Abstract
For the identification of RNA-binding proteins that specifically interact with potato spindle tuber viroid (PSTVd), we subjected a tomato cDNA expression library prepared from viroid-infected leaves to an RNA ligand screening procedure. We repeatedly identified cDNA clones that expressed a protein of 602 amino acids. The protein contains a bromodomain and was termed viroid RNA-binding protein 1 (VIRP1). The specificity of interaction of VIRP1 with viroid RNA was studied by different methodologies, which included Northwestern blotting, plaque lift, and electrophoretic mobility shift assays. VIRP1 interacted strongly and specifically with monomeric and oligomeric PSTVd positive-strand RNA transcripts. Other RNAs, for example, U1 RNA, did not bind to VIRP1. Further, we could immunoprecipitate complexes from infected tomato leaves that contained VIRP1 and viroid RNA in vivo. Analysis of the protein sequence revealed that VIRP1 is a member of a newly identified family of transcriptional regulators associated with chromatin remodeling. VIRP1 is the first member of this family of proteins, for which a specific RNA-binding activity is shown. A possible role of VIRP1 in viroid replication and in RNA mediated chromatin remodeling is discussed.
Viroids are the smallest known pathogens of higher plants. They consist of an infectious, single-stranded, covalently closed circular RNA molecule of ca. 250 to 400 nucleotides (nt). Viroids replicate autonomously, causing symptoms in specific host-viroid combinations of developmental disorders (9). Viroids are divided into two structural and functional distinct families. Potato spindle tuber viroid (PSTVd) is the type member of the Pospiviroidae and replicates in the nucleus. Avocado sunblotch viroid, the type member of the Avsunviroidae, replicates in the chloroplast of plant cells (14). Replication of both groups of viroids proceeds via RNA intermediates of complementary polarity. There is no evidence for viroid-encoded polypeptides. Thus, all genetic functions of the viroid replicon, such as cell entry and movement, replication, host specificity, and pathogenicity must be encoded in the RNA sequence and the resulting secondary structure, which is believed to provide specific signals for protein factors of the host. The predicted secondary structure of most viroids is a rod-like structure with five structural domains and an unusually high content of G and C bases (26). Despite the simple genome of viroids and the extensive knowledge of their structure and domains, little is known about host factors that interact with these structural elements.
PSTVd replicates in the nucleus and accumulates eventually in the nucleolus as a circular single-stranded molecule, which is defined as positive polarity (44). Inhibition of PSTVd replication by alpha-amanitine indicates that viroid RNA strands of positive and negative polarity are most probably synthesized by the DNA-dependent RNA polymerase II (Pol II) (38, 45). This enzyme can also accept PSTVd RNA as a template and “transcribe” it in vitro, initiating the synthesis at specific start sites of the circular PSTVd “plus” RNA template (12, 53). Oligomeric replication intermediates of negative and positive polarity suggests that replication occurs by a rolling-circle-type mechanism (3, 6).
Some viroid-host protein complexes have been described so far; however, the significance or function of most of them for the viroid life cycle and/or pathogenicity is still unclear.
Using in vitro-reconstituted nuclear complexes, PSTVd RNA was found to be associated with histones and two proteins of 41 and 31 kDa, suggesting that a similar complex occurs in nuclei of infected cells (28, 60). Another protein that potentially interacts with viroids was described by Hiddinga et al. (23). These authors identified a host-encoded 68-kDa protein whose phosphorylation depended on the presence of PSTVd. The protein was immunologically related to a similar double-stranded RNA-dependent protein kinase in mammalian cells that has been implicated in the downregulation of protein synthesis after virus infection. Hammond and Zhao (19) have isolated the gene of another serine/threonine protein kinase, which is induced after viroid infection. The kinase, called PKV, has a molecular mass of 55 kDa, a putative nuclear localization signal and autophosphorylating activity. However, there was no evidence for binding or direct activation of the kinase activity after binding to viroid RNA, and the role of these kinases in viroid life cycle or pathogenesis signal transduction remains elusive.
Two chloroplast proteins from avocado were recently shown to interact with avocado sun blotch viroid, and one of them was involved in maturation of the viroid RNA (5).
Recently, it was shown by in situ hybridization that most PSTVd RNA is localized in the nuclei of infected cells, but replicative forms are also present in the phloem (65). In the same study, it was also shown that PSTVd invades only sink leaves but not source leaves. PSTVd is confined to the vascular tissue of sink-source transition leaves. Further, PSTVd could not be localized in some flower organs, such as petals, anthers, and ovaries. Two groups could independently demonstrate an interaction between hop stunt viroid and the phloem protein PP2, an abundant lectin present in phloem exudates (17, 35). It was suggested that this protein, although not exhibiting sequence-specific RNA binding, might be involved in transport of viroids and possibly also other RNAs.
In the present study, we applied an RNA-ligand screening procedure (40) for the identification of a novel tomato RNA-binding protein VIRP1. We show that the protein interacts specifically with PSTVd RNA. Furthermore, we identified an in vivo complex of the VIRP1 with the PSTVd RNA. We examined the expression of VIRP1 in different healthy and PSTVd-infected tissues and discuss the possible function of this tomato protein within the plant cell and during the viroid life cycle.
MATERIALS AND METHODS
Plasmids and in vitro RNA transcription.
The plasmids used in the course of the present studies for in vitro synthesis of the labeled RNA transcripts were as follows. EcoRI-linearized pHa106 (57) served as a template DNA for longer-than-unit-length positive-strand PSTVd. EcoRI-linearized pSP-Av5.8(−) (52) was used for the production of the pentamer form of PSTVd(+). PvuII-linearized pGEM-3Zf(−) (Promega) and XbaI-linearized pBluecript II KS(+) (Stratagene, La Jolla, Calif.) served as templates for the production of control RNAs. For the actin transcript, a DraI/EcoRI fragment from plasmid pSac3 (46) was inserted into the EcoRI/SmaI sites of pT3T7 vector and, after HindIII linearization, it was used as a template DNA for the negative-strand RNA synthesis. An EcoRI-linearized plasmid was used as a template for the synthesis of the 46-mer U1-RNA stem-loop II containing nt 50 to 92 of the human U1-RNA (25). The template used for transcription of potato U1 snRNA was a pU1EH HindIII-linearized plasmid (7).
Transcription reactions were carried out at 37°C in a final volume of 20 μl under conditions recommended by the manufacturers. After incubation for 1.5 h, the template was digested with RNase-free DNase, followed by phenolization after a further 5 min. The synthesized RNA was separated from the unincorporated nucleoside triphosphates by chromatography through a 2-ml column of Biogel A (0.5 M; Bio-Rad) in 10 mM Tris-HCl-1 mM EDTA (pH 8.0). In vitro transcripts were checked prior to use by 5% polyacrylamide gel electrophoresis (PAGE) containing 8 M urea.
Screening of a cDNA expression library by RNA-ligand.
A λ-ZAPII cDNA expression library was constructed from poly(A)+ mRNA isolated from PSTVd viroid-infected Lycopersicum esculentum (cultivar Rentita) leaves. The screening assay was carried out essentially as previously described by Sägesser et al. (40).
cDNA cloning and DNA sequencing.
