BRF (Houston, Tex.) and housed at Gwathmey Incorporated (Cambridge, Mass.) animal facilities, where experiments were performed. Protocols for handling of the macaques, including feeding and euthanasia, were approved by both Gwathmey Inc. and the University of Minnesota Institutional Animal Care and Use Committee and were in conformity with the National Institutes of Health guidelines. Twelve macaques were intraperitoneally inoculated with 2.9 × 108 PFU of EMCV 30/87 in a 1-ml volume, and two macaques were inoculated with sterile buffer as controls. Macaques were monitored daily for clinical signs of EMCV disease, and three animals were sacrificed at days 14, 21, and 45 p.i. for collection of the brain, heart, kidney, liver, spleen, skeletal muscle, and pancreas. Tissues were analyzed for histopathologic and immunohistochemical changes and for the presence of infectious virus and viral RNA.
Sample processing and histopathologic analysis.
Sections of heart, brain, liver, kidney, spleen, skeletal muscle, and pancreas from pigs, macaques, and mice infected with EMCV for 2, 7, 14, 21, or 45 to 50 days were collected for histopathologic analysis, virus isolation, and RNA isolation. Tissues for RNA isolation were snap-frozen in liquid nitrogen, whereas tissues for histopathology and in situ hybridization were fixed in 10% neutral buffered formalin and embedded in paraffin. Paraffin-embedded sections were cut at a thickness of 4 μm and stained with hematoxylin and eosin for histopathologic analysis. Serum samples collected from the animals at sacrifice were used to determine levels of virus-specific immunoglobulin G (IgG) by ELISA.
In situ hybridization.
To detect the presence of viral genomes in pig tissues, in situ hybridization was performed by using a 414-bp VP1-specific probe as described previously for Theiler's murine encephalomyelitis virus (32). Briefly, a VP1 cDNA was subcloned from EMCV 30/87 into plasmid pUC18 by using BamHI and HindIII restriction sites, and the cDNA probe was prepared by digesting the VP1 plasmid with the NcoI and BamHI restriction enzymes (14). The probe was labeled with 35S-dATP by using the Random Primers DNA labeling system (Invitrogen) and was purified by using G-50 Sephadex Quick Spin columns (Roche, Indianapolis, Ind.). To prepare tissue samples, paraffin-embedded sections were deparaffinized by using xylene. Sections were first digested with 10 μg of proteinase K/ml in phosphate-buffered saline for 30 min at 37°C and then treated with 0.1 M triethanolamine containing acetic anhydride. The sections were prehybridized in a buffer containing deionized formamide, Denhardt's solution, sodium chloride, salmon sperm DNA, yeast total RNA, and yeast tRNA for 4 h at room temperature before hybridization with a 35S-labeled 309-bp VP1 probe. Hybridization was performed overnight at 37°C and was followed by extensive washes in reducing buffer at 55°C. Air-dried slides were immersed in an NTB2 film emulsion (Eastman Kodak Co., Rochester, N.Y.) and exposed at 4°C for 5 days.
RT-PCR.
Pig, macaque, and mouse tissues collected at days 2, 7, 14, 21, 45, and 90 p.i. were analyzed for EMCV RNA by nested reverse transcription-PCR (RT-PCR) as described previously (4). Samples of brain, heart, liver, kidney, spleen, or skeletal muscle tissues were homogenized in TRIzol (Invitrogen), followed by chloroform extraction of total RNA. Five micrograms of total RNA was reverse transcribed by using an oligo(dT) primer and the Superscript II reverse transcription kit (Invitrogen) before the outer and nesting PCRs were performed using primer pairs specific for the VP1 or VP2 gene of EMCV 30/87 sequenced in our laboratory. The VP1 primers used for the outer PCR were GCCTCAGTTTGACCCTGCTTATG (5′ primer) and CGGCTCTCGGAGTCATGTCAATC (3′ primer) (product size, 592 bp), and those for the nesting reaction were CGTCTCACAGAAATTTGGGGCAAC (5′ primer) and CCAGGCTTCCTGTGTTGTCAAATC (3′ primer) (product size, 340 bp). The VP2 primers for the outer PCR were CAGTAGGCCGTCTTGTTGGTTATG for the 5′ end and CACTTCAAGATCCACGGTGGTGTTG for the 3′ end (product size, 499 bp), and the nesting primers were GGCACTGTTCATGATGGAGAACAC for the 5′ end and GGTGATCCATAGCAAAGGGACCTTTC for the 3′ end (product size, 350 bp). All primer sequences are given in 5′-to-3′ orientation. The outer PCR was performed on 1 μl of the template cDNA by adding 0.2 mM deoxynucleoside triphosphates, 2 mM magnesium chloride, 10 pmol of each primer, and 1 U of Taq polymerase (Invitrogen). For the nested PCR, 2.5 μl of the first-round PCR mixture was used. PCR products were analyzed by agarose gel electrophoresis.
Nucleotide sequence accession number.
Nucleotide sequences for EMCV 30/87 have been submitted to GenBank (accession number AY296731).
RESULTS
Molecular analysis of EMCV 30/87 and comparison with other EMCV strains.
The entire EMCV 30/87 genome was sequenced for comparison with other EMCV strains and correlated with the pathogenesis of virus infection in different animal species. The sizes of the LP, VP1, VP2, VP3, VP4, 2A, 2B, 2C, 3A, 3B, 3C, and 3D genes of EMCV 30/87 (GenBank accession number AY296731) were similar to those of strains 2887A/91 (accession number AF356822), EMCV R/45 (accession number M81861), PV21/58 (accession number X74312), EMC-B/58 (accession number M22457), EMC-D/58 (accession number M22458), and PV2/58 (accession number X87335). However, Mengo virus M/48 (accession number L22089) had an extra amino acid (proline) in the 2B protein at position 10 from the N terminus of the protein. Comparison of the nucleotide and predicted amino acid sequences for the coding regions encompassing the 12 genes demonstrated that EMCV 30/87 shared the highest overall nucleotide sequence identity (84.2%) and predicted amino acid identity (96.4%) with strain 2887A/91, isolated from an aborted porcine fetus in Belgium, and with strain PV21/58, a laboratory isolate from a virus isolated from a pig myocardium in Panama (Tables 1 and 2). EMCV 30/87 also had high nucleotide (84.0%) and amino acid (96.6%) sequence identities with EMCV R/45, isolated from the myocardium of a chimpanzee. Of the 12 EMCV genes, 2B was the most conserved among the eight EMCV strains, demonstrating 90.4 to 94.2% nucleotide sequence identity and 94.7 to 97.3% predicted amino acid identity (Tables 1 and 2).
