Abstract
Transforming growth factor β1 (TGF-β1) signaling is compromised in many tumors, thereby allowing the tumor to escape the growth-inhibitory and proapoptotic activities of the cytokine. Human adenoviruses interfere with a number of cellular pathways involved in cell cycle regulation and apoptosis, initially placing the cell in a “tumor-like” state by forcing quiescent cells into the cell cycle and also inhibiting apoptosis. We report that adenovirus-infected cells resemble tumor cells in that TGF-β1 signaling is inhibited. The levels of TGF-β1 receptor II (TβRII) in adenovirus-infected cells were decreased, and this decrease was mapped, by using virus mutants, to the E1A gene and to amino acids 2 to 36 and the C-terminal binding protein binding site in the E1A protein. The decrease in the TβRII protein was accompanied by a decrease in TβRII mRNA. The decrease in TβRII protein levels in adenovirus-infected cells was greater than the decrease in TβRII mRNA, suggesting that downregulation of the TβRII protein may occur through more than one mechanism. Surprisingly in this context, the half-lives of the TβRII protein in infected and uninfected cells were similar. TGF-β1 signaling was compromised in cells infected with wild-type adenovirus, as measured with 3TP-lux, a TGF-β-sensitive reporter plasmid expressing luciferase. Adenovirus mutants deficient in TβRII downregulation did not inhibit TGF-β1 signaling. TGF-β1 pretreatment reduced the relative abundance of adenovirus structural proteins in infected cells, an effect that was potentiated when cells were infected with mutants incapable of modulating the TGF-β signaling pathway. These results raise the possibility that inhibition of TGF-β signaling by E1A is a means by which adenovirus counters the antiviral defenses of the host.
Transforming growth factor β1 (TGF-β1) is a prototype member of a family of multifunctional cytokines (36). Originally discovered as a fibroblast growth factor, TGF-β1 was soon found to play an important role in a variety of physiological processes including immunoregulation, the cell cycle, apoptosis, and formation of the extracellular matrix (14). In epithelial cells, TGF-β1 negatively affects the cell cycle primarily through transcriptional upregulation of cyclin-dependent kinase inhibitors (37). In the immune system, TGF-β1 along with interleukin-10 functions to control and limit the extent of the adaptive immune response (14, 23).
The signaling pathways for all members of the TGF-β1 family are similar (36). Intracellular signaling is initiated upon the binding of the active cytokine to the TGF-β receptor II (TβRII) homodimer and the assembly of a heterotetrameric complex consisting of receptors I and II. TβRII is a ubiquitously expressed constitutively active serine/threonine kinase (60, 61). Once the heterotetrameric receptor complex is formed, TβRII phosphorylates TβRI and thereby greatly enhances TβRI serine/threonine kinase activity. The Smad family of proteins includes secondary mediators of TGF-β signaling (39). Receptor-specific Smads that are phosphorylated by activated TβRI associate with Smad 4 and other factors to form a transcriptionally competent complex that enters the nucleus and modulates gene expression.
The TGF-β1 signaling pathway is inactivated in many tumors, presumably allowing the tumors to escape TGF-β1-mediated growth inhibition and apoptosis (13, 37). Frequently, inhibition of TGF-β1 signaling occurs by either abolition of the function of a common mediator, Smad 4, or by interference with TβRII function. Some of the reported mechanisms of TβRII downregulation include inhibition of promoter activity (33), decrease in mRNA stability (29), and intracellular retention (8).
Human adenovirus causes a number of benign diseases (26) and may establish persistency in lymphoid cells (21). Quiescent epithelial cells are believed to be the main target of acutely replicating adenovirus in vivo. Infection is divided into two stages, early and late. Early genes begin to be expressed prior to viral DNA replication and encode proteins that usurp the cell (58). Progression into the late stage of infection and successful completion of the viral life cycle require replication of the viral DNA genome. The cellular DNA synthesis machinery may facilitate viral genome replication; upon infection, cell cycle perturbations in quiescent epithelial cells, primarily due to the adenovirus E1A proteins, are observed (17). In addition, the adenovirus genome encodes a number of proteins that counteract host cell apoptosis, whether it is induced by unscheduled cell cycle progression or mediated by the immune system (25, 38, 58). Because of unscheduled entry into the cell cycle and protection against apoptosis, adenovirus-infected cells are forced into a tumor-like state.
Increased levels of active TGF-β1 are created at the site of inflammation through the release of active cytokines by a subpopulation of macrophages and regulatory T cells and by local activation of extracellular matrix-associated latent complexes (24). It seems possible that the growth-inhibitory and proapoptotic functions of TGF-β1 would be both inhibitory to viral DNA replication and detrimental to the survival of infected epithelial cells. In addition or alternatively, the immunoregulatory function of TGF-β1 could diminish the antiadenovirus immune response and accordingly may be beneficial for in vivo adenovirus replication and possibly persistence. Therefore, it is reasonable to consider whether disruption of the TGF-β1 signaling pathway might occur in adenovirus-infected cells.
Here we report that adenovirus mediates a decrease in TβRII protein levels and that the E1A proteins are responsible for the effect. This decrease is accompanied by a reduction in steady-state TβRII mRNA levels. TGF-β1-mediated signaling in infected cells is inhibited; adenovirus mutants that fail to downregulate TβRII do not inhibit TGF-β1-mediated signaling. Finally, activation of the TGF-β1 pathway decreases the abundance of adenovirus structural proteins in infected cells.
MATERIALS AND METHODS
Cell lines.
Human A549 lung adenocarcinoma and human HepG2 hepatocellular carcinoma cell lines were purchased from the American Type Culture Collection. A549 cells were grown in Dulbecco's modified Eagle medium (DMEM; JRH Biosciences, Lenexa, Kans.) supplemented with 10% fetal calf serum (FCS; HyClone, Logan, Utah). HepG2 cells were grown in DMEM-F12 medium supplemented with 10% FCS.
Adenovirus mutants.
Ad2, Ad5, and rec700 were used as wild-type controls. rec700 is a recombinant virus derived from Ad2 and Ad5 with an E1A region from Ad5 (59). E1A.2-36 and E1A.81-120 (47) are adenovirus mutants that lack amino acids 2 to 36 and 81 to 120, respectively, in the E1A proteins (kindly provided by Elizabeth Moran, Temple University). pm975 and 12Swt express only the 13S and 12S E1A isoforms, respectively (40, 48). dl808 lacks the E4 region (deletion encompasses map units 92.0 to 97.1) (7). dl7001 and dl327 lack the entire E3 region, with the exception of the gene for E3-12.5K present in dl327 (44). dl764 and dl753, mutants, derived from rec700, lack the RIDβ and RIDα genes, respectively (5, 52). 12S.2-36 and 12S.928 are 12Swt-based mutants with an E1A N terminus deletion and a point mutation, respectively. dl313 expresses E1A proteins lacking amino acids 220 to 289. dl312 lacks the E1A region (30). Ad/E3 is an E1-negative replication-defective adenovirus vector expressing E3 proteins under the control of the cytomegalovirus (CMV) promoter (54). 176-9 (dC-term) is a 12S mutant with a deletion of amino acids 224 to 284 in the E1A protein (49). dl118 is an E1B deletion mutant (2). Mutants E1A.2-36, E1A.81-120, pm975, 12Swt, 12S.2-36, 12S.928, dl313, dl312, and dl118 are all in a dl309 genetic background. dl309 is an Ad5 mutant that lacks the genes for the E3 RIDα, RIDβ, and 14.7K proteins.
Antibodies and reagents.
Rabbit anti-TβRII antibodies used for Western analysis were purchased from Santa Cruz Biotechnology (catalog numbers sc-400 and sc-220; Santa Cruz, Calif.). Affinity-purified goat anti-TβRII raised against an extracellular receptor domain was purchased from R&D Systems (catalog number AF-241-NA; Minneapolis, Minn.) and used for immunoprecipitations. A rabbit anti-Ad5 antibody was purchased from the American Type Culture Collection. The anti-phospho-Smad 2 antibody was from Upstate Biotechnology (Charlottesville, Va.). Recombinant human TGF-β1 was purchased from R&D Systems; it was reconstituted and stored in accordance with the manufacturer's instructions.
Western analysis.