For further analysis of the λ-ZAPII phage clones, an excision procedure using the R408 helper phage to generate subclones in pBluescript SK(−) (Stratagene) phagemid vector was performed (47). Templates for DNA sequence analysis were purified by using Qiagen plasmid kit according to manufacturer's protocols. Dideoxynucleotide chain termination reactions (43) were conducted with deoxyadenosine 5′-[γ-[35S]thio]triphosphate (Amersham) for DNA sequence analysis on both strands or by automatic sequencing. The sequencing reactions were done with the U.S. Biochemicals T7 Sequenase version 2.0 kit as previously described by Rouer (39). Introduction of sequencing priming sites throughout the cDNA cloned in pBluescript SK(−) was generated by using the transposon γδ (48). Sequence analysis and database searches were done by using FASTA, ALIGNMENT, MAP, and BESTFIT routines of the University of Wisconsin Genetic Computer Group Package, version 8.0 (8), and tools available through National Center for Biotechnology Information (NCBI), The Arabidopsis Information Resource, and European Bioinformatics Institute websites.
The nucleic acid sequences reported here were submitted to the EMBL database under accession numbers AJ249592, AJ249593, AJ249594, and AJ249595.
Northern and Southern blot analysis.
RNA was purified from PSTVd-infected and noninfected tomato leaves as described previously (51). Poly(A)+ mRNA purification was carried out with a Dynabead mRNA purification kit (Dynal, Oslo, Norway) according to the manufacturer's instructions. For RNA gel blot analysis, 15 μg of total RNA and poly(A)+ mRNA extracted from 100 μg of total RNA were electrophoresed on 1% agarose gel containing formaldehyde (42) or guanidine thiocyanate (16) and then blotted onto Nytran membrane (Schleicher & Schuell). Blots were prehybridized at 65°C in 50% formamide as described earlier (37) with an α-32P-labeled 0.7-kb transcript specifying an internal portion of the VIRP1 cDNA (nt 695 to 1435) or a labeled DNA probe containing the RsaI fragment of the cDNA (nt 136 to 2339). In cases where a DNA probe was used, formamide was omitted from the hybridization solution, and in all cases an additional washing step with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) was performed. Equal loading of RNA was verified by ethidium bromide staining of the gel before transfer to the membrane. Alternatively, the RNA blot was hybridized with a radioactively labeled RNA probe for the constitutively expressed actin gene.
Genomic DNA from L. esculentum (cultivar Rentita) leaves was extracted as described by Saghai-Maroof et al. (41). For DNA gel blot analysis, 10 μg of genomic DNA was digested with restriction enzymes, fractionated on 0.7% agarose gels, and transferred onto Nytran membranes (Schleicher & Schuell). Hybridization was performed at 50°C with an internal VIRP1 BamHI fragment (encompassing nt 695 to 1435) RNA probe as described above for Northern blot analysis. Membranes were washed at a final stringency of 1% saline sodium citrate buffer and 1% sodium dodecyl sulfate (SDS) at 50°C for 1 h before autoradiography.
Protein expression and purification.
Two oligonucleotides, His1 (5′-TAT GCA TCA CCA TCA CCA TCA CGT GTT AAC CCG G-3′) and His2 (5′-GAT CCC GGG TTA ACA CGT TAT GGT GAT GGT GAT GCA-3′) were synthesized, annealed, and ligated into the NdeI/BamHI site of pET-3a (49), producing pHis1. This vector includes six histidine residues following the initiator methionine. The pVIRP1 (nt 1 to 2770) and pVIRP1Δ (nt 1091 to 2403) cDNAs were excised from the pBluescript SK(−) vector by SmaI/XhoI digestion and then inserted into the HpaI site of the pHis1 expression vector (the XhoI site had been blunted with the Klenow fragment), yielding pHis-VIRP1 and pHis-VIRP1Δ, respectively. The original library clone λVIRP1Δ contained a stretch of nine GA nucleotides derived from the cloning procedure, in addition to several nucleotides from the vector; library clone λVIRP1 contained several nucleotides from the vector. Expression of recombinant protein was performed in Escherichia coli BL21(DE3) cells by IPTG (isopropyl-β-d-thiogalactopyranoside) induction in Luria-Bertani (LB) medium supplemented with 100 μg of ampicillin/ml. Expression and purification of the proteins was essentially as described previously (7). E. coli-expressed fusion protein pHis-VIRP1Δ was further purified under denaturing conditions by using Ni-nitrilotriacetic acid (NTA) columns according to the manufacturer's instructions (Qiagen, Hilden, Germany). The protein was renatured by stepwise dialysis against native buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10% [vol/vol] glycerol, and 1 mM dithiothreitol).
Northwestern blot analysis.
Recombinant E. coli BL21(DE3) harboring plasmids pHIS-VIRP1 and pHis-VIRP1Δ was grown at 30°C in LB medium containing 100 μg of ampicillin/ml. RNA-protein interactions analyzed by Northwestern assay were conducted essentially as reported previously (40, 63).
Electrophoretic mobility shift assay (EMSA).
Protein-RNA interaction analyses were performed by mixing protein and nucleic acid in binding buffer (10 mM HEPES-NaOH [pH 8.0], 50 mM KCl, 100 mM EDTA, and 5% glycerol) in a final volume of 10 μl. All samples contained 1 μg of yeast tRNA and ∼6 × 103 cpm of the radioactively labeled RNA. Unless otherwise indicated, the RNA-protein mixtures were kept for 60 min at room temperature (22°C). The complexes were separated by electrophoresis through a 6% nondenaturing polyacrylamide gel (29:1 [acrylamide/bisacrylamide ratio]) by using 0.5× TBE buffer (50 mM Tris base, 50 mM boric acid, and 1 mM EDTA [pH 8.3]) at a constant voltage of 10 V cm−1 and room temperature. Gels were prerun for 1 h before samples were loaded. Gels were dried and exposed to X-ray film.
Purification of extracts and IP.
Tissue irradiation and homogenization was performed essentially as described previously (5). Briefly, leaf tissue from PSTVd-infected tomato was irradiated with 20 J of UV light/cm2. After irradiation, crude extract was obtained by homogenizing the tissue in immunoprecipitation (IP) buffer (0.1 M Tris-HCl [pH 9], 0.1 M NaCl, 0.1 M 2-mercaptoehanol, 10 mM EDTA) at a ratio of 5 ml/g (fresh weight) of tissue. The extract was clarified by low-speed centrifugation. IP was performed with an extract volume corresponding to 0.2 g of fresh tissue. Then, 5 mg of protein A-Sepharose (Amersham) was equilibrated in IP buffer by rotating the mixture at 4°C for 1 h. The beads were treated with 10 μl of VIRP1 rabbit polyclonal antibody (prepared from purified HIS-VIRP1Δ protein at the Department of Applied Biochemistry and Diagnostics, Institute of Molecular Biology and Biotechnology) in a volume of 200 μl, incubated for an additional hour, and subsequently incubated with the PSTVd-infected tomato extracts. After incubation overnight at 4°C, protein A-Sepharose was pelleted and washed four times with IP buffer.