TABLE 1.
Percent nucleotide sequence identity between the genes of EMCV 30/87 and those of other EMCV strains and Mengo virusa
| Virus | % Nucleotide sequence identity with EMCV 30/87
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | LP | VP1 | VP2 | VP3 | VP4 | 2A | 2B | 2C | 3AB | 3C | 3D | |
| 2887A/91 | 84.2 | 83.1 | 85.0 | 82.3 | 83.8 | 80.5 | 73.7 | 94.2 | 83.7 | 80.9 | 82.8 | 86.4 |
| EMCV R/45 | 84.0 | 84.1 | 84.6 | 82.3 | 83.8 | 80.0 | 73.0 | 94.0 | 83.8 | 80.6 | 82.3 | 86.4 |
| PV21/58 | 84.2 | 84.6 | 85.0 | 82.0 | 84.0 | 80.0 | 73.7 | 93.8 | 83.7 | 81.8 | 82.6 | 86.6 |
| Mengo-M/48 | 80.2 | 79.1 | 79.8 | 80.2 | 80.8 | 79.0 | 65.7 | 90.4 | 79.7 | 76.2 | 77.4 | 82.2 |
| EMC-B/58 | 82.5 | 82.1 | 81.7 | 80.1 | 81.8 | 80.0 | 74.8 | 90.4 | 83.5 | 81.5 | 77.1 | 85.5 |
| EMC-D/58 | 82.5 | 83.1 | 81.9 | 79.9 | 81.8 | 80.0 | 75.1 | 90.4 | 83.5 | 81.5 | 77.1 | 85.5 |
| PV2/58 | 82.6 | 83.1 | 81.8 | 79.9 | 81.8 | 80.0 | 75.1 | 90.4 | 83.7 | 81.5 | 77.1 | 85.7 |
The translated region consisted of 6,876 nucleotides for all EMCV strains and 6,879 nucleotides for Mengo virus M/48.
TABLE 2.
Percent predicted amino acid sequence identity between proteins of EMCV 30/87 and those of other EMCV strains and Mengo virusa
| Virus | % Predicted amino acid sequence identity with EMCV 30/87
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | LP | VP1 | VP2 | VP3 | VP4 | 2A | 2B | 2C | 3AB | 3C | 3D | |
| 2887A/91 | 96.4 | 88.1 | 99.3 | 97.7 | 99.6 | 97.1 | 86.0 | 97.3 | 99.1 | 90.7 | 95.6 | 95.9 |
| EMCV R/45 | 96.6 | 91.0 | 97.8 | 98.0 | 99.6 | 95.7 | 84.6 | 97.3 | 99.4 | 88.9 | 94.6 | 95.7 |
| PV21/58 | 96.4 | 91.0 | 99.3 | 97.3 | 99.6 | 95.7 | 86.0 | 96.7 | 99.4 | 91.7 | 95.6 | 95.9 |
| Mengo-M/48 | 94.0 | 86.6 | 96.8 | 98.0 | 98.3 | 94.3 | 83.9 | 94.7 | 96.3 | 89.8 | 89.3 | 93.3 |
| EMC-B/58 | 95.1 | 88.1 | 97.1 | 98.4 | 99.6 | 95.7 | 83.2 | 94.7 | 97.8 | 92.6 | 90.7 | 95.0 |
| EMC-D/58 | 95.2 | 89.6 | 97.8 | 98.4 | 99.6 | 95.7 | 83.2 | 94.7 | 97.8 | 92.6 | 90.7 | 95.0 |
| PV2/58 | 95.4 | 89.6 | 97.5 | 98.4 | 99.6 | 95.7 | 83.9 | 94.7 | 98.5 | 93.5 | 90.7 | 95.2 |
Each of the 12 proteins was the same length in all the viruses, except for Mengo virus protein 2B, which had 1 more amino acid (151 amino acids) than 2B proteins from other viruses.
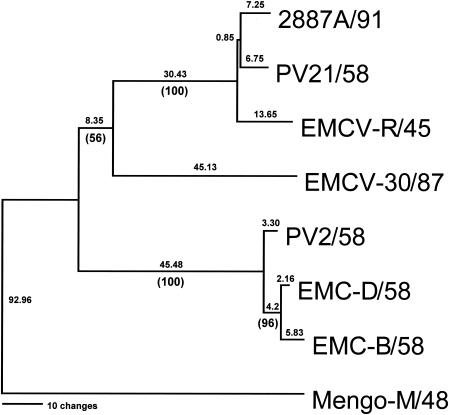
Phylogenetic comparison of the seven EMCV strains and Mengo virus revealed the closeness of EMC-B/58, EMC-D/58, and PV2/58 (Fig. 1), all of which are variant mutants derived from EMCV-M. EMCV R/45 and 2887A/91 segregated together, and EMCV 30/87 appeared to segregate between the two clusters. Mengo virus M/48 segregated far from the seven EMCV strains. Perhaps the most intriguing finding was that EMCV 30/87 had the shortest poly(C) tract (Table 3), which was observed both in a mixed population of low-passage-number EMCV 30 used to inoculate mice and in two plaque-purified strains of the virus (EMCV 30/PA and EMCV 30/PB). EMCV 30/87 had a poly(C) tract consisting of 14 C's (CUC5UC8), whereas the older EMCV isolates (EMCV R/45, PV2/58, PV21/58, EMC-B/58, and EMC-D/58) had 118 to 142 C's and Mengo virus M/48 had 61 C's. Strain 2887A/91, another recent isolate from an aborted pig fetus, also had a short poly(C) tract (C10UCUC3UC10) and was highly pathogenic. The 5′ NTR of EMCV 30/87 consisted of approximately 724 nucleotides, a size comparable to that of strain 2887A/91 (728 nucleotides) but 135 nucleotides shorter than that of PV21/58, the virus with the longest poly(C) tract (Table 3). The 10 to 21 nucleotides at the immediate 5′ side of the poly(C) tract consisted of 60 to 75% A residues, leading to a highly conserved region in all EMCV strains and Mengo virus (Table 3). Another conserved region was observed 4 to 6 nucleotides downstream of the poly(C) tract (Table 3). Analysis of the 3′ NTR of EMCV 30/87 revealed no major differences among eight EMCV strains. The 3′ NTR consisted of 125 nucleotides in addition to a short poly(A) tail, which was comparable to the 3′ NTRs of all other EMCV strains (107 to 126 nucleotides) and Mengo virus M/48 (125 nucleotides).
FIG. 1.
