Abstract
The “Spanish” influenza pandemic of 1918 was characterized by exceptionally high mortality, especially among young adults. The surface proteins of influenza viruses, hemagglutinin and neuraminidase, play important roles in virulence, host specificity, and the human immune response. The complete coding sequence of hemagglutinin was reported last year. This laboratory has now determined the complete coding sequence of the neuraminidase gene of the 1918 virus. Influenza RNA fragments were isolated from lung tissue of three victims of the 1918 flu; complete sequence was generated from A/Brevig Mission/1/18, with confirmatory sequencing carried out on A/South Carolina/1/18 and A/New York/1/18. The 1918 neuraminidase gene sequence was compared with other N1 subtype neuraminidase genes, including 9 N1 strains newly sequenced for this study. The 1918 neuraminidase shares many sequence and structural characteristics with avian strains, including the conserved active site, wild-type stalk length, glycosylation sites, and antigenic sites. Phylogenetically, the 1918 neuraminidase gene appears to be intermediate between mammals and birds, suggesting that it was introduced into mammals just before the 1918 pandemic.
The influenza pandemic of 1918–19 was exceptional in the extent of its mortality, especially among 15–45 year olds, killing over 20 million people. To understand the origins of the pandemic virus and to seek clues to its exceptional virulence, this laboratory has undertaken a project to determine the complete genetic sequence of the 1918 virus by using influenza RNA fragments preserved in lung tissues of victims of the pandemic.
The neuraminidase (NA) protein of the influenza A virus is a glycoprotein expressed on the viral surface. Its principal biological role is the cleavage of the terminal sialic acid residues that are receptors for the virus' hemagglutinin (HA) protein. Removal of these residues from the surface of infected cells and from newly formed viruses prevents the budding viruses from clumping to each other or the cell surface (1). The ability to cleave sialic acid is also thought to help the virus penetrate mucus (2). Nine subtypes of NA have been identified, all of which are tetrameric and share a common structure consisting of a globular head, a thin stalk region, and a small hydrophobic region that anchors the protein in the virus membrane. The active site consists of a pocket on the surface of each subunit formed by 15 charged amino acids. These amino acids are conserved in all influenza A viruses (3).
NA, as a surface protein, is also targeted by the human immune system. Because NA functions largely in the release of newly formed viral particles, antibodies against it do not prevent initial infection. However, they sharply limit its spread and therefore, in humans, selection favors NA variants with mutations that hinder antibody recognition (antigenic drift). Along with the other major influenza surface protein, HA, NA demonstrates more genetic variability than other influenza genes. The globular head of the protein presents at least two antigenic sites; drift in the amino acids making up these sites can allow the virus to escape previous immunity (4). In humans, NA is known to have shifted in 1957, when a novel NA of the N2 subtype replaced the previously circulating N1 subtype. Shift in NA is not critical in the initiation of a pandemic, as the pandemic strain of 1968 retained the previously circulating N2. However, widespread immunity to N2 is thought to have lessened the severity of the 1968 pandemic (5, 6).
We have analyzed the 1918 strain's NA gene in a continuation of our work to characterize the 1918 influenza virus. Previous phylogenetic analyses of the complete 1918 HA gene (7) place the 1918 strain within and near the root of the mammalian clade. This placement suggests that the 1918 HA shares many characteristics with subsequent human and swine strains. Nevertheless, the 1918 HA is more closely related to avian HAs than any other mammalian strain, and it shares many structural characteristics with avian isolates. It has only the four glycosylation sites conserved in all H1 HAs and none of the additional sites that have accumulated by antigenic drift in human strains over time. In the areas of H1 that have been identified as antigenic in humans, the 1918 HA is virtually identical to the consensus sequence of avian strains, whereas human strains from 1933 on show extensive drift at these sites. Overall, the results of the HA analyses suggest either that the 1918 strain entered the human population directly from birds (but that modern bird strains have drifted from their 1918 sequences) or that the 1918 strain entered a mammalian host in the years immediately preceding 1918 and adapted there before emerging as a pandemic virus. A similar scenario appears to be true for the virus' NA gene.
Materials and Methods
Cases and Strains.