For proteinase K digestion after IP, immunoprecipitates were mixed with 60 μg of proteinase K in a volume of 50 μl of 10 mM Tris-HCl (pH 7.8)-5 mM EDTA-1 mM 2-mercaptoethanol-0.5% SDS, followed by incubation at 37°C for 45 min. After digestion, the preparations were treated with phenol, and RNAs were recovered by ethanol precipitation. RNAs were separated by denaturing 5% PAGE, electroblotted onto nylon membranes (Hybond-N; Amersham), and fixed by exposure to UV irradiation. Membranes were hybridized at 70°C in the presence of 50% formamide with a α-32P-labeled PSTVd RNA probe.
RESULTS
Isolation of a PSTVd RNA-binding protein.
We made use of a cDNA expression library cloned in λ-ZAPII and originating from PSTVd-infected tomato leaf tissues. This library was subjected to the RNA-ligand screening procedure that we developed previously (40). As RNA probe we used a linear α-32P-labeled PSTVd positive-strand RNA derived from plasmid pHa106 (57). The RNA transcript Ha106 represents a full monomeric unit of PSTVd, flanked by terminal repeats of the central conserved region that is highly conserved among viroids of the PSTVd group. The RNA transcript Ha106 is highly infectious and assumes a secondary structure that allows in vitro processing to a circular molecule (54, 57), thus representing a functional viroid RNA.
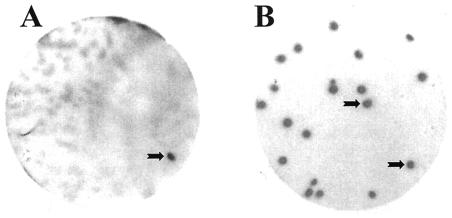
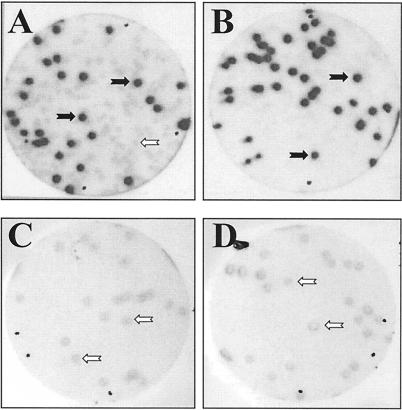
A total of ca. 600,000 plaques were screened for filter-absorbed recombinant proteins that are able to bind to the synthetic PSTVd positive-strand RNA ligand Ha106. This primary screening delivered 18 plaques exhibiting RNA binding (Fig. 1A). Since plaques were confluent and contaminated with phages from different clones, bacteriophages were collected from the signaling plaques and subjected to a secondary screening (Fig. 1B), during which they were purified to homogeneity. This was confirmed in a tertiary screening, which delivered only signaling plaques. In this way we obtained 14 independent cDNA clones that maintained binding affinity after a third round of purification. It is noteworthy that we had only a very low number of false-positive clones, which demonstrates the reliability of the screening procedure.
FIG. 1.
Detection of PSTVd RNA-binding protein VIRP1. A total of about 600,000 plaques from a cDNA expression library from tomato leaves were subjected to a primary screening, by using a longer-than-unit-length PSTVd RNA transcript (57) as a radioactively labeled probe. (A) The tomato cDNA expression library was plated at ca. 5,000 PFU/plate. The black arrow shows the signal corresponding to the viroid binding protein 1 clone after a primary screening. (B) The clone picked from the primary screening was plated at a density of 25 PFU/plate, and the PSTVd RNA-binding properties of positive clone 1 were confirmed by secondary screening. Plaque lift and binding assays with α-32P-labeled RNA transcripts were performed as described in Materials and Methods.
The size of the cDNA inserts varied between 1.3 and 2.7 kb. However, partial nucleotide sequencing of the inserts revealed an identical overlapping nucleotide sequence. The cDNA with the longest insert, now designated λVIRP1, was used for further analysis. The λ clone was converted to plasmid DNA by phage rescue excision and then sequenced on both strands. The cDNA insert consisted of 2,770 bp. Sequence analysis revealed that the insert begins with a 5′-untranslated region (5′-UTR) of 224 bases, followed by an AUG codon at position 225 and an open reading frame (ORF) of 1,809 bases. An in-frame terminator UAA codon 6 bp upstream of the AUG triplet suggests that the AUG codon at position 225 serves as the initiation codon. The predicted ORF, which encodes a polypeptide with 602 amino acids, is followed by a 3′-untranslated region (3′-UTR) of 716 bp.
Sequences of the 14 clones started at various sites of the 5′-UTR or ORF but had the same sequence. However, their 3′-UTRs showed differences. Four classes of 3′-UTRs designated 3′-UTR-a, 3′-UTR-b, 3′-UTR-c, and 3′-UTR-d were detected, with lengths of 341, 370, 659, and 716 nt, respectively. All four 3′-UTRs ended with an 18- to 25-nt poly(A) tail. At position 2136, a sequence motif UUGUAA was located in all four 3′-UTRs; however, an A-rich noncanonical positioning signal is localized only at position 2596 of 3′-UTR-c and 3′-UTR-d (accession no. AJ249592, AJ249593, AJ249594, and AJ249595).
None of the four 3′-UTRs contained a full mRNA polyadenylation control signal, as determined by analysis of more than 5,000 Arabidopsis thaliana genes (18).
Analysis of VIRP1 sequence.
The λVIRP1-encoded protein (VIRP1) of 602 amino acids has a calculated molecular mass of 65,586 kDa and a predicted basic pI of 8.24. VIRP1 contains some functional domains or regions of biased composition. This includes a proline-rich domain (amino acids 322 to 404) and a serine-rich domain (amino acids 566 to 595). Residues 302 to 305 provide possible sites for amidation, and there are several potential sites for phosphorylation, N myristoylation, and N glycosylation. An ATP/GTP-binding site motif A (residues 21 to 28) can be found and—most importantly—a bipartite nuclear localization signal (residues 328 to 345) (10, 62).
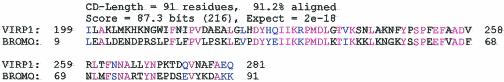
VIRP1 shows limited sequence similarity with a number of proteins, including the gene products of brm (56) and fsh (22), RING3 (2) and SNF2 (29). However, in each case the similarity is limited to the same single domain. This is a conserved domain of 110 amino acids rich in aromatic amino acids, which is called bromodomain and is found in several eukaryotic transcription factors (21, 24). Recently, it was shown that the bromodomain binds acetylated lysines (11, 64) (Fig. 2). Also, the Arabidopsis protein At1g05910 has homology to VIRP1 in the bromodomain region. At1g05910 itself is homologous to yeast YTA7, a protein, which belongs to the AAA family. The bromodomain has been proposed to be a “chromatin tag,” which targets proteins to the dynamic chromatin in the nucleus (13).
FIG. 2.