Phylogenetic relationship between EMCV 30/87 and other EMCV strains. Following alignment of the contiguous predicted amino acid sequences for the translated regions of the viruses (LP, VP4, VP2, VP3, VP1, 2A, 2B, 2C, 3A, 3B, 3C, and 3D), a rooted phylogram was generated by maximum parsimony analysis using Mengo virus M/48 as the outgroup. Absolute distances are listed above each branch, with bootstrap confidence levels given below in parentheses. GenBank accession numbers for the viruses are given in Materials and Methods.
TABLE 3.
Comparison of the length of the poly(C) tract and the nucleotide sequences of the adjoining regions in EMCV 30/87 with those in other EMCV strains and Mengo virus M/48
| Straina | Species from which original strain was isolated | Disease associated with original isolate | Sequences of the poly(C) tract and adjoining regionsb |
|---|---|---|---|
| EMCV-30/87 | Pig | Reproductive failure | GUGCCACCCCAAACAACAACAACAAAACAAA-16(CUC5UC8)-UUACUAUACUGGCCGAAG |
| 2887A/91 | Pig | Reproductive failure | GUGCCACCCCAAAACAACAACAAA-27(CCCCC6UCUC3UC10)-UAACGUUACUGGCCGAAG |
| Mengo-M/48 | Monkey | Myocarditis | GUGCCACCCCAAAACCACAUAA-63(CCCCCCCCCCC40UC10UC)-ACAUUACUGGCCGAAG |
| PV2/58 | Pig | Myocarditis | GUGCCACCCCAACCAACAACAAAACAAAAA-118(CCCCCC113)-AACGUACUGGCCGAAG |
| EMC-B/58 | Pig | Myocarditis | GUGCCACCCCAACAUAACAACAGA-127(CCCCCCCCCCCC116)-AACGUUACUGGCCGAAG |
| EMC-D/58 | Pig | Myocarditis | GUGCCACCCCAACAUAACAACAGA-130(CCCCCCCCCCCC119)-AACGUUACUGGCCGAAG |
| EMCV-R/45 | Chimpanzee | Myocarditis | GUGCCACCCCAAAAUAACAACAGA-132(CCC113UCUC3UCC9)-UAACGUUACUGGCCGAAG |
| PV21/58 | Pig | Myocarditis | GUGCCACCCCAAAACAACAACAGA-158(CCCC138UCUC3UU9)-UAACGUUACUGGCCGAAG |
GenBank accession numbers of strains are given in Materials and Methods.
For the poly(C) tract, the number before the parentheses represents the entire length of the tail (C's plus U's), whereas subscript numbers are the numbers of C's in that area. Conserved sequences flanking the poly(C) tract on both the 5′ and 3′ ends are in bold.
EMCV-induced clinical disease in mice, pigs, and macaques.
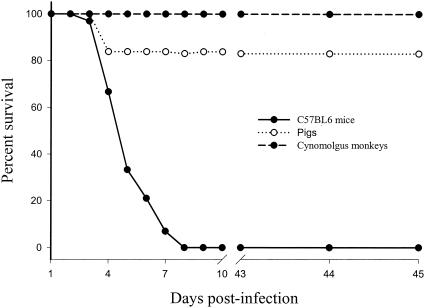
All mice (n = 25) inoculated with ≥4 PFU of EMCV 30/87 died between days 2 and 7 p.i. from acute encephalitis (Fig. 2). The clinical signs in mice included hunched posture, ruffled fur, lethargy, anorexia, and hind limb paralysis. The clinical signs lasted for 24 to 72 h before mice became moribund with signs of severe respiratory failure. No mice died of sudden cardiac failure, which was observed in 16% of EMCV 30/87-infected pigs. Mice inoculated with 104 to 105 PFU of EMCV 30/87 developed disease 2 to 4 days p.i., whereas mice inoculated with 4 to 100 PFU of virus developed disease between days 5 and 7 p.i. Terminally, mice developed posterior limb paresis or complete paralysis before rapidly becoming moribund.
FIG. 2.
Survival curves of EMCV-infected mice, pigs, and macaques. Six-week-old C57BL/6 mice (n = 30), 5-week-old pigs (n = 35), and 2- to 4-year-old cynomolgus macaques (n = 12) were intraperitoneally inoculated with EMCV 30/87 and monitored daily for clinical signs and fatality for 45 days. Sham-inoculated pigs (n = 2), mice (n = 4), and macaques (n = 2) remained asymptomatic and had no fatalities.
Six of the 37 pigs inoculated with 2.9 × 108 PFU of EMCV 30/87 (16.2%) died of acute myocardial failure 2 to 3 days later. The rest of the pigs did not develop any overt clinical signs. None of the 12 cynomolgus macaques inoculated with 2.9 × 108 PFU of EMCV 30/87 (the same virus dose as that given to pigs) developed detectable clinical signs. Electrocardiographic profiles were not determined for mice, pigs, or macaques. A summary of the survival curves of mice, pigs, and macaques following EMCV infection is presented in Fig. 2. Sham-inoculated pigs (n = 2), mice (n = 4), and macaques (n = 2) remained asymptomatic throughout the experimental period and had no fatalities.
Pathogenesis of EMCV 30/87 in mice.