The cases used for this study were as described previously (7). Briefly, the first case was an Inuit woman exhumed from permafrost in Brevig Mission, on the Seward Peninsula of Alaska. Historical records show that influenza spread through the village of Brevig Mission in about 5 days, killing 72 people, representing 85% of the adult population (8). Histologically, this case showed evidence of acute massive pulmonary hemorrhage. The second case was a 21-year-old male who died of influenza and pneumonia at the Ft. Jackson, SC, camp hospital on 26 September 1918, 6 days after appearance of symptoms. The third case was a 30-year-old male stationed at Camp Upton, NY, who died of influenza with massive pulmonary edema on 26 September 1918, after being ill only 3 days. The strains sequenced from these cases have been named: A/Brevig Mission/1/18 (H1N1), A/South Carolina/1/18 (H1N1), and A/New York/1/18 (H1N1), respectively.
In addition to A/Brevig Mission/1/18 (henceforth referred to as Brevig/18), the complete NA coding sequences of nine additional influenza strains were determined for this study. Four of the strains [A/Sw/Iowa/30 (H1N1), A/Weiss/43 (H1N1), A/FM/1/47 (H1N1), and A/Duck/Alberta/35/76 (H1N1)] were obtained from the American Type Culture Collection. One strain, A/NJ/11/76 (H1N1), was kindly provided by E. Kilbourne (New York Medical College, Valhalla, NY). Four strains [A/Duck/Ohio/175/86 (H1N1), A/Duck/Ohio/30/86 (H1N1), A/Duck/Ohio/194/86 (H1N1), and A/Duck/Ohio/118C/93 (H1N1)] were kindly provided by R. Slemons (Ohio State University, Columbus, OH). A further N1 sequence, from A/Swine/England/92 (H1N1) (hereafter referred to as Sw/England/92), was kindly provided by Ian Brown (Veterinary Laboratories Agency, Weybridge, UK). In addition to the strains sequenced for this study, phylogenetic analyses used 22 other complete N1 subtype NA genes and one N8 subtype NA gene (as an outgroup). The source, abbreviation, and GenBank accession numbers for all 33 strains are listed in Table 1.
Table 1.
Viral strains used in this study
| Strain | Abbreviation | GenBank number | Source |
|---|---|---|---|
| A/Brevig Mission/1/18 (H1N1) | Brevig Mission18 | AF250356 | This paper |
| A/Chicken/Hong Kong/220/97 (H5N1) | Ch/HK220/97 | AF046081 | GenBank |
| A/Chile/1/83 (H1N1) | Chile83 | X15281 | GenBank |
| A/Duck/Alberta/35/76 (H1N1) | Dk/Alberta76 | AF250362 | This paper |
| A/Duck/Ohio/118C/93 (H1N1) | Dk/Ohio93 | AF250361 | This paper |
| A/Duck/Ohio/175/86 (H1N1) | Dk/Ohio175/86 | AF250358 | This paper |
| A/Duck/Ohio/194/86 (H1N1) | Dk/Ohio194/86 | AF250360 | This paper |
| A/Duck/Ohio/30C/86 (H1N1) | Dk/Ohio30/86 | AF250359 | This paper |
| A/Equine/Kentucky/1/81 (H3N8) | Eq/Kentucky81 | M14917 | GenBank |
| A/FM/1/47 (H1N1) | FM47 | AF250357 | This paper |
| A/FPV/Rostock/34 (H7N1) | FPV/Rostock34 | X52226 | GenBank |
| A/Hokkaido/11/88 (H1N1) | Hokkaido88 | D31944 | GenBank |
| A/Hokkaido/2/92 (H1N1) | Hokkaido92 | D31945 | GenBank |
| A/Hong Kong/156/97 (H5N1) | HK/156/97A | AF046089 | GenBank |
| A/Hong Kong/156/97 (H5N1) | HK/156/97B | AF028708 | GenBank |
| A/Kiev/59/79 (H1N1) | Kiev79 | M38355 | GenBank |
| A/Leningrad/1/54 (H1N1) | Leningrad54 | M38309 | GenBank |
| A/NJ/11/76 (H1N1) | NJ/11/76 | AF250363 | This paper |
| A/NJ/8/76 (H1N1) | NJ/8/76 | M27970 | GenBank |
| A/Parrot/Ulster/73 (H7N1) | Parrot/Ulster73 | K02252 | GenBank |
| A/Puerto Rico/8/34 Cambridge (H1N1) | PR/34 | J02146 | GenBank |
| A/Swine/England/195852/92 (H1N1) | Sw/England92 | AF250366 | Ian Brown |
| A/Swine/Iowa/30 (H1N1) | Sw/Iowa30 | AF250364 | This paper |
| A/Swine/Obihiro/5/92 (H1N1) | Sw/Obihiro92 | D31947 | GenBank |
| A/USSR/90/77 (H1N1) | USRR77 | K02018 | GenBank |
| A/Weiss/43 (H1N1) | Weiss43 | AF250365 | This paper |
| A/WI/4754/94 (H1N1) | WI94 | U53166 | GenBank |
| A/WS/33 (H1N1) | WS/33 | L25816 | GenBank |
| A/WSN/33 (H1N1) | WSN/33A | L25817 | GenBank |
| A/WSN/33 (H1N1) | WSN/33B | J02177 | GenBank |
| A/Yamagata/120/86 (H1N1) | Yamagata86 | D31948 | GenBank |
| A/Yamagata/32/89 (H1N1) | Yamagata89 | D31950 | GenBank |
RNA Extraction.