Bromodomain of VIRP1. Consensus protein motif pfam00439 (protein motif data collection Pfam 8.0) is aligned with a region of VIRP1. The data were retrieved from http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. Identical amino acids are in red, and the conservative changes are in blue.
Except for the partial similarity to proteins from other organisms, there is similarity to two Arabidopsis proteins of unknown function (At5g10550 and At5g65330). In this case, the homology is extended to amino- and carboxy-terminal parts of the proteins, including regions from positions 98 to 145 and from 185 to 290 and also the regions from positions 390 to 470 and from 520 to 602 of VIRP1 (data not shown).
Structure and transcript levels of the VIRP1 gene.
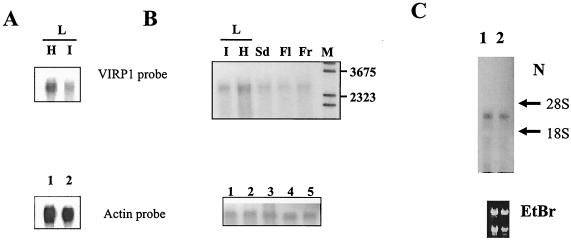
VIRP1 expression was readily detected in flower, fruit, seeds, and in the leaves (Fig. 3). In viroid-infected plants expression of VIRP1 mRNA was repressed at least twofold compared to healthy controls. Separate examination of flower revealed the presence VIRP1 mRNA in sepals as well as in petals.
FIG. 3.
Expression and tissue distribution of VIRP1 mRNA in tomato. mRNA extracted from 100 μg of total RNA was size separated in 1% agarose-formaldehyde (A and C) or agarose-guanidine thiocyanate (B) gels. RNA was blotted and hybridized with an α-32P-labeled 0.7-kb VIRP1 antisense RNA probe. (A) Northern blot of RNAs from healthy (lane H) and PSTVd-infected (lane I) tomato leaves. The same blot was also probed with an actin RNA transcript for loading controls as shown in the lower panel (lanes 1 and 2, respectively). (B) Northern blots of RNAs from various tissues. Poly(A)+ mRNA from leaf (L, lanes I [infected] and H [healthy]), seed (lane Sd), flower (lane Fl), and fruit (Fr) were hybridized with antisense VIRP1 RNA. M, DNA molecular weight marker. (C) Northern blot of total RNA from petals (lane 1) and sepals (lane 2) of infected plants hybridized with VIRP1 DNA. Equal loading was verified by visualization of 28S and 18S rRNAs on the blot.
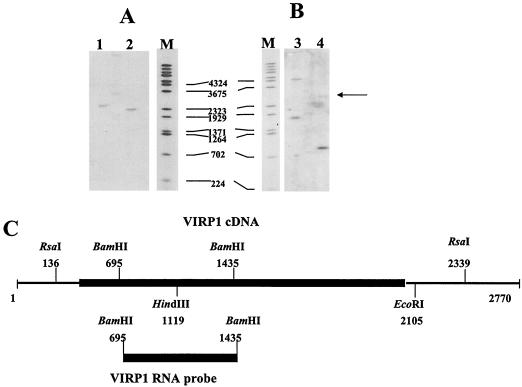
To identify the copy number of the gene, genomic DNA isolated from tomato was hybridized with a VIRP1-specific RNA probe spanning nt 695 to 1435 at stringent hybridization conditions. After digestion of genomic tomato DNA with RsaI, a single signal of ∼2.4 kb was detected. Mixed digestion with RsaI and EcoRI delivered a unique hybridization signal at ∼2.2 kb. Digestion of genomic DNA with BamHI was expected to give only one fragment but delivered an additional weaker fragment (arrow). This suggests the existence of a second related gene with homology to VIRP1 (Fig. 4).
FIG. 4.
Southern analysis of tomato genomic DNA. (A and B) Southern analysis of tomato total DNA. Lane 1, digestion with RsaI; lane 2, digestion with RsaI/EcoRI; lane 3, digestion with HindIII; lane 4, digestion with BamHI; lane M, lambda DNA molecular weight marker digested with BstEII. (C) Restriction map of VIRP1 and the probe used for hybridization. The coding region is shown by a heavy black box.
Specificity of binding of protein VIRP1 to PSTVd RNA.
To verify the RNA-protein interaction and to determine the degree of specificity we used three different methods. The first method used was the plaque lift assay, which represents a variation of the screening procedure. In this assay, the phage λVIRP1 expressing the recombinant VIRP1 protein was mixed with a nonrecombinant phage at a known ratio, and the resulting plaques transferred to filters (plaque lift). As in the screening procedure, the filters were allowed to react with an RNA ligand. However, we used here various kinds of radioactively labeled RNA transcripts (Fig. 5). Only when PSTVd RNA (monomeric or multimeric form) was used as ligand could plaques originating from recombinant phages be discriminated from those generated by control phages.
FIG. 5.
Binding of VIRP1 by plaque lift assay. Binding specificity of VIRP1 and PSTVd RNA was tested by plaque lift assay. A mixture (1:1) of λVIRP1 and λ-ZAPII phage was plated out and tested for binding with different RNAs. (A and B) Only when monomeric (A) or multimeric (B) PSTVd positive-strand RNA forms (for details see Materials and Methods) were used as RNA probes could positive signals be discriminated from those due to background. (C and D) When the same mixture was plated at a lower density of 25 PFU/plate and allowed to interact with either potato U1 RNA (C) or human U1 RNA (D), only background signals were visible. In panels C and D the numbers of signals visible on the autoradiograph were the same as the numbers of plaques per plate. The black arrows show the signals corresponding to VIRP1 plaques, which interact with PSTVd (positive signals), while the open arrows indicate the background signals.
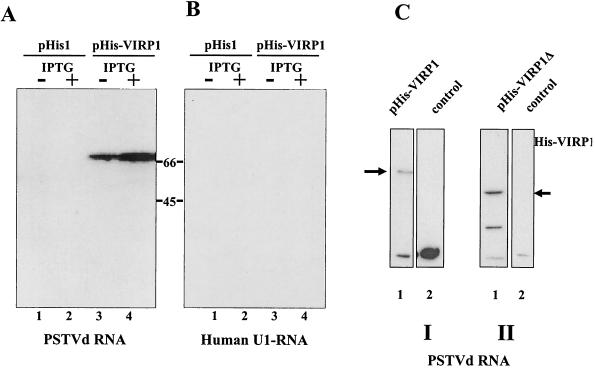
The two other methods required a different mode of expression of the recombinant VIRP1. Therefore, the cDNAs of VIRP1 and VIRP1Δ (encompassing the full coding region or the C-terminal part from nt 1091 to 2403, the smallest cDNA clone isolated but still capable of binding) were subcloned in a modified pET3a expression vector, pHis1, which allows expression of the recombinant protein at high levels under the control of T7 RNA polymerase promoter (49). The modification of the pHis1 vector allows the fusion of six histidine residues to the amino terminus. Northwestern assays were performed with crude protein extracts from E. coli cells in order to confirm the binding capacity of the protein to PSTVd RNA. Protein extracts from E. coli cells containing plasmids pHis-VIRP1 or pHis-VIRP1Δ were separated on SDS-10% PAGE gels and transferred to a nitrocellulose membrane. A signal could be detected after the membrane was probed with radioactively labeled PSTVd positive-strand RNAs; however, no signal was visible when a transcript of U1 RNA (Fig. 6) or other unrelated RNAs were used (not shown). Both the full-length and the C-terminal part of VIRP1 were binding in this assay, thus confirming the specific interaction of the expressed protein, once immobilized on nitrocellulose membranes, with PSTVd RNA.