HepG2 or A549 cells were washed three times with phosphate-buffered saline and lysed on ice in radioimmunoprecipitation assay buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA, 1 mM NaF) for 30 min. Aspirated buffer was centrifuged at 12,000 rpm in an Eppendorf centrifuge (model 5415C) for 10 min, and the protein concentration of the supernatant was measured by the Bio-RadDC protein assay (Bio-Rad Laboratories, Hercules, Calif.). Equal protein amounts of 50 (TβRII) or 30 μg (adenovirus late proteins) were loaded into each lane of the SDS-8% polyacrylamide gel electrophoresis (PAGE) gel. Proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.) and incubated with a combination of anti-TβRII antibodies (1:300 dilution each) or the anti-Ad5 antibody (1:1,600 dilution). Following application of the secondary horseradish peroxidase-conjugated antibody and subsequent washes, a signal was generated with a commercial chemiluminescence substrate (LumiGLO; KPL, Gaithersburg, Md.). The signal was detected by autoradiography and quantified by densitometry using FluorChem software (Alpha Innotech Corporation, San Leandro, Calif.).
Analysis of late protein synthesis.
A549 cells were maintained under 10 or 0.2% FCS or 0.2% FCS plus 5 ng of TGF-β1/ml for 3 days prior to infection and throughout the infection. During 3 days of pretreatment, medium was refreshed once to sustain the activity of the cytokine. Fresh medium was also supplied at the beginning of the infection.
RPA.
HepG2 cells were mock infected or infected with adenovirus mutants and maintained in the presence of AraC. Total RNA was isolated from infected and mock-infected HepG2 cells with TRIzol reagent (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Radioactively labeled RNA antisense probes were generated by the Riboprobe Combination System Sp6/T7 (Promega, Madison, Wis.). The TβRII probe was synthesized with T7 polymerase from a pBluescript TβRII probe plasmid linearized with HindIII. To generate the pBluescript TβRII probe, a HindIII-PstI TβRII fragment recovered from the mycTβRII plasmid (kindly provided by Yoav Henis, Tel Aviv University) was cloned into the pBluescript (−) multiple cloning site. The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe was synthesized with Sp6 polymerase from an XbaI-linearized template (kindly provided by Rob Fleming, Saint Louis University). Thirty (TβRII) or 6 (GAPDH) μg of total RNA was hybridized to the freshly made radioactively labeled probes and processed with a commercially available RNase protection assay (RPA) kit (RPAIII; Ambion, Dallas, Tex.) according to the manufacturer's instructions.
Immunoprecipitations.
A549 cells were mock infected or infected with rec700 at a multiplicity of infection of 50 PFU/cell and maintained in the presence of 20 μg of AraC/ml. At 18.5 h postinfection (p.i.), cells were washed and incubated with DMEM lacking cysteine and methionine for 30 min. At 19 h p.i., the preincubation medium was changed to fresh cysteine- and methionine-negative DMEM supplemented with 100 μCi of a [35S]methionine-cysteine mixture (EasyTag Express protein labeling mixture; Perkin-Elmer, Boston, Mass.)/ml. After 15 min of incubation in radioactive medium, cells were either immediately placed on ice (0-h chase) or washed three times with prewarmed cold DMEM and incubated in DMEM containing 10% FCS for the time period shown in Fig. 4A and D. At the end of a chase period, the cells were placed on ice, washed four times with ice-cold phosphate-buffered saline, and scraped into 100 μl of 0.5% NP-40 lysis buffer (20 mM sodium phosphate, 250 mM NaCl, 30 mM sodium pyrophosphate, 5 mM EDTA, 10 mM NaF). Cell lysates were normalized to the amount of protein in the lysate and combined with 0.25 μg of the antibody and 20 μl of 50% protein G-agarose (Roche, Indianapolis, Ind.). Following overnight incubation, the agarose beads were washed with 0.5% SDS lysis buffer; immunoprecipitated proteins were dissociated from the agarose in 2× Laemmli buffer and separated on SDS-8% PAGE gels. Gels were dried, and the signal was visualized by autoradiography and phosphorimaging with a STORM phosphorimager, with subsequent quantitation by ImageQuant, version 4.0, software.
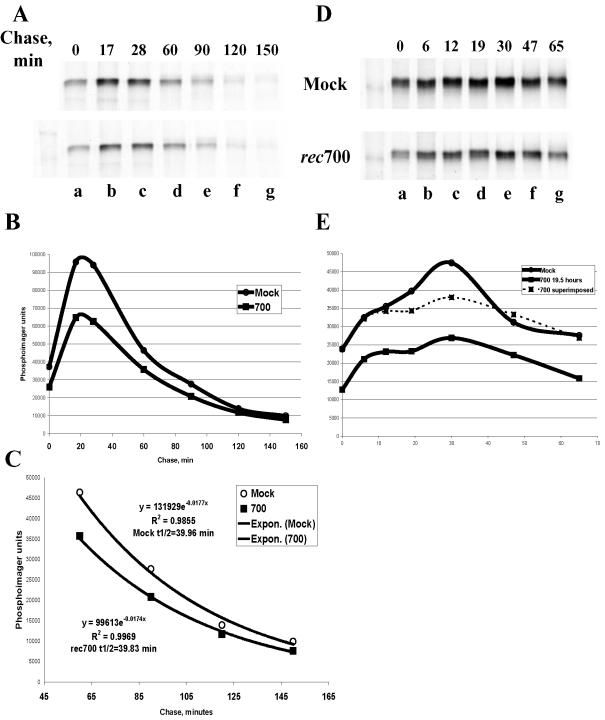
FIG. 4.
Stability of the TβRII protein as determined by pulse-chase analysis in adenovirus-infected cells. (A and D) In two independent experiments, A549 cells were mock infected or infected with rec700 at 50 PFU/cell and maintained in the presence of AraC throughout the infection. At 19 h p.i., cells were pulsed with [35S]methionine-cysteine for 15 min and chased in 10% FCS-supplemented nonradioactive medium for the indicated time periods. Immunoprecipitated, radioactively labeled TβRII was quantified by phosphorimager at each time point of chase (B and E, respectively). (C) Calculations of the TβRII half-life (t1/2).
Luciferase assay.
HepG2 cells seeded in 24-well dishes were transfected with 0.5 μg of 3TP-lux (kindly provided by J. Massague, Sloan-Kettering Institute) and 0.3 μg of pCMV-β Gal plasmid. At 6 h posttransfection, HepG2 cells were mock infected or infected with rec700 at 50 PFU/cell. Infections were maintained in the presence of freshly supplied AraC at 20 μg/ml. At 12 h p.i., medium was replaced with serum-free medium containing 5 ng of human recombinant TGF-β1 (R&D Systems)/ml. At 26 h p.i., cells were lysed, and luciferase activity was measured with a luminometer by using the luciferase assay system (Promega). To determine β-Gal activity, the same volume of cell lysate was incubated in assay buffer (200 mM sodium phosphate [pH 7.3], 2 mM MgCl2, 100 mM β-mercaptoethanol, 1.33 mg of o-nitrophenyl-β-d-galactopyranoside [ONPG]/ml) and measured on a spectrophotometer at 405 nm. In place of infection, in some experiments cells were cotransfected with 0.5 μg of either 13S E1A or 12S E1A plasmids (kindly provided by G. Chinnadurai, Saint Louis University).
RESULTS
TβRII is downregulated in the course of adenovirus infection.
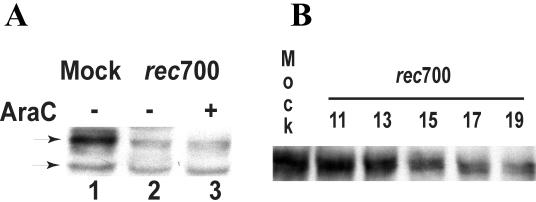
Figure 1A illustrates TβRII protein levels observed in human A549 cells infected with rec700, an Ad5-Ad2-Ad5 recombinant that functions as a wild-type adenovirus (59). Two forms of TβRII are shown: the mature form of the receptor and the short-lived TβRII precursor. (This precursor has been shown to be sensitive to endoglycosidase H digestion in vitro in extracts obtained from Mv1Lu cells [57]). TβRII levels in adenovirus-infected cells were significantly decreased (Fig. 1A); in multiple experiments, only 5 to 20% of TβRII proteins remained at 19 h p.i.
FIG. 1.
Adenovirus infection downregulates the TβRII protein. (A) A549 human lung adenocarcinoma cells were infected with rec700 at 50 PFU/cell. Some of the infected cells were maintained in 20 μg of AraC/ml. Cell lysates collected at 19 h p.i. were subjected to Western blotting using anti-TβRII antibodies (Santa Cruz Biotechnology). Upper and lower arrows, mature and immature forms of TβRII, respectively. (B) A549 cells were infected with rec700 at 10 PFU/cell. Cell lysates were collected at the indicated hours p.i. and analyzed by Western blotting. Equal protein amounts were loaded per lane.