RNA lysates from the paraffin-embedded tissues were produced as previously described (9). RNA was isolated from the frozen lung tissue and from cultured virus strains by using RNAzol (Tel-Test, Friendswood, TX), following the manufacturer's instructions.
Reverse Transcription–PCR (RT-PCR).
RT was carried out at 37°C for 45 min in 20 μl by using 300 units MMLV-reverse transcriptase/1 × RT buffer (GIBCO/BRL)/5 μM random hexamers/200 nM dNTPs/10 mM DTT. RT reaction (2 μl) was added to an 18 μl PCR reaction mixture containing 50 mM KCl, 10 mM Tris⋅HCl, 2.5 mM MgCl2, 1 μM each primer, 100 nM dNTPs, 1 unit Amplitaq Gold (Perkin–Elmer), and 2 μCi 32P dATP (3,000 Ci/mmol) (20 μl total volume). The entire NA coding sequence of Brevig/18 (1,407 nucleotides) was amplified in 22 overlapping fragments such that the sequences corresponding to primers could be confirmed. The primers were designed as degenerate N1 consensus primers by using alignments of human, swine, and avian N1 sequences, or as 1918-specific primers once partial sequence was available. Four overlapping primer sets were used to amplify the entire NA coding sequence of nine strains obtained from the American Type Culture Collection, E. Kilbourne, and R. Slemons. Primer sequences used for these amplifications are available on request.
PCR cycling conditions were: 9 min at 94°C; 40 cycles of 94°C for 30 sec, 50°C for 30 sec, 72°C for 30 sec; 72°C, 5 min. One-sixth of the reaction was separated on a 7% denaturing polyacrylamide gel, dried, and visualized by autoradiography. Bands were excised, electroeluted, and ethanol precipitated. One-fourth of the eluted product was added to a 50 μl PCR reaction (50 mM KCl/10 mM Tris⋅HCl/2.5 mM MgCl2/200 nM dNTPs/1 μM each primer/1 unit Amplitaq Gold) and cycled as above. Reaction product (2 μl) was cloned into the PCR 2.1 vector (Invitrogen), following the manufacturer's instructions.
Replicate RT-PCR reactions from independently produced RNA preparations gave identical sequence results, except at position 788 (see Results).
DNA Sequencing.
Direct PCR by using M13 sequencing primers was done on white bacterial colonies, and the products were sequenced on an Applied Biosystems 377 automated sequencer by using standard protocols.
Phylogenetic Analyses.
Phylogenetic analyses of the NA gene used parsimony [Phylogenetic Analysis using Parsimony (paup, Version 3.1.1)] (10) and neighbor-joining [Molecular Evolutionary Genetics Analysis (mega, Version 1.1)] (11). The optimization method used in paup was acctran.
The software package macclade (12) was used to follow NA character evolution and accumulated changes in the influenza NA sequence family and to evaluate the effect of different tree topologies on branch lengths, tree lengths, and character transformation.