FIG. 6.
Northwestern analysis of VIRP1. Specificity of binding of VIRP1 protein with PSTVd positive-strand RNA was confirmed by Northwestern assay. Crude extracts of E. coli BL21 cells harboring either pHIS, pHis-VIRP1 or pHis1-VIRP1Δ or without plasmid (see the text for details) were separated by SDS-10% PAGE, transferred to nitrocellulose membranes and probed with radioactively labeled transcripts. In panels A and C, hybridization was done with PSTVd positive-strand RNA; in panel B, hybridization was done with U1 RNA. (A and B) BL21(pHIS) or BL21(pHIS-VIRP1) crude cell extracts. Lanes 1 and 3, noninduced cells; lanes 2 and 4, IPTG-induced cells. (C) BL21 cell extracts. Lane I1, BL21(pHis-VIRP1) cell extracts; lane I2, BL21 cell extracts; lane II1, BL21(pHis1-VIRP1Δ) cell extracts; lane II2, BL21 cell extracts. The arrows indicate the VIRP1 and VIRP1Δ proteins, respectively. In panel C, parts I and II present results from two different experiments.
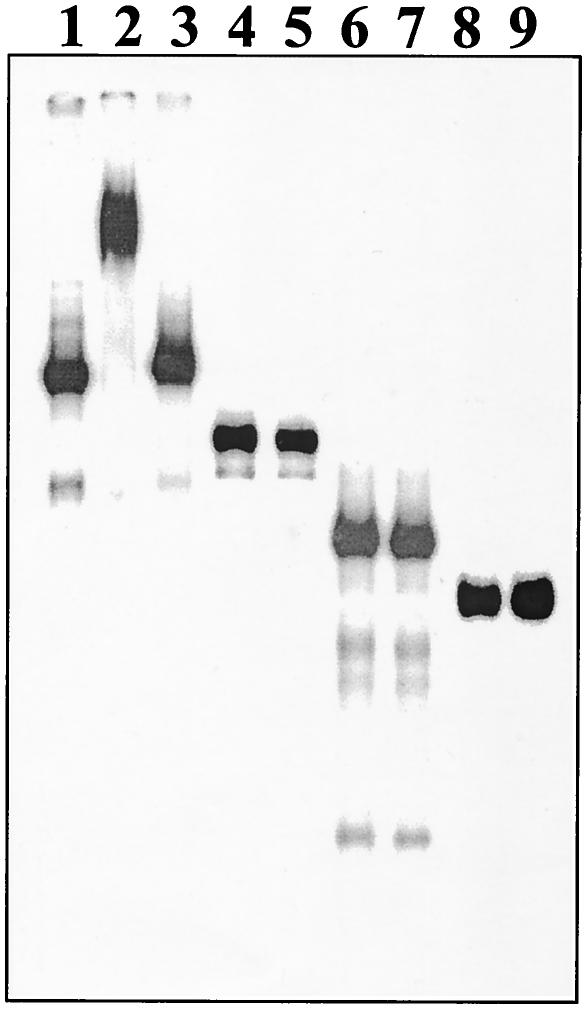
To further test the specificity of interaction, the VIRP1Δ protein was purified after overexpression in E. coli BL21(DE3) and subsequent chromatography on a Ni-NTA-agarose column. The purified protein was used to test the interaction of VIRP1Δ and PSTVd in solution by EMSAs. The protein was incubated with radioactively labeled monomeric PSTVd positive-strand RNA in the presence of tRNA as competitor, and the complexes were analyzed on native polyacrylamide gels (Fig. 7). Retardation of PSTVd RNA was observed in the presence of VIRP1Δ protein. Using unlabeled PSTVd RNA as a competitor, the retardation of the radioactively labeled RNA could be reversed. No retardation of VIRP1Δ was observed when U1 RNA or plasmid-derived RNA transcripts were used (Fig. 7). The collective data of Fig. 5 to 7 demonstrated that interaction of VIRP1 and PSTVd RNA occurs in a sequence-specific manner.
FIG. 7.
Specificity of binding of PSTVd RNA by EMSA. Radioactively labeled RNAs were incubated with or without purified VIRP1Δ protein for 60 min at room temperature. Mixtures were electrophoresed on 6% native polyacrylamide gels and subjected to autoradiography. When PSTVd RNA (lanes 1 to 3) was incubated with VIRP1Δ protein (lane 2), retardation due to RNA-protein complex formation could be observed, which could be competed for in the presence of 100-fold excess nonlabeled PSTVd RNA (lane 3). In contrast, when other RNA transcripts, e.g., pGEM-3Zf(−) (lanes 4 and 5), potato U1 snRNA (lanes 6 and 7), or pBluescript II KS(+) (lanes 8 and 9), were incubated together with VIRP1Δ (lanes 5, 7, and 9), no such retardation was detected.
VIRP1 protein and PSTVd RNA form a complex in vivo.
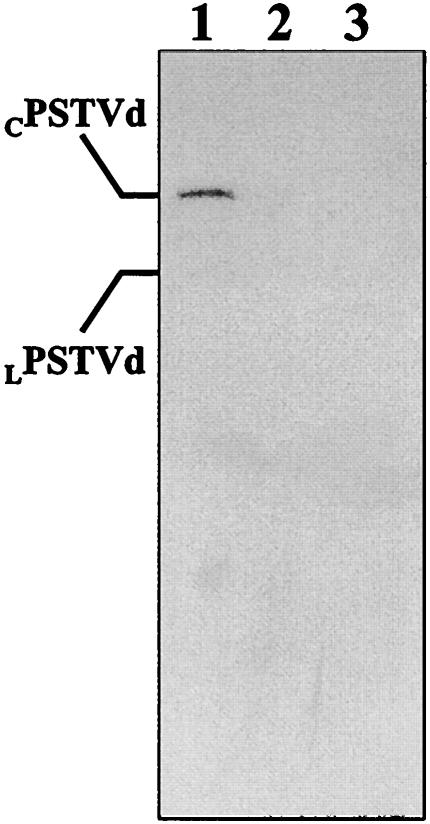
The search for cellular factors that assist steps in the biological cycle of viroids in the present work has been done by applying the in vitro RNA-ligand screening. Although the interaction between VIRP1 and PSTVd RNA is specific, as demonstrated by three independent approaches, its possible functional significance depends on whether the observed interaction occurs also in vivo. To identify the in vivo complex, IP assays were performed with a VIRP1-specific and a nonspecific polyclonal antibody. PSTVd-infected tissue was UV irradiated in order to “freeze” the interaction between the VIRP1 protein and the PSTVd RNA (5). Irradiation of the plant tissue prevents possible disruption of the complex during the homogenization procedure, thus resulting in enrichment in PSTVd RNA-protein complexes. The immunoprecipitates were treated with proteinase K, and the digestion products were analyzed by denaturing polyacrylamide gel and by Northern blot hybridization with a probe for detecting positive-strand PSTVd forms. A signal corresponding to the circular positive-strand RNA (and traces of linear positive-strand RNA) could be detected in the sample immunoprecipitated with VIRP1-specific polyclonal antibody, but no signal was observed when an unrelated antibody was used (Fig. 8).