Blocking DNA synthesis with AraC maintains adenovirus infection in the early stage (50). As shown in Fig. 1A (lane 3), TβRII levels in AraC-treated infected cells were decreased, indicating that the decrease is mediated by an adenovirus early protein. Similar results were obtained with HepG2 cells (data not shown).
A time course of TβRII downregulation in adenovirus-infected cells is presented in Fig. 1B. Only the mature form of TβRII is shown. The decrease in TβRII levels first became evident between 13 and 15 h p.i.
E1A is responsible for the decrease in TβRII protein levels in adenovirus-infected cells.
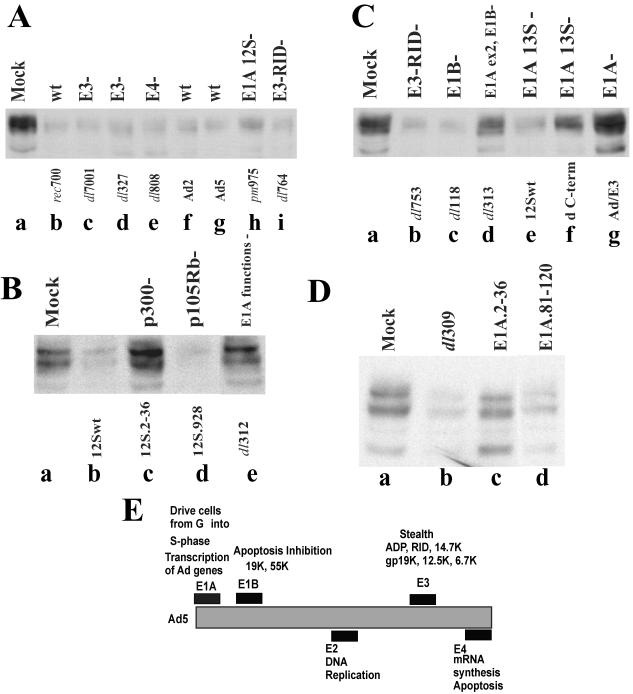
A set of adenovirus mutants was used to map TβRII downregulation to an early protein function. Wild-type rec700, Ad2, and Ad5 were highly effective in decreasing TβRII levels (Fig. 2A, lanes b, f, and g). Virus mutants lacking most of the E4 (dl808) or E1B (dl118) region were similar to wild-type virus in clearing TβRII (Fig. 2A, lane e, and C, lane c). Thus, the E1B and E4 regions are not required to downregulate TβRII.
FIG. 2.
The E1A 13S or 12S protein, including amino acids 2 to 36 and the CtBP binding site, is required to downregulate the TβRII protein in adenovirus-infected cells. A549 (A and B) or HepG2 (C and D) cells were infected with wild-type (wt) or mutant adenoviruses at 50 PFU/cell. Cell lysates were harvested at 24 h p.i. for the wild type and most mutants and at 48 h p.i. for dl313, 12Swt, dC-term, Ad/E3, 12S.2-36, 12S.928, and dl312. Cell lysates were subjected to Western blotting with anti-TβRII antibodies (Santa Cruz Biotechnology). (E) Schematic of the adenovirus (Ad) genome. The E1A proteins activate the transcription of adenovirus genes and deregulate the cell cycle by suppressing or activating cellular proteins and genes. E1B proteins suppress cellular apoptosis. E3 proteins confer a stealth function to the virus by inhibiting immune cell-mediated apoptosis. E4 proteins function in gene regulation, in part by facilitating degradation of p53; they are also required for viral mRNA transport from the nucleus. Virus DNA replication is necessary for late protein synthesis derived from the major late transcription unit. At about 24 h p.i., virions begin to assemble in the cell nucleus, and after 2 to 3 days cell lysis begins to occur, with the release of virions.
Several mutants with lesions in the E3 region were examined inasmuch as E3 proteins downregulate a number of cell surface receptors, including epidermal growth factor receptor, Fas, and TRAIL receptors 1 and 2 (3, 6, 16, 46, 51, 53). Somewhat surprisingly, deletion of all or nearly all of the E3 region (dl7001 and dl327; Fig. 2A, lanes c and d) or deletion of the gene for RIDα (dl753; Fig. 2C, lane b) or RIDβ (dl764; Fig. 2A, lane i) did not affect TβRII downregulation. In addition, expression of all E3 genes from the deleted E1 region of replication-defective adenovirus (Ad/E3) (54) failed to remove TβRII at 2 days p.i. (Fig. 2C, lane g). These results indicate that E3 proteins are not required to decrease TβRII levels.
Given that downregulation of TβRII is an early function and that the early E1B, E3, and E4 regions are not required to mediate the decrease in TβRII, the immediate-early E1A region was examined. Mutant dl312 lacks most of the E1A region (30); TβRII levels in cells infected with dl312 were not decreased (Fig. 2B, lane e), establishing that E1A expression is required for TβRII downregulation. There are two interpretations of this result: first, the E1A protein itself could be responsible for TβRII downregulation; second, considering that E1A is required for efficient induction of adenovirus genes, an E1A-inducible viral protein(s) could be responsible. We favor the first interpretation because under the conditions of the experiment, namely, high multiplicity of infection (50 PFU/cell) and a long period p.i. (48 h), it is known that adenovirus early genes are expressed in an E1A-independent manner (22, 41).
The E1A gene is expressed as two major alternatively spliced 13S and 12S mRNAs, which encode proteins of 289 (289R) and 243 (243R) amino acids, respectively. A mutant expressing either the 13S or 12S isoform of E1A (pm975 or 12Swt, respectively) was effective in downregulating TβRII (Fig. 2A, lane h, and C, lane e) (pm975 was not quite as effective as the wild type). The finding that 12S E1A alone is functional excludes a rigorous requirement for the E1A CR3 domain, which is present in the 289R protein but absent from the 243R protein, in suppressing TβRII levels.
Interestingly, when adenoviruses with mutations in E1A were examined, a defect in TβRII downregulation was observed. Mutant dl313 was mostly defective in clearing TβRII (Fig. 2C, lane d). Mutant dl313 expresses E1A proteins that are truncated at their C termini (deletion of amino acids 220 to 289), suggesting that the C-terminal portion of the E1A proteins is required for downregulation of TβRII. In accord with this suggestion, a phenotype similar to that of dl313 was observed in a 12Swt mutant (dC-term) with an E1A whose C terminus lacked amino acids 224 through 284 (including the C-terminal binding protein [CtBP] binding site) (Fig. 2C, lane f). (Mutant dl313 also lacks the E1B region, but given that the E1B mutant dl118 was not defective in downregulating TβRII [Fig. 2C, lane c], we presume that the defect is due to the deletion in the C-terminal region of E1A.)
In addition to the C-terminal region of E1A, the N-terminal region is also important because a 12Swt mutant (12S.2-36) with an E1A lacking amino acids 2 to 36 was completely incapable of TβRII downregulation (Fig. 2B, lane c). On the other hand, the absence of the pRB binding site in another 12Swt mutant (12S.928) had no effect on the efficiency of TβRII downregulation (Fig. 2B, lane d). The importance of the N-terminal E1A domain for effective TβRII downregulation was further confirmed by examining TβRII levels in cells infected with E1A mutants that express both the 13S and 12S forms of E1A (Fig. 2D). Mutants E1A.2-36 and E1A.81-120 (derived from dl309) express mutant E1A proteins lacking amino acids 2 to 36 and 81 to 120, respectively. As expected, the E1A.2-36 mutant was unable to downregulate TβRII, whereas E1A.81-120 was as efficient as the parent virus. All infections were monitored in parallel for expression of E1A and the E2 DNA binding protein (DBP) by immunofluorescence; typically, 90 to 95% of cells stained positive for E1A when applicable, and 60 to 70% were DBP positive, with a homogenous pattern of staining in cells incubated with AraC (data not shown).
We conclude that the 13S (289R) and 12S (243R) forms of E1A force downregulation of TβRII protein, that the N-terminal (amino acids 2 to 36) and C-terminal (amino acids 224 to 284) regions of the E1A proteins are required for TβRII downregulation, and that the pRb-binding region of the E1A protein is not required.
Adenovirus's ability to decrease TβRII protein levels correlates with its ability to downregulate TβRII mRNA.