Neighbor-joining analyses routinely used the proportion of differences between the sequences as the distance measure (p distance). Analyses were performed on 32 complete N1 subtype genes. All analyses were bootstrapped 100 replications.
Results
Sequence Analyses.
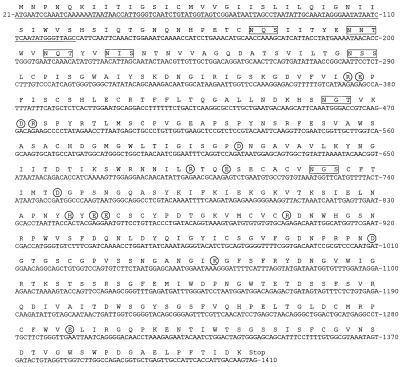
The entire NA coding sequence (1,407 nucleotides) was determined from the frozen sample obtained from Brevig Mission, AK. The sequence is shown in Fig. 1. One nucleotide (at position 788) was found to be heterogeneous within Brevig/18 with two-thirds of clones having a C (and therefore a phenylalanine at amino acid 255) and the rest an A (leucine at 255) at this site. Because A/South Carolina/1/18 also had a C at this site, nucleotide 788 is listed as a C in Fig. 1. The two archival cases were partially sequenced, with 283 bases (409–467, 539–698, 1,019–1,085) sequenced (20% of the coding region) from A/New York/1/18 and 452 bases (21–163, 292–387, 409–467, 785–876, 1019–1085) sequenced (32% of the coding region) from A/South Carolina/1/18. No nucleotide differences were found among the cases. The lack of sequence variation is similar to the results obtained when the entire HA1 domain of HA was sequenced in all three cases (7).
Figure 1.
Complete coding sequence of the NA gene of the 1918 influenza virus. The sequence of A/Brevig Mission/1/18 is shown with a theoretical translation. The numbering of the gene is aligned to A/NJ/8/76 and refers to the sequence in the sense (mRNA) orientation. The sequence coding for the signal peptide is underlined. Boxed amino acids indicate potential glycosylation sites as predicted from the sequence. Circled amino acids indicate the active site residues (3).
In addition to the 1918 cases, the NA coding sequences from a number of other influenza strains were determined for this study. Two human strains, A/Weiss/43 (H1N1) and A/FM/1/47 (H1N1), were included to bridge the gap between the early human strains of the 1930s and the next available sequence from 1954. A/Swine/Iowa/30 (H1N1) was included as it is known to be closely related antigenically to the 1918 virus (13). A/Duck/Alberta/35/76 (H1N1) and four duck strains from Ohio were added to provide strains representative of those found in wild birds; previously, the only avian N1 sequences available were from domesticated birds. The sequence of A/Swine/England/92 was included in phylogenetic analyses to represent an independent introduction of an avian H1N1 into mammals.
Catalytic Site.
The active site of the NA protein consists of a pocket in the top surface of each subunit of the tetrameric protein. The pocket contains 15 charged amino acids that are conserved in all influenza A viruses (3). All of these charged amino acids are conserved in Brevig/18.
Glycosylation Sites.
In HA, avian H1 strains have four conserved glycosylation sites, and there is a distinct accumulation of glycosylation sites in human H1 strains over time. The novel glycosylation sites are thought to benefit the virus in humans by masking antigenic sites (14). In NA, this strategy appears to be less common. Avian N1 viral strains, from both domesticated birds and the wild birds sequenced here, have seven conserved glycosylation sites. Additional glycosylation sites that might serve in antigenic masking are found in only two instances: at amino acids 365–367 in human strains from 1947–1986 and at amino acids 454–456 from 1954 to the present. Brevig/18 NA has only the seven glycosylation sites shared by avian N1 strains.
The glycosylation site containing residue 146 has been studied extensively. This site is conserved in all N1 subtype NAs except two independently derived strains that are neurotropic in mice, A/WSN/33 (H1N1) and A/NWS/33 (H1N1). The loss of this glycosylation site is thought to contribute to the extended tropism of these strains (15). All of the strains newly sequenced for this study, including A/Sw/Iowa/30, retain the wild-type glycosylation site. The nucleotides coding for amino acid 146 were sequenced in all three 1918 cases, and all retain the glycosylation site.