FIG. 8.
VIRP1-PSTVd RNA in vivo complexes. PSTVd-infected extracts from irradiated tissue were used for IP assays. After IP, the samples were treated with proteinase K, and RNAs were recovered by ethanol precipitation after phenolization. Lane 1, RNAs extracted from IP reaction with a VIRP1Δ-specific polyclonal antibody; lane 2, nonspecific polyclonal antibody used for the IP; lane 3, no antibody was used. Extracted RNAs were separated by PAGE, electroblotted, and hybridized with a α-32P-labeled negative-strand PSTVd RNA probe.
DISCUSSION
Expression and posttranscriptional modification of VIRP1.
In this work, we present the sequence of a cDNA, which encodes a protein binding specifically to PSTVd viroid RNA. It is the first host factor identified for this nuclearly replicating family of viroids. Three proteins have been isolated in our laboratory by this method: VIRP1 (described here); a second, as-yet-uncharacterized viroid-binding protein (VIRP2 [unpublished results]); and a novel RNA-binding protein from Triturus carnifex (7). A fourth protein was isolated at another laboratory by this approach (50), thus also verifying the specificity of this method for RNA-protein interactions that may be transient in vivo and not easily detected biochemically.
An interesting finding regarding the expression of VIRP1 is that it is repressed at least two- to threefold in leaves of viroid-infected plants. This observation was verified in several experiments. At the moment, it is unclear whether this is an indirect interaction (VIRP1 is among a subset of genes that are indirectly downregulated upon viroid infection) or whether this is a direct interaction involving the binding of PSTVd to VIRP1. Ectopic expression of VIRP1 will clarify this question.
Another interesting finding is that VIRP1 is expressed in petals in infected (Fig. 3C) and noninfected (data not shown) plants, an organ that has been found to be free of PSTVd RNA in in situ hybridization experiments in tomato and Nicotiana benthamiana plants (65). Zhu et al. (66) have shown that PSTVd RNA can in principle replicate independently in petals, if the initial RNA is produced from a transgene expressed in this organ, but most probably it cannot be transported to petals after a normal mechanical infection of leaves. The presence of VIRP1 in petals indicates that it might have a role in replication and that is not always in the same cells and tissues with viroid RNA in a complex.
In the course of the RNA-ligand screening, we have identified four identical VIRP1 ORFs that display an unusual organization of their 3′-UTRs. All four UTRs share the first 343-bp sequence; the next 26 nt are shared only by UTR-b, UTR-c, and UTR-d; the next 289 nt are shared by UTR-c and UTR-d and, finally, UTR-d has a unique terminal region before the poly(A) sequence. Therefore, it seems that the termination of transcription and/or polyadenylation of VIRP1 are subject to some mechanism of control. Indeed, in some cases we could detect multiple transcripts around the expected length; however, we do not know yet the mechanism underlying this complexity (unpublished results). It is possible that different signals on these UTRs regulate differential transcript stability, export, or translatability. Recently, a new mechanism of translational regulation has been discovered that involves small nearly perfect or perfect complementary RNAs, the so-called micro-RNAs, which are also present in plants (30, 31). Interestingly, one of these micro-RNAs matches perfectly with transcripts of transcription factors involved in signaling cascades, indicating that this flexible regulation system is applied for regulatory proteins. It will be interesting to investigate whether VIRP1 is also subject to such regulation. Alternatively, VIRP1 mRNA might be subject to stability regulation through other RNA-binding proteins.
Possible function of VIRP1.
The most striking feature of the identified VIRP1 is the presence of a bromodomain. This is a conserved domain of ca. 110 amino acids that is characterized by four α-helices and is present in several eukaryotic nuclear proteins (21, 24). The conserved domain can be arranged as a single, double, or multiple units and frequently occurs in combination with a wide spectrum of various other functional protein domains. At least 40 different motifs can be found in bromodomain-containing proteins, e.g., AAA motifs and MYB, SAND, SET, or WD40 domains (data retrieved from the CDART-NCBI programme; http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi). The complexity of the bromodomain-containing proteins is indicative of their multiple roles. However, a unifying role is emerging: the bromodomain is thought to be a “tag” for localization of the corresponding protein in the nucleus and, in particular, in dynamic chromatin (13).
Further, it is clear that a number of bromodomain-containing proteins have a role in development in different organisms. For example, Drosophila brahma and its murine homologue Brg1 are components of chromatin remodeling complexes of the so-called SWI/SNF class (27; reviewed in reference 55). Bromodomain-containing proteins such as the Drosophila trx-G group of genes (ash1, brm, and fsh) and Caenorhabditis elegans genes lin-49 and lin-59 are important for somatic or embryonic development, indicating how chromatin dynamics may influence cell fate. Some of these genes (trx-G) act as positive regulators; for example, lin-49 and lin-59 are thought to regulate Hom-C genes, the counterparts of Hox genes in C. elegans (4). GCN5 interacts with acetylated histone H3 and H4 peptides, and this interaction could potentially induce repression of Gcn5p acetyltransferase activity (34). It has also been reported that the bromodomain of human Gcn5p interacts with Ku-DNA-dependent protein kinase, a kinase activated by DNA and important in DNA repair and replication (1).
It is possible that bromodomain-containing proteins act to initiate and maintain an epigenetic mark, resulting in a specific transcriptional state during development, independent of the specific function of the regulated genes, as more or less “structural” chromatin state maintainers. This would also explain the relatively high expression levels of these proteins. In accordance with this view, VIRP1 also has a relatively high expression level.
Since viroid symptoms include developmental disorders (such as leaf malformation and dwarfism), it is possible that the VIRP1-viroid interaction is involved in symptom formation. It is likely that VIRP1 has another cellular (RNA) target so that viroid RNA could compete for VIRP1, thus removing the protein at least in part from its natural substrate and/or location. This interaction might cause some or all of the pleiotropic symptoms, which characterize viroid infection.
In this context it is interesting that PSTVd RNA has been found to be in a complex with nuclear histones (60).
VIRP1 is the first protein that has the bromodomain associated with an RNA-binding domain. In view of the importance of RNA-mediated methylation in plants as an additional regulatory and defense mechanism, this type of protein gains a special interest. Since PSTVd is capable of initiating methylation of homologous DNA sequences in the nucleus (59), it is conceivable that this bromodomain-containing protein may play a role in transmitting the RNA-mediated sequence specificity to the methylation machinery of the nucleus. In this case, host genome sequences that share the necessary extent of homology with parts of viroid RNA will be methylated. This mechanism has also been discussed as a possible pathogenicity mechanism (36).