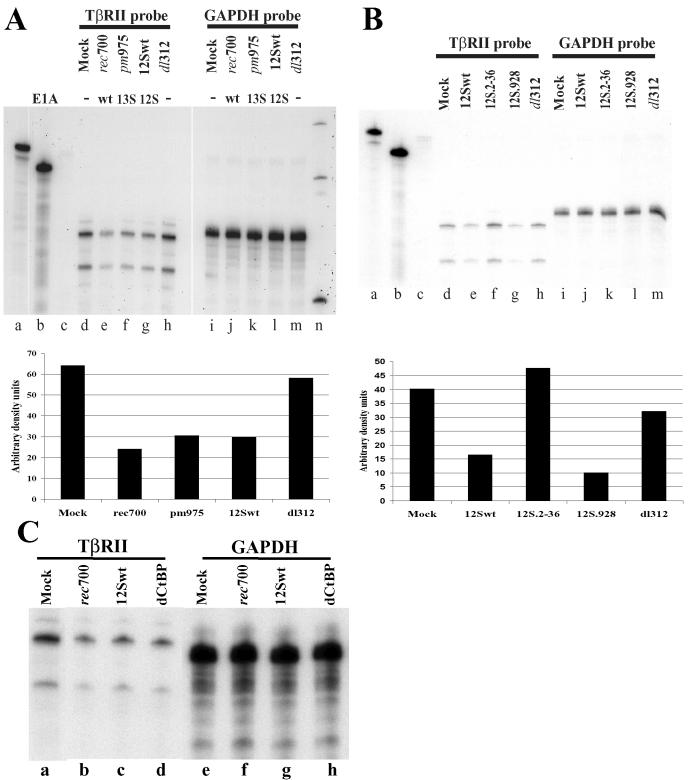
As a means to examine whether the decrease in TβRII protein levels was due to a decrease in TβRII mRNA levels, an RNase protection assay was employed to determine TβRII mRNA levels in mock-infected cells and cells infected with a limited set of E1A mutants. The densities of the RNase-protected fragments were analyzed with FluorChem software (Alpha Innotech Corporation) and are presented in the graphs under the corresponding autoradiography data (Fig. 3A and B). The density of each TβRII fragment was normalized against the density of the corresponding GAPDH signal. Figure 3A shows that the decrease in TβRII mRNA (seen as two RNase-protected bands) was most apparent in rec700-infected cells (compare lane e with lane d) (2.7- and 2.4-fold decreases in the TβRII signal compared to signals for mock infection and infection with E1A-negative mutant dl312, respectively). TβRII mRNA levels were also decreased in cells infected with a mutant expressing only the 13S or 12S isoform of E1A (pm975 or 12Swt, respectively; lanes f and g), but the decrease in TβRII mRNA due to these two mutants was consistently less than that due to the wild-type adenovirus (Fig. 3A and B). TβRII mRNA levels in cells infected with dl312 (Fig. 3A, lane h) were similar to those in mock-infected cells (lane d). Under the conditions of infection used (50 PFU/cell, 43 h p.i.), early and late virus genes are expressed by dl312 (22; our unpublished observations). Therefore, it is likely that TβRII mRNAs are not downregulated by dl312 because it does not make the E1A protein although it makes other adenovirus proteins.
FIG. 3.
E1A is required to reduce TβRII mRNA levels in adenovirus-infected cells. HepG2 cells were infected with wild-type (wt) or mutant adenoviruses at 50 PFU/cell and maintained in the presence of AraC throughout the infection. Total RNA was isolated at 19 (mock, rec700, and pm975) and 43 h p.i. (12Swt, 12S.2-36, 12S.928, dl312, and dCtBP). An RNase protection assay was performed using 30 and 6 μg of total RNA for detection of TβRII and GAPDH mRNA, respectively. Lanes a and b (A and B), full-length GAPDH and TβRII probes, respectively, hybridized with yeast (Torula sp.) total RNA without subsequent RNase treatment; lanes c (A and B), both probes hybridized with yeast total RNA and treated with RNase. Radioactively labeled DNA markers (A, lane n) are 300, 200, and 100 bp. RNase-protected TβRII and GAPDH mRNA fragments were quantified by densitometry (bar graphs). The densities of TβRII fragments normalized against the density of GAPDH fragments are presented under the corresponding data.
Figure 3B illustrates the TβRII mRNA levels with two 12Swt-based mutants. Mutant 12S.928 (Fig. 3B, lane g), which is defective in binding pRB, decreased TβRII mRNA levels similarly to the parent virus (12Swt; Fig. 3B, lane e), whereas 12S.2-36 had no effect on TβRII mRNA levels (Fig. 3B, lane f). GAPDH mRNA, which was used as a control, was not affected by these viruses (Fig. 3A and B, lanes i to m). Interestingly, a ΔCtBP mutant that is partially defective in decreasing TβRII protein levels (Fig. 2C, lane f) was as efficient as the parent 12Swt virus in decreasing the corresponding mRNA levels (Fig. 3C, compare lanes c and d), suggesting that E1A second-exon functions, including encoding the CtBP binding domain, are dispensable for the TβRII mRNA decrease in infected cells. In conclusion, in accord with the results of the previous section, E1A proteins, and particularly the N-terminal amino acids of E1A, decrease TβRII mRNA in adenovirus-infected cells.
Dynamics of the TβRII protein as determined by pulse-chase analysis are affected in adenovirus-infected cells.
There is a quantitative difference in steady-state TβRII mRNA and protein levels in infected cells: whereas TβRII protein levels were decreased 10-fold by 19 h p.i. (Fig. 1A), there was only a 2.4- to 2.7-fold decrease in steady-state TβRII mRNA levels at this time compared to levels in mock-infected cells (Fig. 3). This difference was not due to the host cell shutoff that is observed during the late stages of infection inasmuch as experiments were done under conditions where infection was limited to the early phase.
A TβRII protein pulse-chase experiment was performed to examine potential differences in the rates of synthesis and the half-lives of TβRII in adenovirus- and mock-infected cells. A549 cells were infected at 50 PFU/cell and maintained in freshly supplied AraC throughout the experiment. At 19 h p.i., cells were metabolically labeled with a [35S]cysteine-methionine mixture for 15 min and chased in cold medium (Fig. 4A and D). TβRII was immunoprecipitated, resolved by SDS-PAGE, and detected by a STORM phosphorimager as well as autoradiography. The phosphorimager signal was quantified with ImageQuant software (Fig. 4B and E). Representative autoradiograms and phosphorimager quantitations of the TβRII-specific signal from two independent experiments are shown in Fig. 4.
Since a 2.4- to 2.7-fold decrease in TβRII mRNA was observed in infected cells, the decrease in the levels of TβRII protein in infected cells detected at 0 min of chase, compared to the level in mock-infected cells, should not be more than this, unless the translation step is affected. TβRII mRNA possesses extended 5′ and 3′ untranslated regions; therefore, translational regulation of TβRII abundance is conceivable. In multiple experiments (more than five), there was approximately a twofold difference in the TβRII between the adenovirus- and mock-infected cells at 0 min of chase (Fig. 4A and D; quantitation in Fig. 4B and E); this suggests that TβRII translation was not markedly affected by adenovirus infection.
Curiously, for both the mock-infected and infected samples, there was an increase in detectable TβRII following the chase, peaking at about 30 min of chase. This increase was reproducible and was seen in A549 and HepG2 cells. Further, it occurred even when the chase was conducted in the presence of cycloheximide to inhibit the elongation of translation (data not shown). One explanation is that the antibody used does not efficiently detect the initial form of TβRII; it detects only a form that arises from posttranslational modifications. In any event, this increase in detectable TβRII was reproducibly lower in infected cells than in mock-infected cells; this is apparent in Fig. 4E, in which the curves for the infected and mock-infected samples are superimposed.
To calculate the TβRII half-life, data points obtained for both mock-infected and infected cells were fitted to an exponential equation with Excel. The fitness of the exponential trend was confirmed by the corresponding R2 values. Because of the delay in peak accumulation of TβRII, only data points taken from the 60- to 150-min chase interval were used to determine the TβRII half-life. A TβRII half-life of 40 min in mock-infected A549 cells was calculated (Fig. 4C). This calculated half-life is shorter than the reported TβRII half-life of 1.5 to 2 h in mink lung epithelial cells (57), which may be a species or cell line phenomenon. Importantly, no significant difference in TβRII half-life between the mock- and adenovirus-infected cells was found (Fig. 4C).
At this point we cannot explain why, if the half-lives of the protein in mock- and adenovirus-infected cells are the same and the rate of precursor synthesis is decreased 2-fold, a 3-fold decrease in TβRII mRNA produces a 10-fold decrease in TβRII protein in infected cells, as determined by Western blotting. We have repeated pulse-chase and immunoprecipitation of TβRII using two additional antibodies raised against the N-terminal and C-terminal sequences of the protein; results were similar to those presented in the Fig. 4. In addition, we have tried two alternative lysis buffers: radioimmunoprecipitation assay buffer (used in a study by Wells et al. [57] to immunoprecipitate TβRII from mink cells in a pulse-chase experiment) and Laemmli buffer (containing 1% SDS). Both methods of lysing cells yielded results similar to the one shown in Fig. 4A. The events occurring during the initial TβRII processing in the infected cells that lead to decreased peak detection of TβRII (as shown in Fig. 4A and D) could potentially contribute to TβRII downregulation.