Stalk Region.
The functional NA protein is configured as a homotetramer in which the active sites are found on a terminal knob carried on a thin stalk (3). The stalk region is made up of approximately 50 amino acids beginning about 40 amino acids from the N terminus of the protein and is characterized by at least one cysteine residue and one or more glycosylation sites. Other than these common features, the amino acids making up the stalk are highly variable (16). The stalk region of the 1918 NA has one cysteine and four glycosylation sites, as do all wild avian strains and most mammalian strains. Some early human strains have deletions of 11–16 amino acids in the stalk region, as do many strains isolated from chickens. The deletions vary in length and exact location in the stalk. The 1918 NA has a full-length stalk, as do A/Sw/Iowa/30 (H1N1), A/Weiss/43 (H1N1), and all of the wild avian strains sequenced for this study. A/FM/1/47 (H1N1), however, does have a small deletion, distinct from the deletions found in human strains from the 1930s.
Antigenic Sites.
Although the N1 NA has not been characterized antigenically, it is possible to align the N1 sequence with an N2 subtype NA and examine the N2 antigenic sites for evidence of drift in N1. Colman et al. identify seven regions, comprising 22 amino acids, on the N2 protein that may interact with antibody (3). The homologous 22 amino acids in N1 are unvaried in all avian N1 strains, including the wild avian strains sequenced for this study. Fifteen of the twenty-two show variation in human N1 isolates. Brevig/18 matches the avian consensus at 14 of the 15 sites.
Phylogenetic Analyses.
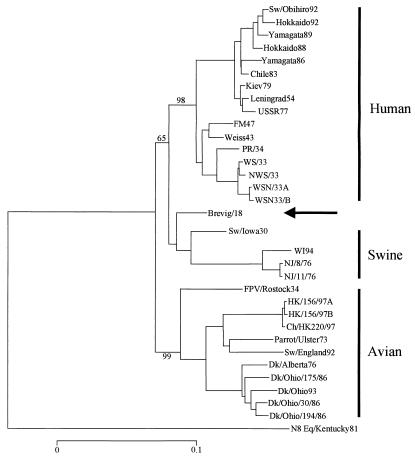
Neighbor-joining analysis of 32 N1 nucleotide sequences by using mega produced three clades: human, swine, and avian. Brevig/18 was placed within and near the root of the swine clade (Fig. 2). Analysis of synonymous substitutions placed the 1918 NA within and near the root of the older swine isolates. However, when nonsynonymous substitutions were analyzed, or when N1 protein sequences were analyzed, Brevig/18 was placed within and near the root of the avian clade. These results suggest that the 1918 NA sequence shares many characteristics with both mammalian and avian viruses.
Figure 2.
Phylogenetic analysis of influenza N1 NA genes. The tree was produced with mega by using neighbor-joining and p distance. Influenza strain abbreviations are given in Table 1. Brevig/18 is identified by an arrow. Bootstrap values are given for selected nodes, and a distance bar is shown below the tree (0.1 p distance is approximately 132 nucleotide differences).
An analysis of avian nucleotide sequences plus Brevig/18 resulted in two clades, North American avian and Eurasian avian, with the 1918 NA extremely close to the root of the North American avian clade. However, because Brevig/18 differed from its closest North American avian relative by 168 nucleotides and from its closest Eurasian relative by only 174 nucleotides, this suggests strongly that Brevig/18 is essentially equidistant from both the North American and Eurasian avian viruses.
Thirty-two N1 nucleotide sequences were analyzed by parsimony (paup), together with one N8 sequence as outgroup. Two trees of 1843 steps were produced (consistency index of 0.63) with Brevig/18 within and near the root of the human clade in both trees. When protein sequences were analyzed, four trees of 621 steps (consistency index of 0.78) were produced, all of which placed Brevig/18 within and near the root of the avian clade. Forcing Brevig/18 to be within and near the root of the human clade (i.e., making the protein tree look like the nucleotide tree) resulted in a tree that was only two steps longer. These results suggest, as found with neighbor-joining, that the 1918 virus shares many characteristics with both mammalian and avian viruses.