Based on examinations by protein motif analysis programs, VIRP1 does not appear to contain a typical RNA-binding recognition motif. It will be interesting to characterize the RNA-binding domain, which is localized at the carboxy-terminal part of the protein, as well as to determine the specific primary and secondary structure of the RNAs that can interact with VIRP1.
The homology of VIRP1 with a bromodomain-containing protein from Arabidopsis, which also contains an AAA domain and is the yeast and human homologue of Tat-binding protein 7 (YTA-7 or TBP-7), is interesting. TBP-7 has been shown to interact in human cells with TBP-1 (33), a protein with possibly dual roles in transcription and 26S protease complexes and a regulator of retroviral transcription (15, 32). Interestingly, the tomato homologue to TBP-1, LeMa-1 (37), has been isolated earlier in our laboratory by using a different screening method. Although no RNA-binding capacity of bacterially expressed LeMa-1 could be verified thus far in vitro, it is conceivable that the three proteins—VIRP1, TBP-7 and LeMA-1—might exist as a complex in vivo since two (VIRP1 and TBP-7) could interact through their bromodomains and the third, LeMa-1, possibly interacts with TBP-7. The complex of TAT-binding and associated proteins is involved in elongation of retroviral transcripts in mammalian cells (67). In some plants, retroviroid-like elements have been described (58). It is therefore possible that viroids are derived from retroviral elements. Further, PSTVd has been shown to be replicated by Pol II (45). VIRP1 is a good candidate to be connected with PSTVd transcription by Pol II.
Possible role of VIRP1 in viroid life cycle.
It has been proposed that multimeric PSTVd, which is assumed to be replicated via DNA-dependent RNA Pol II in the nucleoplasm, is transported into the nucleolus to be processed into monomeric molecules, thereby accounting for the high concentration of PSTVd detected in the nucleolus (20). However, it has been recently reported that direct transport of fluorescent PSTVd monomer transcripts into the nucleolus is also possible (61).
VIRP1 is the ideal candidate for several possible steps in viroid life cycle: (i) transferring viroid RNA to the nucleus, since it contains nuclear localization signals; (ii) exporting it from the nucleus, e.g., through phosphorylation-dephosphorylation of the serine-rich domain at the carboxy terminus; (iii) tethering viroid RNA to the Pol II transcription complex through interactions of its bromodomain; and (iv) transferring the RNA-mediated methylation signal, thereby mediating a possible mechanism of viroid-induced repression of genes. The presence of circular viroid in the immunoprecipitated complex indicates a possible nucleolar localization of the protein or a role in transporting the viroid monomer there in order to be stored until it is transferred to other cells.
Also, a possible role in cell-to-cell internuclear or long distance transport cannot be excluded. Mutational analysis of VIRP1, as well as specification of the viroid domain(s) which bind to it, will elucidate this interesting viroid-host interaction.
Acknowledgments
This work was supported by grants BIO2-CT93-0400, BIO4-CT-2300, and PENED 1324 of the E.C. and the Ministry of Development (Greece). A. E. Martínez de Alba was partially supported by a fellowship of the Basque Country.
We are grateful to Sergia Tzortzakaki for excellent technical assistance, R. Flores for providing time and space for the IP experiment, and K. Kalantidis and M. A. Denti for many discussions and critical reading of the manuscript.
REFERENCES
- 1.Barlev, N. A., V. Poltoratsky, T. Owen-Hughes, C. Ying, L. Liu, J. L. Workman, and S. L. Berger. 1998. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol. Cell. Biol. 18:1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, S., I. Hanson, A. Kelly, D. J. Pappin, and J. Trowsdale. 1992. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 2:203-210. [DOI] [PubMed] [Google Scholar]
- 3.Branch, A. D., B. J. Benenfeld, and H. D. Robertson. 1988. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 85:9128-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamberlin, H. M., and J. H. Thomas. 2000. The bromodomain protein LIN-49 and trithorax-related protein LIN-59 affect development and gene expression in Caenorhabditis elegans. Development 127:713-723. [DOI] [PubMed] [Google Scholar]
- 5.Daros, J. A., and R. Flores. 2002. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daros, J. A., J. F. Marcos, C. Hernandez, and R. Flores. 1994. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 91:12813-12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denti, M. A., A. E. Martínez de Alba, R. Sägesser, M. Tsagris, and M. Tabler. 2000. A novel RNA-binding protein from Triturus carnifex identified by RNA-ligand screening with the newt hammerhead ribozyme. Nucleic Acids Res. 28:1045-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diener, T. O. 1987. Potato spindle tuber viroid: biological properties, p. 9-35. In T. O. Diener (ed.), The viroids. Plenum Press, Inc., New York, N.Y.
- 10.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences: a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 11.Dyson, M. H., S. Rose, and L. C. Mahadevan. 2001. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front. Biosci. 6:D853-865. [DOI] [PubMed] [Google Scholar]
- 12.Fels, A., K. Hu, and D. Riesner. 2001. Transcription of potato spindle tuber viroid by RNA polymerase II starts predominantly at two specific sites. Nucleic Acids Res. 29:4589-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filettici, P., P. Ornaghi, and P. Ballario. 2001. The bromodomain: a chromatin browser? Front. Biosci. 6:D866-D876. [DOI] [PubMed] [Google Scholar]
- 14.Flores, R., J. A. Daros, and C. Hernandez. 2000. Avsunviroidae family: viroids containing hammerhead ribozymes. Adv. Virus Res. 55:271-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, H., N. Reis, Y. Lee, M. H. Glickman, and R. D. Vierstra. 2001. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goda, S. K., and N. P. Minton. 1995. A simple procedure for gel electrophoresis and northern blotting of RNA. Nucleic Acids Res. 23:3357-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez, G., and V. Pallas. 2001. Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant-Microbe Interact. 14:910-913. [DOI] [PubMed] [Google Scholar]
- 18.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 96:14055-14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond, R. W., and Y. Zhao. 2000. Characterization of a tomato protein kinase gene induced by infection by potato spindle tuber viroid. Mol. Plant-Microbe Interact. 13:903-910. [DOI] [PubMed] [Google Scholar]
- 20.Harders, J., N. Lukacs, M. Robert-Nicoud, T. M. Jovin, and D. Riesner. 1989. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 8:3941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haynes, S. R., C. Dollard, F. Winston, S. Beck, J. Trowsdale, and I. B. Dawid. 1992. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20:2603. [DOI] [PMC free article] [PubMed]
- 22.Haynes, S. R., B. A. Mozer, N. Bhatia-Dey, and I. B. Dawid. 1989. The Drosophila fsh locus, a maternal effect homeotic gene, encodes apparent membrane proteins. Dev. Biol. 134:246-257. [DOI] [PubMed] [Google Scholar]