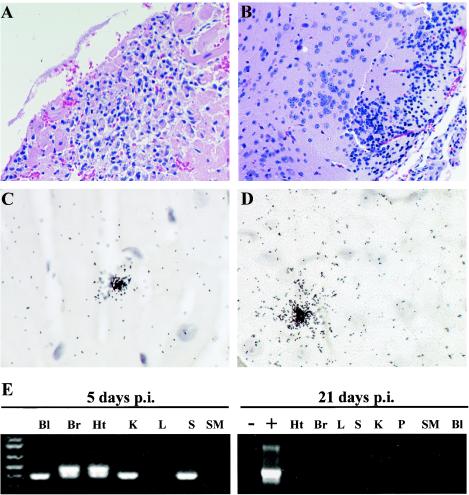
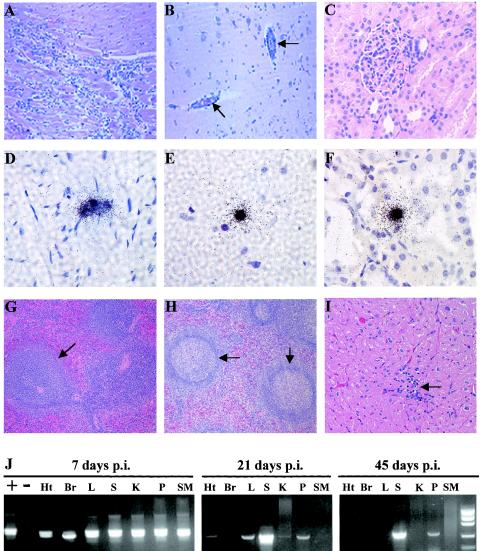
All EMCV 30/87-infected mice died within 2 to 7 days. Following intraperitoneal inoculation, there was widespread virus distribution, as demonstrated by detection of EMCV RNA in the blood, spleen, kidneys, liver, heart, brain, and skeletal muscle by RT-PCR (Fig. 3E; Table 4). Viral RNA could also be localized by in situ hybridization, particularly in tissue sections from the heart (Fig. 3C) and brain (Fig. 3D). Histopathologic changes were observed in the heart and brain but not in the liver, spleen, kidney, pancreas, or skeletal muscle. In the heart, there were large foci of lymphocytic infiltration in the myocardium (Fig. 3A), accompanied by degeneration and necrosis of cardiac myocytes and sarcoplasm. In the brain, similar foci of lymphocytic infiltration were observed in the cerebral cortices (Fig. 3B), hypothalami, and hippocampi. To investigate whether EMCV persistence and chronic disease can occur in mice, we inoculated mice (n = 8) with lower titers (2 to 10 PFU), but they did not develop either clinical disease or anti-EMCV antibodies, indicating that the virus titers were too low to establish an infection. In addition, no EMCV RNA was detected in the brain, heart, spleen, pancreas, blood, kidney, liver, or skeletal muscle in these mice 21 days p.i. (Fig. 3E), further indicating that no infection was established.
FIG. 3.
Pathological changes and detection of viral RNA in mice infected with EMCV 30/87. Five-week-old C57BL/6 mice were intraperitoneally inoculated with 102 to 103 PFU of EMCV 30/87, and histopathologic changes and the presence of EMCV RNA in the blood (Bl), heart (Ht), brain (Br), spleen (S), pancreas (P), liver (L), kidney (K), and skeletal muscle (SM) were analyzed at 2 to 8 days p.i. All EMCV 30/87-infected mice inoculated with more than 102 PFU of virus died between days 2 and 8 p.i. Histopathologic changes were confined to the brain and heart. In the heart, there were large foci of lymphocytic infiltration in the myocardium (A), accompanied by degeneration and necrosis of cardiac myocytes, whereas in the brain, similar lymphocytic infiltrations were observed in the cerebral cortices (B), hypothalami, and hippocampi. EMCV RNA was detected in both the heart (C) and brain (D) by in situ hybridization, but also in the blood, kidney, spleen, and skeletal muscle by RT-PCR (E). In the brain and heart, which had high levels of EMCV RNA, nested RT-PCR using VP2 primers followed by electrophoresis resulted in two visible bands, the larger (499 bp) representing the primary reaction product and the smaller (350 bp) representing the nested reaction product. For histopathology, heart and brain sections were embedded in paraffin and 4-μm-thick sections were stained with hematoxylin and eosin. In situ hybridization was performed using a 35S-labeled VP1 cDNA probe. Results for mice inoculated with 2 to 10 PFU are shown in the right portion of panel E.
TABLE 4.
Percent EMCV RNA-positive animals following EMCV 30/87 infectiona
| Animals | Days p.i. | % of animals positive for EMCV RNA in:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Brain | Heart | Spleen | Liver | Kidney | Skeletal muscle | Pancreas | ||
| Mice | 2-7 | 57.9 | 73.7 | 76.9 | 52.6 | 73.7 | 68.4 | NDb |
| Macaques | 7 | 75 | 100 | 100 | 100 | 100 | 100 | 100 |
| Mice | 14-21 | 18.2 | 27.3 | 9.1 | 0 | 0 | 18.2 | 18.2 |
| Macaques | 21 | 25 | 75 | 75 | 75 | 50 | 25 | 75 |
| Macaques | 45 | 0 | 0 | 75 | 0 | 25 | 0 | 25 |
At days 2 to 7 p.i., 19 mice were analyzed for each tissue. At days 14 to 21 p.i., 11 mice were analyzed. No viral RNA was detected at day 21 p.i. in mice. Four cynomolgus macaques were analyzed at each time point (total = 12). Sham-infected mice (n = 4) and macaques (n = 2) were negative for EMCV RNA.
ND, not done.
Pathogenesis of EMCV 30/87 in pigs.
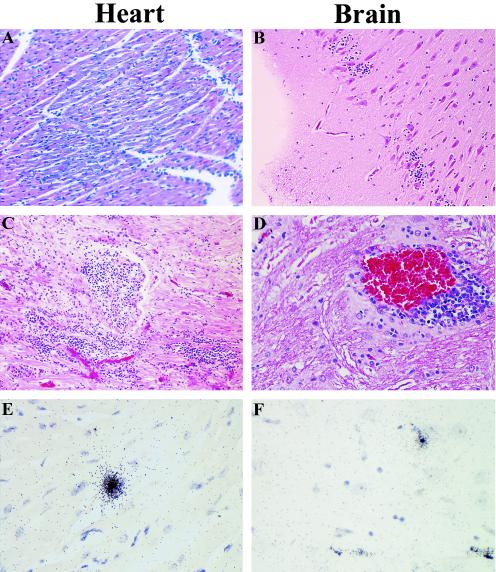
The detailed clinical and histopathologic profile of EMCV 30/87 in swine has been described previously (4). Limited experiments performed to provide data for direct comparison with viral pathogenesis in mice and cynomolgus macaques reaffirmed that the virus replicated primarily in the brain, heart, and spleen, rarely in the liver, and not in the kidneys and skeletal muscles during the first 7 days. However, in the chronic phase of the disease, persistent viral RNA was more widespread and was detected in the brain, liver, heart, spleen, kidney, pancreas, and skeletal muscles. Histopathologic changes in both the acute and chronic phases were confined to the heart and brain. Pigs sacrificed in the acute phase of the disease (days 7 to 21 p.i.) exhibited severe myocardial lesions, including multiple foci of lymphocytic infiltration, degeneration, and necrosis (Fig. 4A). Brain changes included infiltration of lymphocytes in the cerebral cortices (Fig. 4B) and meninges, as well as perivascular cuffing in the cerebral cortex and hippocampi. In the chronic phase of the disease (days 45 to 90 p.i.), multiple discrete foci of myocardial mineralization and lymphocytic infiltration were observed (Fig. 4C), which were larger and more numerous than the lesions observed in macaques. Brain lesions in the chronic stages of disease were characterized by perivascular cuffing in the cerebral cortex (Fig. 4D), medulla, and cerebellum. EMCV persistence in the heart, brain, liver, kidney, spleen, skeletal muscle, pancreas, and mesenteric lymph nodes during the acute phase, but primarily in the heart (Fig. 4E) and brain (Fig. 4F) in the chronic phase, was demonstrated by using in situ hybridization and RT-PCR. There was mild lymphocytic infiltration in the spleen in the acute phase, but no pathological changes were detected in the liver, pancreas, kidney, skeletal muscles, or mesenteric lymph nodes at any stage of the disease.