TGF-β1 signaling in adenovirus-infected cells is inhibited.
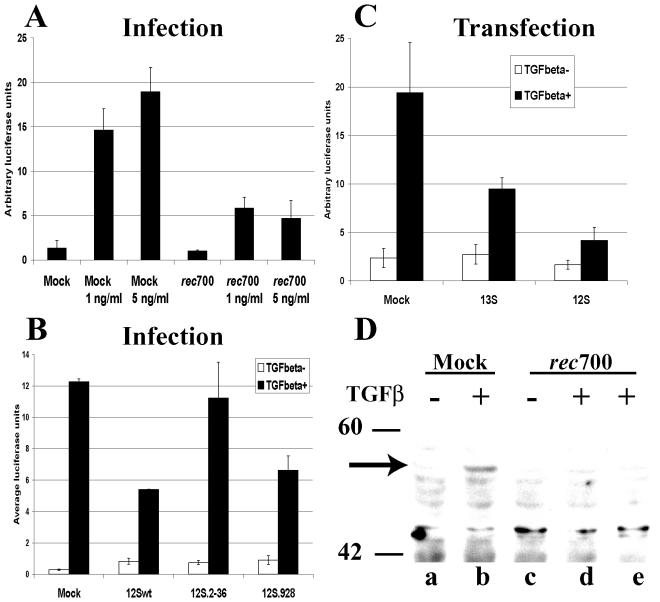
Given that TβRII levels in infected cells are decreased and that the protein required for the decrease is E1A, then E1A should inhibit TGF-β1-induced signaling. To examine this prediction, we used a transient-transfection system in which HepG2 cells were transfected with p3TP-lux, a TGF-β1-responsive reporter plasmid (60), and pCMV-βgal as a transfection control. HepG2 cells were used because they had a higher efficiency of transfection than A549 cells. Subsequently, cells were either mock-infected or infected with the wild-type adenovirus or mutants. As shown in Fig. 5A, upon treatment of mock-infected cells with 1 or 5 ng of active human recombinant TGF-β1/ml, a robust activation of the signaling pathway occurred, as is evident from the increased luciferase activity. In contrast, very little activation of the TGF-β1 signaling pathway occurred in rec700-infected cells, as illustrated by a minimal increase in luciferase production (Fig. 5A).
FIG. 5.
E1A inhibits TGF-β-induced signal transduction in adenovirus-infected cells as determined by using the 3TP-lux reporter plasmid. (A) HepG2 cells were transfected with 3TP-lux and pCMV-βGal plasmids. After 6 h of transfection, cells were infected with rec700 at 50 PFU/cell or mock infected. Infections were maintained in the presence of AraC. Cells were treated with TGF-β1 from 12 to 26 h p.i.; subsequently, cells were lysed and luciferase and β-Gal activities were measured. Luciferase values were normalized against β-Gal activity for each sample. Each experimental condition was done in triplicate, and the average values are shown. (B) HepG2 cells were transfected with 3TP-lux and CMV β-Gal plasmids and an empty vector or a plasmid expressing either the 13S or 12S isoform of E1A. At 18 h posttransfection, cells were treated with recombinant TGF-β1 (3 ng/ml) for 8 h. (C) HepG2 cells were treated as described for panel A. Cells were maintained from 12 to 48 h p.i. in the presence of 5 ng of TGF-β1/ml. (D) A549 cells were infected with 50 PFU/cell of rec700 or mock infected and maintained in the presence of AraC. At 20 (lane d) or 24 (lane e) h p.i. cells were mock treated or treated with 5 ng of TGF-β1/ml for 20 min. Levels of phospho-Smad 2 (arrow) were determined by Western analysis. Molecular weight marker positions are shown.
Adenovirus mutants expressing only the 12S E1A wild-type or mutant isoform were analyzed for TGF-β1 signaling. Signaling was inhibited in cells infected with 12Swt and the 12S mutant deficient in pRB binding (12S.928), but not in cells infected with the mutant harboring a deletion in the N terminus (12S.2-36) (Fig. 5B). These results are consistent with results in Fig. 2B, which showed that TβRII protein levels were downregulated by 12Swt and by the 12S.928 mutant but not by the 12S.2-36 mutant.
As a further check for the ability of E1A to block TGF-β1 signaling, we cotransfected the p3TP-lux reporter with plasmids expressing the 13S and 12S forms of E1A. As shown in Fig. 5C, both E1A isoforms reduced TGF-β-induced luciferase synthesis, confirming a role for E1A in inhibiting TGF-β1 signal transduction and indicating that E1A can suppress TGF-β1 signaling in the absence of other adenovirus proteins.
Finally, we examined the levels of phosphorylated Smad 2 in cells infected with rec700. Levels of phospho-Smad 2 in mock-infected cells increased upon TGF-β1 treatment, as determined by Western analysis using the anti-phospho-Smad 2 antibody (Fig. 5D, lanes a and b). Little Smad 2 phosphorylation was induced in rec700-infected cells at 20 h p.i. (Fig. 5D, lane d), and no phosphorylated Smad 2 was detected in infected cells at 24 h p.i. (Fig. 5D, lane e). The lack of TGF-β1-inducible Smad 2 phosphorylation in infected cells suggests that TβRII downregulation by adenovirus contributes to the loss of TGF-β1 signaling observed in infected cells.
TGF-β1 reduces the accumulation of adenovirus late proteins and virus yields.
Large amounts of structural proteins are produced during the late stage of infection, and expression is dependent on adenovirus genome replication. Synthesis of late proteins is followed by virion assembly and release. Therefore, the levels of structural proteins in infected cells can be used to evaluate progression through the viral life cycle. To determine whether TGF-β1 signaling has an effect on adenovirus life cycle progression, the accumulation of adenovirus late proteins as well as E1A proteins in TGF-β1 treated cells was examined.
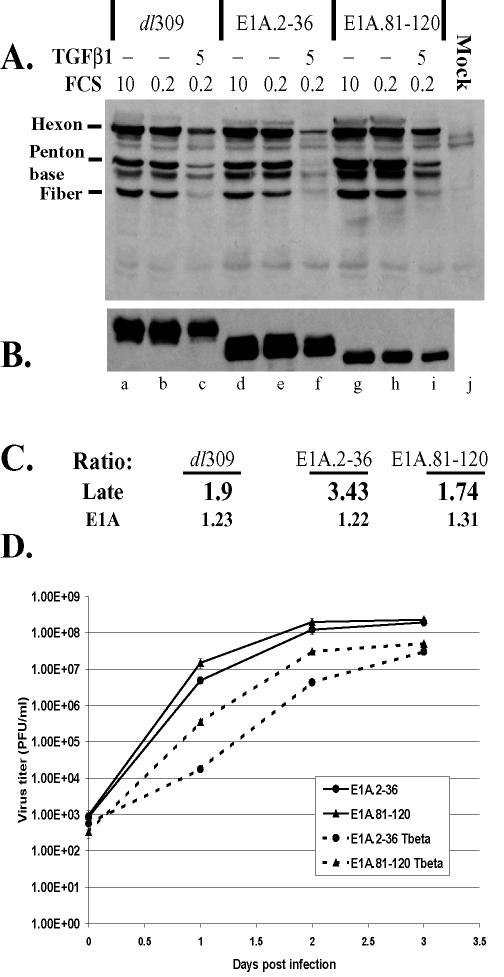
A549 cells undergo growth arrest, but not apoptosis, upon treatment with active TGF-β1 (43) (data not shown). In our experiments, prior to infection, A549 cells were maintained in 10 or 0.2% FCS or 0.2% FCS containing 5 ng of active TGF-β1/ml for 3 days. Cells were subsequently infected at 35 PFU/cell with wild-type dl309 or mutant E1A.2-36 or E1A.81-120 adenoviruses. Infected cells were kept under conditions identical to the pretreatment environment. At 24 h p.i., levels of adenovirus structural and E1A proteins were examined by Western analysis using anti-Ad5 and anti-E1A antibodies, respectively. The relative abundances of structural and E1A proteins were quantified by densitometric analysis. In addition, the ratio of signal in serum-starved cells to that in TGF-β1-treated cells was calculated for structural and E1A proteins (Fig. 6A and B, ratios of lanes b to c, e to f, and h to i; summarized in Fig. 6C).
FIG. 6.