To identify amino acids that might be involved in mammalian adaptation, the macclade program was used to determine which amino acids make Brevig/18 appear mammalian and not avian. Thirteen such amino acids were identified, none of which are active site residues (Table 2). Interestingly, Sw/England/92, which remains avian-like phylogenetically, despite circulation in mammals since 1979, differs from the avian consensus at five of these thirteen sites. This suggests that changes at these sites might be some of the minimal adaptive changes necessary to go from an avian-adapted to a mammalian-adapted form.
Table 2.
Predominant amino acids at selected positions in N1 neuraminidase proteins
| Strain† | Amino acid position*
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 74 | 77 | 79 | 81 | 84 | 188 | 264 | 267 | 285 | 287 | 307 | 344 | 352 | |
| Avian | F/L | E | A | A | T | I | V | V | A | E | N | Y | K |
| Sw/Eng/92 | F | E | T | V | K | I | V | V | S | E | N | N | K |
| Brevig/18 | V | G | D | T | I | M | T | I | T | K | D | N | R |
| Human, old | V | G | D | T | I | M | T | I | T | K | D | N | R |
| Swine, old | I | G | D | T | I | M | I | I | T | K | D | N | R |
| Human, new | V | G | D | T | T | M | T | I | T | T | N | D | R |
| Swine, new | I | G | D | T | I | M | I | I | T | K | D | N | R |
| Location‡: | St | St | St | St | S | I | S | S | S | I | S | I | S |
Amino acid numbering is aligned to the N1 NA of A/NJ/8/76 (H1N1).
Predominant amino acid residues are shown for avian strains, 1930s H1N1 strains (old human and old swine), and recent H1N1 strains (new human and new swine).
Theoretical structural location on N1 as aligned to N9 crystal structure by using the rasmol program (29). St, stalk; S, surface of globular head; I, buried internally in globular head.
Rate of Amino Acid Replacement.
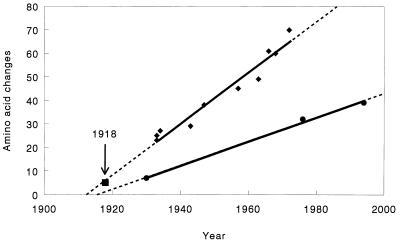
The most parsimonious paup protein tree was imported into the macclade program, and the total numbers of unambiguous amino acid changes from node to node within the human and swine branches of the tree were determined. These changes, minus the 1918 data point, were then plotted vs. year of isolation, and regression lines were determined (Fig. 3). It is apparent that both human and swine proteins accumulate phylogenetically informative amino acid replacements in a linear fashion over time, and that the 1918 data point falls very close to the extrapolated human regression line. Both human and swine regression lines extrapolate to a common ancestor that appeared to exist sometime around 1910–1915. Phylogenetically informative changes occur in the human NA at a rate of about one change per year, whereas the rate for swine NA is about one change every 2 years.
Figure 3.
Rate of change of influenza NA genes. One most parsimonious paup N1 protein tree was imported into macclade, and unambiguous amino acid changes were traced in the human (excluding Brevig/18) and swine lineages. The number of changes was plotted vs. year of isolation and regression lines calculated and drawn by using Microsoft Excel (Microsoft, Redmond, WA) and Slide Write (Advanced Graphics, Carlsbad, CA). The regression lines were then extrapolated, and the Brevig/18 data point was plotted. Human viral isolation times were corrected for the 20-year gap in circulation of H1N1 viruses from 1957–1977 (28).
Discussion
Analysis of the genetic sequence of the 1918 influenza has been undertaken to elucidate its possible origin and the reasons for its virulence (17). The NA gene is important both because of its functional role in promoting the dissemination of the virus during infection and because, like HA, it is a principal target of the human immune system.
In many ways, analyses of the 1918 NA sequences give results similar to that of the 1918 HA study (7). In both cases, phylogenetic analyses show that of all mammalian isolates, the 1918 sequences are the most closely related to avian isolates, but also suggest that the 1918 sequences share enough characteristics with mammalian isolates to distinguish them from the avian clade. The placement of the Brevig/18 NA nucleotide sequence in the phylogenetic trees is usually within and near the root of the mammalian clade, suggesting that the 1918 NA is very similar to the ancestor of all subsequent swine and human isolates. The number of differences between the Brevig/18 sequence and early human strains, or between the Brevig/18 sequence and swine strains, is also consistent with its being the ancestor of all subsequent mammalian N1s (Fig. 3). At the same time, and in contrast to the results with HA, phylogenetic analyses of the NA protein place Brevig/18 within the avian clade. In these cases, branch lengths are very short, and bootstrap values are low, suggesting that there are not enough differences among the sequences to place them unambiguously.