- 23.Hiddinga, H. J., C. J. Crum, J. Hu, and D. A. Roth. 1988. Viroid-induced phosphorylation of a host protein related to a dsRNA-dependent protein kinase. Science 241:451-453. [DOI] [PubMed] [Google Scholar]
- 24.Jeanmougin, F., J. M. Wurtz, B. Le Douarin, P. Chambon, and R. Losson. 1997. The bromodomain revisited. Trends Biochem. Sci. 22:151-153. [DOI] [PubMed] [Google Scholar]
- 25.Jessen, T. H., C. Oubridge, C. H. Teo, C. Pritchard, and K. Nagai. 1991. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 10:3447-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keese, P., and R. H. Symons. 1985. Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 82:4582-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 28.Klaff, P., R. Gruner, R. Hecker, A. Sattler, G. Theissen, and D. Riesner. 1989. Reconstituted and cellular viroid-protein complexes. J. Gen. Virol. 70:2257-2270. [Google Scholar]
- 29.Laurent, B. C., X. Yang, and M. Carlson. 1992. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol. Cell. Biol. 12:1893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llave, C., K. D. Kasschau, M. A. Rector, and J. C. Carrington. 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14:1605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llave, C., Z. Xie, K. D. Kasschau, and J. C. Carrington. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053-2056. [DOI] [PubMed] [Google Scholar]
- 32.Nelbock, P., P. J. Dillon, A. Perkins, and C. A. Rosen. 1990. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science 248:1650-1653. [DOI] [PubMed] [Google Scholar]
- 33.Ohana, B., P. A. Moore, S. M. Ruben, C. D. Southgate, M. R. Green, and C. A. Rosen. 1993. The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionarily conserved genes. Proc. Natl. Acad. Sci. USA 90:138-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ornaghi, P., P. Ballario, A. M. Lena, A. Gonzalez, and P. Filetici. 1999. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol. 287:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Owens, R. A., M. Blackburn, and B. Ding. 2001. Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant-Microbe Interact. 14:905-909. [DOI] [PubMed] [Google Scholar]
- 36.Pelissier, T., and M. Wassenegger. 2000. A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA 6:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prombona, A., M. Tabler, M. Providaki, and M. Tsagris. 1995. Structure and expression of LeMA-1, a tomato protein belonging to the SEC18-PAS1-CDC48-TBP-1 protein family of putative Mg2+-dependent ATPases. Plant Mol. Biol. 27:1109-1118. [DOI] [PubMed] [Google Scholar]
- 38.Rackwitz, H. R., W. Rohde, and H. L. Sanger. 1981. DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature 291:297-301. [DOI] [PubMed] [Google Scholar]
- 39.Rouer, E. 1994. Direct neutralization of alkaline-denatured plasmid DNA in sequencing protocol by the sequencing reagent itself. Nucleic Acids Res. 22:4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sägesser, R., E. Martinez, M. Tsagris, and M. Tabler. 1997. Detection and isolation of RNA-binding proteins by RNA-ligand screening of a cDNA expression library. Nucleic Acids Res. 25:3816-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saghai-Maroof, M. A., K. M. Soliman, R. A. Jorgensen, and R. W. Allard. 1984. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81:8014-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanger, H. L. 1987. Viroid function: viroid replication, p. 117-166. In T. O. Diener (ed.), The viroids. Plenum Press, Inc., New York, N.Y.
- 45.Schindler, I.-M., and H.-P. Muhlbach. 1992. Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 84:221-229. [Google Scholar]
- 46.Shah, D. M., R. C. Hightower, and R. B. Meagher. 1982. Complete nucleotide sequence of a soybean actin gene. Proc. Natl. Acad. Sci. USA 79:1022-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strathmann, M., B. A. Hamilton, C. A. Mayeda, M. I. Simon, E. M. Meyerowitz, and M. J. Palazzolo. 1991. Transposon-facilitated DNA sequencing. Proc. Natl. Acad. Sci. USA 88:1247-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 50.Ta, M., and S. Vrati. 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 74:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabler, M., I. Gunther, R. Kern, and H. L. Sanger. 1989. A microscale procedure for isolating and sequencing the viroid RNA present in one gram of infected leaf tissue. J. Virol. Methods 23:111-126. [DOI] [PubMed] [Google Scholar]
- 52.Tabler, M., and H. L. Sanger. 1985. Infectivity studies on different potato spindle tuber viroid (PSTV) RNAs synthesized in vitro with the SP6 transcription system. EMBO J. 4:2191-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabler, M., and M. Tsagris. 1990. Viroid replication mechanisms, p. 185-205. In R. S. Fraser (ed.), Recognition and response in plant virus interactions, vol. H41. Springer-Verlag, Berlin, Germany.
- 54.Tabler, M., S. Tzortzakaki, and M. Tsagris. 1992. Processing of linear longer-than-unit-length potato spindle tuber viroid RNAs into infectious monomeric circular molecules by a G-specific endoribonuclease. Virology 190:746-753. (Erratum, 192:397, 1993.) [DOI] [PubMed]
- 55.Tamkun, J. W. 1995. The role of brahma and related proteins in transcription and development. Curr. Opin. Genet. Dev. 5:473-477. [DOI] [PubMed] [Google Scholar]
- 56.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68:561-572. [DOI] [PubMed] [Google Scholar]
- 57.Tsagris, M., M. Tabler, and H. L. Sanger. 1991. Ribonuclease T1 generates circular RNA molecules from viroid-specific RNA transcripts by cleavage and intramolecular ligation. Nucleic Acids Res. 19:1605-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vera, A., J. A. Daros, R. Flores, and C. Hernandez. 2000. The DNA of a plant retroviroid-like element is fused to different sites in the genome of a plant pararetrovirus and shows multiple forms with sequence deletions. J. Virol. 74:10390-10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wassenegger, M., S. Heimes, L. Riedel, and H. L. Sanger. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76:567-576. [DOI] [PubMed] [Google Scholar]
- 60.Wolff, P., R. Gilz, J. Schumacher, and D. Riesner. 1985. Complexes of viroids with histones and other proteins. Nucleic Acids Res. 13:355-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woo, Y.-M., A. Itaya, R. A. Owens, L. Tang, R. W. Hammond, H.-C. Chou, M. M. C. Lai, and B. Ding. 1999. Characterization of nuclear import of potato spindle tuber viroid RNA in permeabilized protoplats. Plant J. 17:627-635. [Google Scholar]
- 62.Yoneda, Y. 1997. How proteins are transported from cytoplasm to the nucleus. J. Biochem. 121:811-817. [DOI] [PubMed] [Google Scholar]
- 63.Zaidi, S. H., and J. S. Malter. 1995. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J. Biol. Chem. 270:17292-17298. [DOI] [PubMed] [Google Scholar]
- 64.Zeng, L., and M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, Y., L. Green, Y. M. Woo, R. Owens, and B. Ding. 2001. Cellular basis of potato spindle tuber viroid systemic movement. Virology 279:69-77. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, Y., Y. Qi, Y. Xun, R. Owens, and B. Ding. 2002. Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zorio, D. A., and D. L. Bentley. 2001. Transcription elongation: the ‘Foggy' is lifting ellipsis. Curr. Biol. 11:R144-R146. [DOI] [PubMed] [Google Scholar]