TGF-β1 suppresses adenovirus late protein synthesis and virus yields in infected cells. A549 cells were maintained in 10 or 0.2% FCS or 0.2% FCS plus 5 ng of TGF-β1/ml as indicated throughout the experiment. Following 3 days of pretreatment, cells were infected with adenovirus mutants at 10 PFU/cell. At 24 h p.i., cell lysates were collected and analyzed by Western blotting using anti-Ad5 (A) and anti-E1A (B) antibodies. Equal protein concentrations were loaded in all lanes. The signal from every lane was measured and quantified with FluorChem software (Alpha Innotech Corporation). (C) Ratio of the signal in serum-starved cells to that in cytokine-treated cells for each adenovirus mutant. (D) A549 cells were treated and infected as for panels A and B. Cell lysates and supernatants were collected, and total virus yields at the indicated times postinfection were determined.
Serum starvation had a negligible effect on the levels of structural proteins for all three viruses tested (Fig. 6, compare lanes a and b, d and e, and g and h). However, the levels of structural proteins in serum-starved cells maintained in the presence of TGF-β1 were decreased (Fig. 6, lanes c, f, and i). This negative effect was more pronounced in cells infected with E1A.2-36, a mutant (similar to 12S.2-36) incapable of downregulating TβRII and inhibiting TGF-β1 signaling (Fig. 2D). While the ratios of the amounts of structural proteins in serum-starved cells to the amounts in TGF-β1-treated cells for dl309 and E1A.81-120 infections were very similar (1.9 and 1.74, respectively; Fig. 6C), the ratio was higher in cells infected with E1A.2-36 (3.43; Fig. 6C). These results indicate that, although TGF-β1 pretreatment decreases adenovirus structural protein levels in infected cells, a greater effect is observed when the mutant cannot inhibit TGF-β1 signaling. Although the levels of E1A proteins were somewhat lower in TGF-β1-treated cells, the decreases in E1A for all adenovirus mutants were very similar.
To assess the effect of TGF-β1 treatment on virus yields, A549 cells were pretreated as described above and subsequently infected with 10 PFU of E1A.2-36 or E1A.81-120/cell. Following infection, pretreatment conditions were restored. Infected cells together with supernatants were collected at 0, 1, 2, and 3 days p.i. and freeze-thawed three times, and total virus yield was determined by plaque assay for each sample.
E1A.2-36 and E1A.81-120 had similar growth kinetics in control cells but not in TGF-β-treated cells (Fig. 6D). Although E1A.81-120 was as efficient as its wild-type parent virus in decreasing TβRII levels (Fig. 2D), it produced a decrease in late protein synthesis in TGF-β-treated cells at 24 h p.i. that was similar to the decrease observed in cells infected with wild-type virus (Fig. 6A and C). Accordingly, growth of E1A.81-120 was delayed in TGF-β-treated cells compared to that in control cells (about a 1.5-log-unit difference at 24 h p.i. and about a 1-log-unit difference at 48 h p.i.; Fig. 6D). The E1A.2-36 mutant showed greater delays in growth at 24 and 48 h p.i. (2.5 and 2 log units, respectively), correlating with the mutant's inability to downregulate TβRII and to control TGF-β signaling.
DISCUSSION
TGF-β1 is a critical factor in the homeostasis of the immune system. TGF-β1 is produced by regulatory T cells and macrophages to control immune system activation (14, 24). Viruses have evolved to prevent the immune system from premature destruction of infected cells and to limit the antiviral immune response upon infection. Additionally, a TGF-β1-imposed G1/S block in the cell cycle and the induction of apoptosis would probably threaten completion of the viral life cycle. Therefore, it seems reasonable that modulation of TGF-β1 production and activity would be executed by many infectious agents. Indeed, TGF-β1 was reported to partially induce reactivation of latent Epstein-Barr virus (18, 27). TGF-β1 promoter activity was induced by the human papillomavirus E6 oncoprotein (15). TGF-β1 secretion induced by HIV antigens was shown to account for inhibition of gamma interferon induction in response to human immunodeficiency virus (20). Finally, the human T-cell lymphotropic virus type 1 Tax protein was found to inhibit Smad-mediated signaling at the transcriptional level (35).
Our data also support the idea of TGF-β1 pathway modulation by an infectious agent. Our results show that TβRII is downregulated in adenovirus-infected cells. The decrease in TβRII becomes apparent at 13 to 15 h p.i. and is a function of E1A. A limited number of cellular genes undergo negative regulation by E1A, including genes encoding two surface receptors, neu and HER2 (62), major histocompatibility complex class I (MHC-I) genes (31), and some differentiation genes (4). MHC-I downregulation is mediated only by the Ad12 E1A (a member of adenovirus subgroup A) and not Ad5 E1A (subgroup C). Conversely, a number of cellular genes are positively regulated by E1A, including Golgi-associated GP73 (34) as well as genes whose expression is controlled by E2F (17).
According to our mapping studies, amino acids 2 to 36 in the E1A N terminus are particularly important for TβRII downregulation in infected cells. The CtBP binding site in the sequence encoded by the second exon also seems to be involved. On the other hand, the pRB binding site and the CR3 sequences are dispensable.
The ability of adenovirus mutants with E1A lesions to downregulate TβRII protein levels correlates with the ability to decrease TβRII mRNA; the decrease in TβRII mRNA is likely due to transcriptional repression by E1A. The contribution of the E1A N-terminal-binding cellular factors to the TβRII decrease in infected cells is of considerable interest, as CBP/p300, TATA binding protein, and AP-1 all bind to E1A N-terminal sequences (19). The AP-1 complex, in particular, may play a role in positive regulation of the TβRII promoter (1). The ability of E1A to bind CtBP also plays a role in efficient TβRII downregulation in adenovirus-infected cells. CtBP, a transcriptional corepressor, belongs to a family of proteins with both nuclear and cytoplasmic functions (9). As a transcriptional repressor, CtBP has been shown to modulate TGF-β1-mediated transcription. In addition, members of the CtBP family may play a role in Golgi regulation (56).
Modulation of TGF-β1 signaling by E1A has been studied by several groups in the context of cellular transformation. E1A was able to prevent TGF-β1-induced upregulation of p21CIP, p15INK (11), and junB (10). Other groups showed that E1A could relieve TGF-β1-induced growth inhibition, consistent with the role of E1A as a transforming agent (12). The ability to recruit p300 plays an important role in Smad-mediated transactivation (28), and E1A has been shown to displace p300 from Smad-containing complexes (42). The p300 binding site in E1A was shown to be required for E1A's ability to modulate TGF-β1 signaling (45). In a transient-transfection system, E1A was able to interact with Smads 1, 2, and 3, suggesting that E1A may directly modulate the activity of Smad transcription complexes (42). Last, in a mouse cell line stably transfected with E1A, decreased TβRII protein and mRNA levels were observed along with diminished TβRII promoter activity (32).
A 10-fold decrease in TβRII levels, as determined by Western blotting, was observed in adenovirus-infected cells at 19 h p.i. However, at the same time only an ∼2.5-fold decrease in steady-state TβRII mRNA levels was seen, suggesting that some factor(s) in addition to the decrease in TβRII mRNA is responsible for the decrease in TβRII protein. This factor does not appear to be a decrease in the half-life of TβRII, because the half-lives in infected and uninfected cells were similar, as determined in a pulse-chase experiment in which radiolabeled TβRII was analyzed by immunoprecipitation and SDS-PAGE. Also, the factor is probably not a decrease in translation of TβRII because there was at most a twofold reduction due to infection in 35S-labeled TβRII detected by immunoprecipitation at the onset of the chase period and at least a twofold reduction would be expected because the TβRII mRNAs are less abundant in infected cells. The analysis is complicated by the fact that different antibodies were used for immunoprecipitation and Western blotting: the antibody used for immunoprecipitation (from R&D Systems) did not react with TβRII by Western blotting, and the antibody used for Western blotting (from Santa Cruz Biotechnology) produced a large background by immunoprecipitation. Nevertheless, we present the following working hypothesis to explain the quantitative differences. First, the initial rates of translation of TβRII in uninfected and infected cells are the same; the twofold decrease in TβRII seen in infected cells is due to the corresponding decrease in TβRII mRNA levels. Second, in uninfected cells, the TβRII precursor (form X) is processed to the higher-molecular-weight form (form Y), which is more readily detected by the antibody than form X, thereby accounting for the apparent increase in TβRII seen in the first ∼30 min of chase. In infected cells, the development of form Y is inhibited by E1A or its degradation is accelerated. This would account for the decrease in the peak of TβRII observed in infected cells following 30 min of chase (Fig. 4E). Form Y is the species detected in uninfected cells by Western blotting, a method that measures the steady-state levels of TβRII. Since adenovirus inhibits the accumulation of TβRII, a 10-fold difference between infected and uninfected cells is observed by Western blotting. The antibody used in the immunoprecipitations was selected by the manufacturer for its ability to react with and neutralize cell surface TβRII (hypothetical form Y); therefore, the antibody may not have as high of an affinity for the misfolded or incompletely modified TβRII as it does for the mature cell surface receptor.