FIG. 4.
EMCV-induced pathological changes and detection of viral RNA in pigs. Five-week-old pigs were intraperitoneally inoculated with 2.9 × 108 PFU of EMCV 30, and histopathologic changes and the presence of EMCV RNA in the heart, brain, spleen, pancreas, liver, kidney, and skeletal muscle were analyzed 7, 21, 45, and 90 days p.i. In the acute phase (7 days p.i.), inflammation and degenerative changes were observed in the heart (A), and brain (B), but no changes were observed in the other organs. In the chronic phase (21 to 90 days p.i), infiltration of lymphocytes in the heart (C) and perivascular cuffing in the brain (D) were evident throughout the infection period up to day 90 p.i. and were accompanied by persistence, as demonstrated by localization of EMCV RNA by in situ hybridization (E and F). For histopathology, heart and brain sections were embedded in paraffin, and 4-μm-thick sections were stained with hematoxylin and eosin. In situ hybridization was performed by using a 35S-labeled VP1 cDNA probe.
EMCV pathogenesis in cynomolgus macaques.
As shown in Fig. 2, none of the 12 infected macaques developed overt clinical signs, and there were no fatalities among them, even though the same dose of virus (2.9 × 108 PFU) was used to infect both pigs and macaques. Following EMCV 30/87 inoculation, viral RNA was detected in the blood, spleen, liver, heart, kidney, brain, and skeletal muscles (Fig. 5D, E, F, and J; Table 4). The viral RNA localized by in situ hybridization in the heart (Fig. 5D) and brain (Fig. 5E) were associated with several pathological changes. However, although viral RNA was detected in the kidney, liver, spleen, pancreas, or skeletal muscles at day 7 p.i., no histopathologic abnormalities were demonstrated in these organs, as illustrated by the micrograph showing viral RNA in the kidney (Fig. 5F) despite the normal histology of the organ (Fig. 5C). Histopathologic changes in the brain and heart were observed in three of the four macaques at day 7 p.i. In the myocardium, there were numerous large foci of inflammation (Fig. 5A) associated with necrosis and degeneration of cardiac myocytes. The lesions were similar to those observed in acutely infected pigs but were less frequent, suggesting that primate myocytes were not as susceptible to the porcine virus. Histopathologic changes in the brain included numerous foci of perivascular cuffing (Fig. 5B), which were most prominent in the cerebrum and the cerebral cortex.
FIG.5.
EMCV-induced pathological changes and detection of viral RNA in cynomolgus macaques. Adult macaques were intraperitoneally inoculated with 2.9 × 108 PFU of EMCV 30/87, and histopathologic changes were analyzed 7, 21, and 45 days later. At day 7 p.i., there were acute inflammatory and mild degenerative changes in the myocardium (A) and brain (B, arrows), whereas there were no detectable changes in kidneys (C), livers, and spleens from the same animals. In contrast, EMCV RNA was widely distributed in tissues at day 7 p.i., as demonstrated by in situ hybridization (black grains) in the myocardium (D), brain (E), and kidneys (F) and by RT-PCR in the heart (Ht), brain (Br), liver (L), spleen (S), kidney (K), pancreas (P), and skeletal muscle (SM) (J). At days 21 and 45 p.i., mild lymphocytic infiltration was observed in the heart (I, arrow), and the formation of prominent germinal centers was observed in the spleen (H, arrows), in contrast to spleens from 7-day EMCV-infected macaques (G, arrow). EMCV RNA was most consistently observed in the spleen and pancreas at days 21 and 45 p.i. (J). In tissues with high levels of EMCV RNA at days 7 (all tissues), 21 (spleen), and 45 (spleen) p.i., nested RT-PCR using the VP2 primers followed by agarose gel electrophoresis resulted in two visible bands, the larger (499 bp) representing the primary reaction product and the smaller (350 bp) representing the nesting reaction product. For histopathology, macaque tissue sections were embedded in paraffin and 4-μm-thick sections were stained with hematoxylin and eosin. In situ hybridization was performed using a 35S-labeled VP1 cDNA probe.
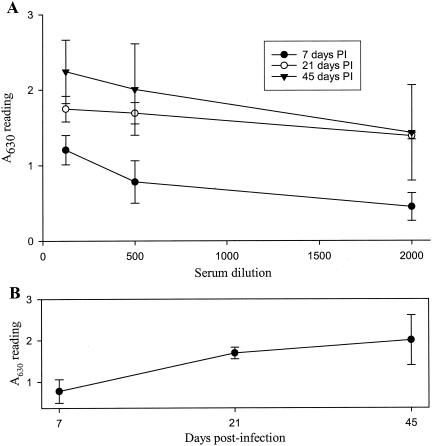
There were histopathologic changes in 7 of 8 spleens (87.5%), 3 of 8 hearts (37.5%), and 2 of 8 brains (25%) for macaques infected for 21 and 45 days. Splenic lesions were characterized by highly prominent germinal centers (Fig. 5H) in the chronically infected macaques compared to those in uninfected or 7-day-infected macaques (Fig. 5G). Myocardial lesions were characterized by small discrete foci of lymphocytic infiltration (Fig. 5I), with minimal disruption of cardiac myofiber arrangement. Brain lesions included small foci of infiltrating lymphocytes in blood vessel walls. Analysis of viral RNA distribution by in situ hybridization and RT-PCR demonstrated EMCV RNA most consistently in the spleen (75% of the cases at day 45 p.i.), but also in the heart (50%), brain (50%), pancreas (25%), and kidney (25%), as demonstrated in Table 4. Serum samples from macaques infected for 7 days had low levels of virus-specific IgG, whereas high levels of EMCV-specific IgG were detected at both days 21 and 45 p.i. (Fig. 6).
FIG. 6.
Detection by ELISA of EMCV-specific IgG in serum samples from experimentally infected cynomolgus macaques. (A) Virus-specific IgG levels in serum samples from macaques infected for 7, 21, or 45 days. Data are presented as mean A630 readings (± standard errors) from four serum samples at each time point performed at serum dilutions between 1:500 and 1:2,000. (B) Profile of virus-specific IgG levels through the 45-day experimental period as determined at a 1:500 serum dilution.