The phylogenetic results suggest that Brevig/18 sequence is intermediate between avian and mammalian sequences, and are consistent with the idea that pandemic viruses acquire their surface proteins directly (with little modification) from avian viruses. Nevertheless, Brevig/18 NA differs at 26 amino acids from its nearest known avian relative (A/Dk/Alberta/76). By contrast, the 1957 pandemic N2 and the N1 from the 1997 Hong Kong H5N1 outbreak differ by only 18 and 2, respectively, from their nearest avian relatives. These results can be interpreted in different ways. Either Brevig/18 NA would more closely resemble a 1918 avian N1, but avian sequences have drifted away from that ancestral sequence over the past 80 years, or the 1918 sequence had acquired mammalian specific changes in a mammalian host in the years preceding the 1918 pandemic. That the ultimate source of the 1918 NA was avian is supported by the phylogenetic analyses, but the precise path of the gene from its avian source to its pandemic form cannot be determined by its sequence alone.
As with HA, the functional and antigenic sites of Brevig/18 NA closely resemble avian isolates. The 15 conserved amino acids making up the active site of the molecule are retained, as are the seven glycosylation sites found in all avian strains. Twenty-two amino acids have been identified as antigenic in the N2 subtype (3). Of the homologous amino acids in N1, 15 have shown variation in human strains. Brevig/18 matches the avian consensus at 14 of the 15 residues, suggesting little or no antigenic pressure on the protein before 1918. Human strains from the 1930s show drift at these sites, with PR/34 differing at 5 of the 15 and WS/33 at 8 of the 15 amino acids.
Several early human strains have deletions of 11 to 16 amino acids in the stalk region of NA (18) that may affect the activity of the protein (19). It has been suggested that the stalk deletions in the earliest human strains, PR/34 and WS/33, may have been inherited from the 1918 pandemic strain (20). However, PR/34 and WS/33 have different stalk deletions, Weiss/43 has a full-length stalk, FM/47 has yet a third distinct deletion, and all subsequent human isolates have full-length stalks. Sw/Iowa/30, shown phylogenetically to be a descendant of the 1918 virus, does not have a stalk deletion. Stalk deletions are common in N1 strains found in chickens (21) but were not found in the five wild-type avian N1s sequenced for this study. The 1918 strain does not have a stalk deletion, suggesting that the various deletions found in early human strains are likely to be artifacts of their extensive culture in various hosts, although naturally occurring stalk deletions in these early human viruses cannot be ruled out.
The amino acid residue 354 in the NA protein is of potential interest. This residue is Asp in Brevig/18. An extensive search of GenBank (in December, 1999) demonstrated that this position is occupied by Gly in nearly all other NA proteins, regardless of subtype. Three exceptions were found, all of which have Glu at this position: A/Equine/Prague/1/56 (H7N7), A/Equine/Cornell/16/74 (H7N7), and A/Swine/England/191973/92 (H1N7). Position 354, which is buried internally in the globular head of NA (as aligned to the N9 crystal structure), is located between two blocks of amino acids that are well conserved in all NA subtypes, and the N-terminal block contains one of the amino acids known to be an active site residue. The change from Gly to Asp (or Glu) at this position could conceivably result in a conformational change in this region of the protein, with consequences affecting NA activity and possibly virulence.
The minimal changes, if any, necessary to allow an avian N1 to function in a mammalian host have not been determined. Differences between Brevig/18 NA and its nearest avian relative are not all necessarily related to mammalian adaptation. Some may reflect differences between the current nearest avian relative and the pandemic's actual 1918 avian ancestor, and some could reflect random mutations that did not diminish the fitness of the virus. One way to distinguish among these possibilities is to compare the 1918 sequence with that of Sw/England/92, a swine-adapted H1N1 that resulted from the independent introduction of an avian H1N1 into European swine in the late 1970s. The H1N1 lineage represented by Sw/England/92, although it was detected in pigs as early as 1979, remains distinctly avian phylogenetically (Fig. 2). Nevertheless, it must by definition have the minimal adaptations necessary to allow it to function in a mammalian host. As shown in Table 2, five amino acids have changed in two separate introductions of avian N1s into mammals (Sw/England/92 and Brevig/18); they may represent the minimal changes necessary for mammalian adaptation. Two of these positions have replacements that are either chemically similar (285) or identical (344) in Brevig/18 and Sw/England/92, suggesting that these two sites may be particularly important.