TGF-β1 signaling in adenovirus-infected cells was decreased, as determined by use of a TGF-β-responsive reporter. Either the 13S or 12S E1A isoform was sufficient for inhibition of TGF-β signaling in a transient-transfection system, as shown in this report and reports from other groups (11, 45). Based on our findings and reports of others, there are at least three potential mechanisms of TGF-β1 signaling inhibition in adenovirus-infected cells: (i) displacement of p300 from Smad-containing transcriptional complexes by E1A, (ii) direct modulation of Smad-mediated transcription by E1A, and (iii) E1A-mediated downregulation of TβRII levels. The ability of E1A to downregulate TβRII levels is dependent on the N-terminal sequences that are also critical for binding transcriptional modulators such as p300 and YY1, as well as S4 and S8 regulatory components of the proteasome (55). The individual contributions of TβRII downregulation and p300 sequestration to the overall inhibition of TGF-β1 signaling by E1A in infected cells should be determined in future studies.
A key question is whether the adenovirus-mediated downregulation of TβRII and TGF-β1 signaling that we had observed in cultured cells is physiologically relevant. Unfortunately, there is not an established animal model to study adenovirus pathogenesis. Accordingly, we explored the effect of TGF-β1 on the adenovirus life cycle in vitro using A549 cells, a cell line commonly used to study adenoviruses. It is known that treatment with TGF-β1 results in growth inhibition but not apoptosis of A549 cells (43). According to our data (Fig. 6), TGF-β1 treatment prior to and throughout the infection decreased the accumulation of adenovirus structural proteins, which correlated with decreased virus yields in cytokine-treated cells. This occurred in cells that were maintained in low FCS. Interestingly, there was more of a decrease with a mutant that does not downregulate TβRII. The decrease is not due to TGF-β1-induced apoptosis inasmuch as A549 cells are resistant. The decrease was not caused by low serum because levels of E1A and structural protein accumulation in cells in 10 and 0.2% serum were equal. These results raise the possibility that TGF-β1 inhibits the adenovirus life cycle and that E1A counteracts this inhibition. One possibility is that there is a delayed transition into the S phase of the cell cycle in cells infected with mutants incapable of inhibiting the TGF-β1 pathway and that this could delay late protein synthesis. Alternatively, adenovirus gene expression may be directly regulated by the Smad-containing transcriptionally active complexes.
In conclusion, we have shown that E1A downregulates TβRII in adenovirus-infected cells and that the activation of TGF-β1 pathway is severely compromised in the course of infection with wild-type adenovirus. The negative effect exerted by TGF-β1 on the accumulation of adenovirus structural proteins and virus growth may have played a role in adenovirus's acquisition of the capability to specifically target TGF-β1 signaling in the host cells.
Acknowledgments
We thank Ann Tollefson, John Tavis, Karoly Toth, and Jung San Huang for helpful discussions and advice and Ajaykumar Vora and Duane Grandgenett for mentorship and technical assistance. We are grateful to Elizabeth Moran for adenovirus mutants, Joan Massague for the 3TP-lux plasmid, G. Chinnadurai for the E1A expression plasmids, Yoav Henis for the myc-tagged TβRII cDNA, and Rob Fleming for the GAPDH plasmid.
This research was supported by a predoctoral fellowship from the American Hearth Association (V.L.T.) and by grants CA58538 and CA71704 from the National Institutes of Health (W.S.M.W.).
REFERENCES
- 1.Adnane, J., E. Seijo, Z. Chen, F. Bizouarn, M. Leal, S. M. Sebti, and T. Munoz-Antonia. 2002. RhoB, not RhoA represses the transcription of the transforming growth factor β type II receptor by a mechanism involving activator protein 1. J. Biol. Chem. 277:8500-8507. [DOI] [PubMed] [Google Scholar]
- 2.Babiss, L. E., P. B. Fisher, and H. S. Ginsberg. 1984. Effect on transformation of mutations in the early region 1b-encoded 21- and 55-kilodalton proteins of adenovirus 5. J. Virol. 52:389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benedict, C., P. Norris, T. Prigozy, J. L. Bodmer, J. A. Mahr, C. Garnett, F. Martinon, J. Tschopp, L. R. Gooding, and C. F. Ware. 2001. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J. Biol. Chem. 276:3270-3278. [DOI] [PubMed] [Google Scholar]
- 4.Bishopric, N. H., G.-Q. Zeng, B. Sato, and K. A. Webster. 1997. Adenovirus E1A inhibits cardiac myocyte-specific gene expression through its amino terminus. J. Biol. Chem. 272:20584-20594. [DOI] [PubMed] [Google Scholar]
- 5.Brady, H. A., and W. S. M. Wold. 1987. Identification of a novel sequence that governs both polyadenylation and alternative splicing in region E3 of adenovirus. Nucleic Acids Res. 15:9397-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlin, C. R., A. E. Tollefson, H. A. Brady, B. L. Hoffman, and W. S. M. Wold. 1989. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell 57:135-144. [DOI] [PubMed] [Google Scholar]
- 7.Challberg, S. S., and G. Ketner. 1981. Deletion mutants of adenovirus 2: isolation and initial characterization of virus carrying mutations near the right end of the viral genome. Virology 114:196-209. [DOI] [PubMed] [Google Scholar]
- 8.Chang, W., M. Parra, C. Ji, Y. Liu, O. Eickelberg, T. L. McCarthy, and M. Centrella. 2002. Transcriptional and post-transcriptional regulation of transforming growth factor β type II receptor expression in osteoblasts. Gene 299:65-77. [DOI] [PubMed] [Google Scholar]
- 9.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 10.Coussens, L. M., K. Yokoyama, and R. Chiu. 1994. Transforming growth factor β1-mediated induction of junB is selectively inhibited by expression of Ad12-E1A. J. Cell. Physiol. 160:435-444. [DOI] [PubMed] [Google Scholar]
- 11.Datto, M. B., P. P. C. Hu, T. F. Kowalik, J. Yingling, and X.-F. Wang. 1997. The viral oncoprotein E1A blocks transforming growth factor β-mediated induction of p21/WAF1/Cip1 and p15/INK4B. Mol. Cell. Biol. 17:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.deGroot, R. P., O. Kranenburg, L. de Wit, J. Eijnden-van Raaji, C. Mummery, A. J. Eb, and A. Zantema. 1995. Adenovirus E1A antagonizes both negative and positive growth signals elicited by transforming growth factor β1. Cell Growth Differ. 6:531-540. [PubMed] [Google Scholar]
- 13.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 14.Derynck, R., and L. Choy. 1998. Transforming growth factor-β and its receptors, p. 593-636. In A. W. Thompson (ed.), The cytokine handbook. Academic Press, San Diego, Calif.