DISCUSSION
Studies have suggested that the poly(C) tract in EMCV strains and Mengo virus may be a determinant in pathogenesis, virulence, and host range (11, 30, 33). Recombinant Mengo virus M/48 variants with shortened poly(C) tracts (C0 or C24) were highly attenuated, producing either no histopathologic changes (C0) or mild meningitis (C24) in 4-week-old mice (33). However, the Mengo virus variants with short poly(C) tracts had growth kinetics and plaque sizes similar to those of wild-type Mengo virus M/48 (C63) in vitro, and they were immunogenic in mice, pigs, and macaques, indicating that the viruses were infectious in these animals (10, 30, 33). After sequencing the genome of EMCV 30/87 and determining that it had the shortest poly(C) tract (CUC5UC8) of all wild-type EMCV strains, we determined the pathogenesis and virulence of the virus in various animal species. Our data demonstrate that EMCV 30/87, a low-passage-number (4 or 5 passages) natural isolate with a short poly(C) tract (CUC5UC8), is highly virulent, resulting in substantial fatalities in mice and pigs and severe pathological changes in mice, pigs, and macaques. Similar findings were observed with the wild-type strain 2887A/91 (C10UCUC3UC10), which induced acute fatal encephalitis characterized by hind limb paralysis as early as day 1 p.i. in mice and had a 50% lethal dose (LD50) of 1 PFU (19). The direct correlation between the length of the poly(C) tract and virulence may be specific to Mengo virus M/48, which is phylogenetically different from most of the EMCVs, as shown in Fig. 1. This hypothesis is supported by studies using poly(C) tract variants from both EMCV R/45 and PV2/58, which demonstrated no correlation between the length of the poly(C) tract and virulence (14, 34, 48).
Another possible explanation for the correlation between virulence and the length of the poly(C) tract in Mengo M variants is that continuous passage and genetic engineering of EMCV strains and Mengo virus result in significant attenuation, as demonstrated with strain 2887A/91 (19). Comparison of the parent strain 2887A/91 and an infectious cDNA clone generated from the parent virus demonstrated that the recombinant 2887A/91 clone had smaller plaque sizes. Mice inoculated with the infectious 2887A/91 cDNA clone developed delayed clinical signs (from day 5 p.i.), with an LD50 of 100 PFU, whereas mice infected with the 2887A/91 parent virus developed clinical signs from day 1 p.i., and the LD50 was 1 PFU (19). A more intriguing fact is that EMCV 30/87 and strain 2887A/91, both recent isolates from pigs, have 4- to 10-fold-shorter poly(C) tracts than the older EMCVs (Table 1), suggesting that evolutionary pressure may favor the emergence of EMCV strains with shorter poly(C) tracts, at least in pigs. A number of more recent EMCV strains have been isolated from pigs and rodents, but their genomic structures, in particular the lengths of their poly(C) tracts, have not been determined (22, 23).
It was demonstrated previously that experimental infection of pigs with EMCV 30/87 resulted in acute fatal cardiac failure for 16% of the pigs and persistent infection in myocardial and CNS cells for the remaining 84% (4). In addition, EMCV 30/87 productively infected primary human cardiomyocytes, suggesting that undetected EMCV in porcine neural or myocardial tissues transplanted into humans may result in severe infection in recipients. The studies described in this report were also undertaken in order to determine the ability of EMCVs to cause interspecies disease outbreaks by comparing the pathogenesis and disease phenotypes induced by EMCV 30/87 in C57BL/6 mice, pigs, and cynomolgus macaques. Infection of mice, pigs, and cynomolgus macaques resulted in similar EMCV 30/87 pathogenesis, characterized by generalized virus distribution followed by localization of virus in the myocardium and CNS, which were the primary sites of virus replication and histopathologic changes in the three animal species. Viral RNA was detected in the blood, spleen, kidneys, liver, heart, and brain in 75 to100% of infected macaques, and in more than 50% of infected mice, within 7 days p.i. Interestingly, the virus spread was slower in pigs, requiring as long as 21 days for detection of viral RNA in all the organs. The most severe lesions (based on the extent of lymphocytic infiltration and the number of lesions) were observed in pigs, followed by mice, with macaques having the least severe disease.
The differences in disease phenotype among the three animal species were intriguing. All infected mice developed acute encephalitis characterized by paresis and flaccid paralysis of the posterior limbs and resulting in death from acute respiratory failure between days 2 and 7 p.i. In contrast, a small proportion of pigs (16%), the original host of the virus, died from sudden cardiac failure 2 days after infection, whereas the rest of the animals remained overtly asymptomatic for as long as 90 days p.i., despite extensive myocardial and CNS histopathologic changes. All EMCV 30/87-infected macaques remained asymptomatic for the entire experimental period (45 days), despite extensive myocardial pathological changes and virus persistence in more than 50% of the animals. Although limited to mice only, previous studies suggested that EMCV strains isolated from pigs or primates could induce severe disease in other animals. EMCV R/45, isolated from a 5-year-old chimpanzee suffering from acute fatal myocarditis in 1945, induced paralysis of the hind legs and interstitial myocarditis in mice (15). Mengo virus M/48, isolated from the cerebrospinal fluid of a paralyzed rhesus macaque, induced paralysis, characterized by lesions in brain and spinal cords, and myocarditis in mice (8). EMC-B/58 and EMC-D/58, variants of EMC-M isolated from a naturally infected domestic pig suffering from severe myocarditis, have been widely used to study the pathogenesis of viral diabetes in a mouse model (18, 29, 46). Strain 2887A/91 induced an acute fatal encephalitis characterized by paralysis of posterior limbs as early as 1 day after infection in mice (19).
Since we have demonstrated that porcine EMCV can productively infect human cardiomyocytes, cynomolgus macaques were infected with EMCV 30/87 as a model of pig-to-human xenozoonosis, designed to provide an indication of porcine EMCV virulence and pathogenesis in humans. The absence of severe clinical disease or death in macaques infected with EMCV 30/87 may suggest that porcine EMCV isolates transmitted during xenotransplantation may not produce severe clinical disease in humans. However, the absence of overt disease may have been due to the fact that the macaques were 2 to 4 years old at the time of EMCV infection, whereas the pigs and mice were 5 weeks old at infection. The finding of widespread virus distribution among internal organs and severe histopathologic changes in the macaques is more significant, particularly because recent studies in our laboratory have demonstrated the transmission of EMCV 30/87 and induction of disease in mice following transplantation (intra-abdominal) of porcine myocardial or pancreatic tissues (3). Using a more clinically relevant model, we have demonstrated that EMCV 30/87-infected porcine islet cells can transmit the virus and induce acute fatal disease in mice (L. A. Brewer, R. S. LaRue, B. Hering, and M. K. Njenga, submitted for publication). Given that long-term immunosuppression will probably accompany xenotransplantation, the findings that a porcine strain of EMCV can establish severe infection in primates and that it can be transmitted during transplantation indicate the need to develop a rapid test to screen pig tissues harvested for transplantation for EMCV RNA.
Acknowledgments
R. LaRue and S. Myers contributed equally to the generation of this report.
National Institutes of Health grant HL04369 supported this research.
We thank Jeremy Alley, Richard Bennett, and Humphrey Lwamba for technical assistance and Michael P. Murtaugh for his critique and valuable discussions.
REFERENCES
- 1.Bae, Y. S., H. M. Eun, and J. W. Yoon. 1989. Genomic differences between diabetogenic and nondiabetogenic variants of encephalomyocarditis virus. Virology 170:282-287. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard, J. L., K. F. Soike, and G. B. Baskin. 1987. Encephalomyocarditis virus infection in African green and squirrel monkeys: comparison of pathological effects. Lab. Anim. Sci. 37:635-639. [PubMed] [Google Scholar]
- 3.Brewer, L. A., C. Brown, M. P. Murtaugh, and M. K. Njenga. Transmission of porcine encephalomyocarditis virus (EMCV) to mice by transplanting EMCV-infected pig tissues. Xenotransplantation, in press. [DOI] [PubMed]
- 4.Brewer, L. A., H. C. M. Lwamba, M. P. Murtaugh, A. C. Palmenberg, C. Brown, and M. K. Njenga. 2001. Porcine encephalomyocarditis virus persists in pig myocardium and infects human myocardial cells. J. Virol. 75:11621-11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerutis, D. R., R. H. Bruner, D. C. Thomas, and D. J. Giro. 1989. Tropism and histopathology of the D, B, K and MM variants of encephalomyocarditis virus. J. Med. Virol. 29:63-69. [DOI] [PubMed]
- 6.Craighead, J. E. 1966. Pathogenicity of the M and E variants of the encephalomyocarditis (EMC) virus. I. Myocardiotropic and neurotropic properties. Am. J. Pathol. 48:333-345. [PMC free article] [PubMed] [Google Scholar]
- 7.Craighead, J. E., and M. F. McLane. 1968. Diabetes mellitus: induction in mice by encephalomyocarditis virus. Science 162:913-914. [DOI] [PubMed] [Google Scholar]
- 8.Dick, G. W. A., K. C. Smithburn, and A. J. Haddow. 1948. Mengo encephalomyelitis virus: isolation and immunological properties. Br. J. Exp. Pathol. 29:547-558. [Google Scholar]
- 9.Duke, G. M., M. A. Hoffman, and A. C. Palmenberg. 1992. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 66:1602-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duke, G. M., and A. C. Palmenberg. 1989. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J. Virol. 63:1822-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duke, G. M., J. E. Osorio, and A. C. Palmenberg. 1990. Attenuation of Mengovirus through genetic engineering of the 5′ non-coding poly(C) tract. Nature 343:474-476. [DOI] [PubMed] [Google Scholar]
- 12.Gajdusek, C. 1955. Encephalomyocarditis infection in childhood. Pediatrics 16:819. [PubMed] [Google Scholar]
- 13.Grobler, D. G., J. P. Raath, L. E. O. Braack, D. F. Keet, G. H. Gerdes, B. J. H. Barnard, N. P. J. Kriek, J. Jardine, and R. Swanepoel. 1995. An outbreak of encephalomyocarditis-virus infection in free-ranging African elephants in the Kruger National Park. Onderstepoort J. Vet. Res. 62:97-108. [PubMed] [Google Scholar]
- 14.Hahn, H., and A. C. Palmenberg. 1995. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their mengovirus counterparts. J. Virol. 69:2697-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helwig, F. C., and E. C. H. Schmidt. 1945. A filter-passing agent producing interstitial myocarditis in anthropoid apes and small animals. Science 102:31-33. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard, G. B., K. F. Soike, T. M. Butler, K. D. Carey, H. Davis, W. I. Butcher, and C. J. Gauntt. 1992. An encephalomyocarditis virus epizootic in a baboon colony. Lab. Anim. Sci. 42:233-239. [PubMed] [Google Scholar]
- 17.Joo, H. S. 1999. Encephalomyocarditis virus, p. 139-144. In B. E. Straw, S. D'Allare, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames.
- 18.Jordan, G. W., and S. H. Cohen. 1987. Encephalomyocarditis virus-induced diabetes mellitus in mice: model of viral pathogenesis. Rev. Infect. Dis. 9:917-924. [DOI] [PubMed] [Google Scholar]
- 19.Kassimi, L. B., A. Boutrouille, M. Gonzague, A. L. Mbanda, and C. Cruciere. 2002. Nucleotide sequence and construction of an infectious cDNA clone of an EMCV strain isolated from aborted swine fetus. Virus Res. 83:71-87. [DOI] [PubMed] [Google Scholar]
- 20.Kim, H. S., W. T. Christianson, and H. S. Joo. 1989. Pathogenic properties of Encephalomyocarditis virus isolated from aborted swine fetuses. Arch. Virol. 109:51-57. [DOI] [PubMed] [Google Scholar]
- 21.Kirkland, P. D., A. B. Gleeson, R. A. Hawkes, H. M. Naim, and C. R. Broughton. 1989. Human infection with encephalomyocarditis virus in New South Wales. Med. J. Aust. 151:176-177. [DOI] [PubMed] [Google Scholar]
- 22.Knowles, N. J., N. D. Dickinson, G. Wilson, E. Cerra, E. Brocchi, and F. De Simone. 1998. Molecular analysis of encephalomyelitis viruses isolated from pigs and rodents in Italy. Virus Res. 57:53-62. [DOI] [PubMed] [Google Scholar]
- 23.Koenen, F., H. Vanderhallen, N. D. Dickinson, and N. J. Knowles. 1999. Phylogenetic analysis of European encephalomyocarditis viruses: comparison of two genomic regions. Arch. Virol. 144:893-903. [DOI] [PubMed] [Google Scholar]
- 24.Koenen, F., K. De Clercq, and R. Strobbe. 1991. Isolation of encephalomyocarditis virus in the offspring of swine with reproductive failure in Belgium. Vlaams. Diergeneeskd. Tijdschr. 60:113-115. [Google Scholar]
- 25.Kruppenbacher, J. P., T. Mertens, H. Muntefering, and H. J. Eggers. 1985. Encephalomyocarditis virus and diabetes mellitus: studies on virus mutants in susceptible and non-susceptible mice. J. Gen. Virol. 66:727-732. [DOI] [PubMed] [Google Scholar]
- 26.Leyssen, P., N. Charlier, P. Lemey, F. Billoir, A. M. Vandamme, E. De Clercq, X. de Lamballerie, and J. Neyts. 2002. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of the Modoc virus, a flavivirus with no known vector. Virology 293:125-140. [DOI] [PubMed] [Google Scholar]
- 27.Low, W. C., J. K. Daniloff, R. P. Bodony, and J. Wells. 1985. Cross-species transplants of cholinergic neurons and the recovery of function, p. 575-584. In A. Bjorkmund and U. Stenevi (ed.), Neural grafting in the mammalian CNS. Elsevier Science Publication, Amsterdam, The Netherlands.