- 15.Dey, A., I. A. Atcha, and S. Bagchi. 1997. HPV16 E6 oncogene stimulates the transforming growth factor-β1 promoter in fibroblasts through a specific GC-rich sequence. Virology 228:190-199. [DOI] [PubMed] [Google Scholar]
- 16.Elsing, A., and H.-G. Burgert. 1998. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. USA 95:10072-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisch, S. M., and J. S. Mymryk. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3:441-452. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda, M., K. Ikuta, K. Yanagihara, M. Tajima, H. Kuratsune, T. Kurata, and T. Sairenji. 2001. Effect of transforming growth factor-β1 on the cell growth and Epstein-Barr virus reactivation in EBV-infected epithelial cell lines. Virology 288:109-118. [DOI] [PubMed] [Google Scholar]
- 19.Gallimore, P. H., and A. S. Turnell. 2001. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene 20:7824-7835. [DOI] [PubMed] [Google Scholar]
- 20.Garba, M. L., C. D. Pilcher, A. L. Bingham, J. Eron, and J. A. Frelinger. 2002. HIV antigens can induce TGF-β1 producing immunoregulatory CD8+ T cells. J. Immunol. 168:2247-2254. [DOI] [PubMed] [Google Scholar]
- 21.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaynor, R. B., and A. J. Berk. 1983. Cis-acting induction of adenovirus transcription. Cell 33:683-693. [DOI] [PubMed] [Google Scholar]
- 23.Gorelik, L., S. Constant, and R. A. Flavell. 2002. Mechanism of transforming growth factor β-induced inhibition of T helper type 1 differentiation. J. Exp. Med. 195:1499-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelik, L., and R. A. Flavell. 2002. Transforming growth factor-β in T-cell biology. Nat. Rev. Immunol. 2:46-53. [DOI] [PubMed] [Google Scholar]
- 25.Horwitz, M. 2001. Adenovirus immunoregulatory genes and their cellular targets. Virology 279:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz, M. S. 2001. Adenoviruses, p. 2301-2326. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 27.Inman, G. J., and M. J. Allday. 2000. Resistance to TGFβ1 correlates with a reduction of TGFβ type II receptor expression in Burkitt's lymphoma and Epstein-Barr virus-transformed B lymphoblastoid cell lines. J. Gen. Virol. 81:1567-1578. [DOI] [PubMed] [Google Scholar]
- 28.Janknecht, R., N. J. Wells, and T. Hunter. 1998. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 12:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, W., M. P. M. Tillekeratne, M. G. Brattain, and S. S. Banerji. 1997. Decreased stability of transforming growth factor β type II receptor mRNA in RER+ human colon carcinoma cells. Biochemistry (Moscow) 36:14786-14793. [DOI] [PubMed] [Google Scholar]
- 30.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 31.Katoh, S., K. Ozawa, S. Kondoh, E. Soeda, A. Israel, K. Shiroki, K. Fujinaga, K. Itakura, G. Gachelin, and K. Yokoyama. 1990. Identification of sequences responsible for positive and negative regulation by E1A in the promoter of H-2Kbm1 class I MHC gene. EMBO J. 9:127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, D. H., J. H. Chang, K. H. Lee, H.-Y. Lee, and S.-J. Kim. 1997. Mechanism of E1A-induced transforming growth factor-β (TGF-β) resistance in mouse keratinocytes involves repression of TGF β type II receptor transcription. J. Biol. Chem. 272:688-694. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S.-J., Y.-H. Im, S. D. Markowitz, and Y.-J. Bang. 2000. Molecular mechanisms of inactivation of TGF-β receptors during carcinogenesis. Cytokine Growth Factor Rev. 11:159-168. [DOI] [PubMed] [Google Scholar]
- 34.Kladney, R. D., A. E. Tollefson, W. S. M. Wold, and C. J. Fimmel. 2002. Upregulation of the Golgi protein GP73 by adenovirus infection required the E1A CtBP interaction domain. Virology 301:236-246. [DOI] [PubMed] [Google Scholar]
- 35.Lee, D. K., B.-C. Kim, J. N. Brady, K. T. Jeang, and S.-J. Kim. 2002. Human T-cell lymphotropic virus type 1 tax inhibits transforming growth factor-beta signaling by blocking the association of Smad proteins with Smad-binding element. J. Biol. Chem. 277:33766-33775. [DOI] [PubMed] [Google Scholar]
- 36.Massague, J. 1998. TGF β signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 37.Massague, J., S. W. Blain, and R. S. Lo. 2000. TGF β signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 38.McNees, A. L., and L. R. Gooding. 2002. Adenovirus inhibitors of apoptotic cell death. Virus Res. 88:87-101. [DOI] [PubMed] [Google Scholar]
- 39.Mehra, A., and J. L. Wrana. 2002. TGF-β and the Smad signal transduction pathway. Biochem. Cell Biol. 80:605-622. [DOI] [PubMed] [Google Scholar]
- 40.Montell, C., E. Fisher, M. Carruthers, and A. Berk. 1982. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature 295:380-384. [DOI] [PubMed] [Google Scholar]
- 41.Nevins, J. R. 1981. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell 26:213. [DOI] [PubMed] [Google Scholar]
- 42.Nishihara, A., J. Hanai, T. Imamura, K. Miyazono, and M. Kawabata. 1999. E1A inhibits transforming growth factor-β signaling through binding to Smad proteins. J. Biol. Chem. 274:28716-28723. [DOI] [PubMed] [Google Scholar]
- 43.Osada, H., Y. Tatematsu, A. Masuda, T. Saito, M. Sugiyama, K. Yanagiasawa, and T. Takahashi. 2001. Heterogenous transforming growth factor (TGF)-β unresponsiveness and loss of TGF-β receptor II expression caused by histone deacetylation in lung cancer cell lines. Cancer Res. 61:8331-8339. [PubMed] [Google Scholar]
- 44.Ranheim, T. S., J. Shisler, T. M. Horton, L. J. Wold, L. R. Gooding, and W. S. M. Wold. 1993. Characterization of mutants within the gene for the adenovirus E3 14.7-kilodalton protein which prevents cytolysis by tumor necrosis factor. J. Virol. 67:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen, X., P. P.-C. Hu, N. T. Liberati, M. B. Datto, J. P. Frederick, and X.-F. Wang. 1998. TGF-β-b-induced phosphorylation of Smad-3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell 9:3309-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shisler, J., C. Yang, B. Walter, C. F. Ware, and L. R. Gooding. 1997. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J. Virol. 71:8299-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 64:4421-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephens, C., and E. Harlow. 1987. Differential splicing yields novel adenovirus 5 E1A mRNAs that encode 30kd and 35kd proteins. EMBO J. 6:2027-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian, T., M. La Regina, and G. Chinnadurai. 1989. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1A protein. Oncogene 4:415-420. [PubMed] [Google Scholar]
- 50.Thomas, G. P., and M. B. Mathews. 1980. DNA replication and the early to late transition in adenovirus infection. Cell 22:523-533. [DOI] [PubMed] [Google Scholar]
- 51.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. M. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726-730. [DOI] [PubMed] [Google Scholar]
- 52.Tollefson, A. E., P. Krajcsi, S. Yei, C. R. Carlin, and W. S. M. Wold. 1990. A 10,400-molecular-weight membrane protein is coded by region E3 of adenovirus. J. Virol. 64:794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tollefson, A. E., A. R. Stewart, S. P. Yei, S. K. Saha, and W. S. M. Wold. 1991. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to down-regulate the epidermal growth factor receptor. J. Virol. 65:3095-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toth, K., M. Kuppuswamy, K. Doronin, O. Doronina, D. L. Lichtenstein, A. E. Tollefson, and W. S. M. Wold. 2002. Construction and characterization of E1-minus replication-defective adenovirus vectors that express E3 proteins from the E1 region. Virology 301:99-108. [DOI] [PubMed] [Google Scholar]
- 55.Turnell, A. S., R. J. Grand, C. Gorbea, X. L. Zhang, W. Wang, J. S. Mymryk, and P. H. Gallimore. 2000. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 19:4759-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weigert, R., M. G. Silletta, S. Spano, G. Turacchio, C. Cericola, A. Colanzi, S. Senatore, R. Mancini, E. V. Pollishchuk, M. Salmona, F. Facchiano, K. N. Burger, A. Mironov, A. Luini, and D. Corda. 1999. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature 402:429-433. [DOI] [PubMed] [Google Scholar]
- 57.Wells, R. G., H. Yankelev, H. Y. Lin, and H. F. Lodish. 1997. Biosynthesis of the type I and type II TGFβ receptors. J. Biol. Chem. 272:11444-11451. [DOI] [PubMed] [Google Scholar]
- 58.Wold, W. S. M., and G. Chinnadurai. 2000. Adenovirus proteins that regulate apoptosis, p. 200-232. In DNA virus replication. Frontiers in molecular biology. Oxford University Press, Oxford, United Kingdom.
- 59.Wold, W. S. M., S. L. Deutscher, N. Takemori, B. M. Bhat, and S. C. Magie. 1986. Evidence that AGUAUAUGA and CCAAGAUGA initiate translation in the same mRNA region E3 of adenovirus. Virology 148:168-180. [DOI] [PubMed] [Google Scholar]
- 60.Wrana, J. L., L. Attisano, J. Carcamo, A. Zentella, J. Doody, M. Laiho, X.-F. Wang, and J. Massaque. 1992. TGF β signals through a heteromeric protein kinase receptor complex. Cell 71:1003-1014. [DOI] [PubMed] [Google Scholar]
- 61.Wrana, J. L., L. Attisano, R. Wieser, F. Ventura, and J. Massaque. 1994. Mechanism of activation of the TGF-β receptor. Nature 370:341-347. [DOI] [PubMed] [Google Scholar]
- 62.Yan, D.-H., L.-S. Chang, and M.-C. Hung. 1991. Repressed expression of the HER-2/c-erb-2 proto-oncogene by the adenovirus E1A gene products. Oncogene 6:343-345. [PubMed] [Google Scholar]