- 28.Murname, T. G. 1981. Encephalomyocarditis, p. 137-147. In G. W. Beran (ed.), CRC handbook series in zoonoses, section B, vol. 2. Viral zoonoses. CRC Press, Boca Raton, Fla.
- 29.Murname, T. G., J. E. Craighead, H. Mondragon, and A. Shelokov. 1960. Fatal disease of swine due to encephalomyocarditis virus. Science 131:498-499. [DOI] [PubMed] [Google Scholar]
- 30.Neal, Z. C., and G. A. Splitter. 1995. Picornavirus-specific CD4+ T lymphocytes possessing cytolytic activity confer protection in the absence of prophylactic antibodies. J. Virol. 69:4914-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelsen-Salz, B., A. Zimmermann, S. Wickert, G. Arnold, A. Botta, H. J. Eggers, and J. P. Kruppenbacher. 1996. Analysis of sequence and pathogenic properties of two variants of encephalomyocarditis virus differing in a single amino acid in VP1. Virus Res. 41:109-122. [DOI] [PubMed] [Google Scholar]
- 32.Njenga, M. K., K. Asakura, S. F. Hunter, P. Wettstein, L. R. Pease, and M. Rodriguez. 1997. The immune system preferentially clears Theiler's virus from the gray matter of the central nervous system. J. Virol. 71:8592-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osorio, J. E., L. R. Martin, and A. C. Palmenberg. 1996. The immunogenic and pathogenic potential of short poly(C) tract Mengo viruses. Virology 223:344-350. [DOI] [PubMed] [Google Scholar]
- 34.Osorio, J. E., Z. C. Neal, M. S. McBride, and A. C. Palmenberg. 2000. Mengovirus and encephalomyocarditis virus poly(C) tract lengths can affect virus growth in murine cell culture. J. Virol. 74:3074-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pakzaban, P., and O. Isaacson. 1994. Neural xenotransplantation: reconstruction of neuronal circuitry across species barriers. Neuroscience 62:989-1001. [DOI] [PubMed]
- 36.Petruccelli, M. A., K. Hirasawa, M. Takeda, S. Itagaki, and K. Doi. 1991. Cardiac and pancreatic lesions in guinea pigs infected with encephalomyocarditis (EMC) virus. Histol. Histopathol. 6:167-170. [PubMed] [Google Scholar]
- 37.Quinn, N. 1990. The clinical application of cell grafting techniques in patients with Parkinson's disease. Prog. Brain Res. 82:619-625. [DOI] [PubMed] [Google Scholar]
- 38.Reddacliff, L. A., P. D. Kirland, W. J. Hartley, and R. L. Reece. 1997. Encephalomyocarditis virus infections in an Australian zoo. J. Zoo Wildl. Med. 28:153-157. [PubMed] [Google Scholar]
- 39.Seaman, J. T., J. G. Boulton, and M. J. Carrigan. 1986. Encephalomyocarditis virus disease of pigs associated with a plague of rodents. Aust. Vet. J. 63:292-294. [DOI] [PubMed] [Google Scholar]
- 40.Secchi, A., C. Socci, P. Maffi, M. V. Taglietti, L. Falqui, F. Bertuzzi, P. De Nittis, L. Piementi, L. Scopsi, V. Di Carlo, and G. Pozza. 1997. Islet transplantation in IDDM patients. Diabetologica 40:225-231. [DOI] [PubMed] [Google Scholar]
- 41.Spadbrow, P. B., and Y. S. Chung. 1970. Hemagglutination-inhibition antibodies to encephalomyocarditis virus in Queensland cattle. Aust. Vet. J. 46:126-128. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thou, J., I. Date, K. Sakai, Y. Yoshimoto, T. Furuta, S. Asari, and T. Ohmoto. 1993. Suppression of immunorejection against rat fetal dopaminergic neurons in a mouse brain by 15-deoxyspergualin. Brain Res. 621:155-160. [DOI] [PubMed] [Google Scholar]
- 44.Verlinde, J. D., and H. E. Van Tangeren. 1953. Human infections with viruses of the Columbia-SK group. Arch. Gesamte Virusforsch. 5:217-230. [DOI] [PubMed] [Google Scholar]
- 45.Wells, S. K., and A. E. Gutter. 1989. Encephalomyocarditis virus: epizootic in a zoological collection. J. Zoo Wildl. Med. 20:291-296. [Google Scholar]
- 46.Yoon, J. W., P. R. McClintock, T. Onodera, and A. L. Notkins. 1980. Virus-induced diabetes mellitus. XVIII. Inhibition by a non-diabetogenic variant of encephalomyocarditis virus. J. Exp. Med. 152:878-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmerman, J. J., E. E. Hill, K. E. Smith, B. L. Kneeland, K. B. Platt, H. T. Hill, G. W. Beran, W. R. Clark, and L. D. Miller. 1994. Encephalomyocarditis virus infection in raccoons (Procyon lotor). J. Zoo Wildl. Med. 25:233-239. [Google Scholar]
- 48.Zimmermann, A., A. Botta, G. Arnold, H. J. Eggers, and B. Nelsen-Salz. 1997. The poly(C) region affects progression of encephalomyocarditis virus infection in the Langerhans' islets but not in the myocardium. J. Virol. 71:4145-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmermann, A., B. Nelsen-Salz, J. P. Kruppenbacher, and H. J. Eggers. 1994. The complete nucleotide sequence and construction of an infectious cDNA clone of a highly virulent encephalomyocarditis virus. Virology 203:366-372. [DOI] [PubMed] [Google Scholar]