One of the characteristics of the 1918 pandemic was its unusual virulence, reflected in the heightened severity of illness and the prevalence of pneumonic complications. The virulence of influenza viruses is a complicated function of genetic characteristics of the virus itself, the immune status of the infected individual, and the dose and route of transmission. Pandemic viruses generally exhibit higher virulence than interpandemic strains, probably in large part because of their antigenic novelty. If both HA and NA are replaced, as occurred in 1957, the pandemic strain will likely be more virulent than if only one of these antigenic targets is replaced, as occurred in 1968 when only HA was replaced. The severity of the 1918 pandemic suggests that both HA and NA were antigenically novel and that the virus had not circulated widely in the human population before the spring of 1918. This is supported by sequence and phylogenetic analyses of both 1918 HA (7) and NA.
The relationship between virulence and the genetic structure of the virus is complex. There are a few examples of simple changes in a single gene resulting in dramatic changes in virulence. One of these is the insertion of basic amino acids in the HA cleavage site, which allows the virus to grow in many tissues outside its normal host cells. Although this change has been found in the H5 and H7 subtypes in birds, it was not found in the 1918 HA (7, 22). In the NA gene, the loss of a glycosylation site at amino acid 146 in WSN/33 (15) contributes to making the virus exceptionally virulent as well as neurotropic in mice (23, 24). This change also was not found in the 1918 strains. Future research may reveal other single gene changes with a dramatic effect on virulence.
It is clear that successful virus replication and spread depends on tightly coordinated interactions among the virus' genes. Reassortment is an important source of variation in influenza viruses, but not all gene combinations result in functional virus, and several genes appear to contribute to the adaptation of a virus to a new host. A pandemic virus must, by definition, be both well adapted to replication in and spread among humans and antigenically novel. In 1957 and 1968, this dual requirement was met by the combination of novel surface protein(s) and PB1 from avian sources with other internal proteins from the previously circulating human-adapted virus (25–27). Analysis of the surface proteins of the 1918 virus suggests that it also acquired its HA and NA from avian viruses, although it is possible that these genes may have been adapting in a mammalian host for some time before emerging in pandemic form.
Acknowledgments
We are grateful to Dr. Johan V. Hultin and the residents of Brevig Mission, AK, for their generosity in supporting this project. We also thank Drs. Edwin D. Kilbourne, Richard D. Slemons, and Ian H. Brown for their contributions of strains and sequences. This project was supported by grants from the American Registry of Pathology, the Department of Veterans Affairs, and intramural funds of the Armed Forces Institute of Pathology.
Abbreviations
- HA
hemagglutinin
- NA
neuraminidase
- RT-PCR
reverse transcription–PCR
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF250356–AF250365).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100140097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100140097
References
- 1.Palese P, Compans R W. J Gen Virol. 1976;33:159–163. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- 2.Burnet F M. Aust J Exp Biol Med Sci. 1948;25:227–233. doi: 10.1038/icb.1947.33. [DOI] [PubMed] [Google Scholar]
- 3.Colman P M, Varghese J N, Laver W G. Nature (London) 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 4.Webster R G, Hinshaw V S, Laver W G. Virology. 1982;117:93–104. doi: 10.1016/0042-6822(82)90510-4. [DOI] [PubMed] [Google Scholar]
- 5.Jahiel R I, Kilbourne E D. J Bacteriol. 1966;92:1521–1534. doi: 10.1128/jb.92.5.1521-1534.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kilbourne E. J Am Med Assoc. 1977;237:1225–1228. [Google Scholar]
- 7.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosby A. America's Forgotten Pandemic. Cambridge, U.K.: Cambridge Univ. Press; 1989. [Google Scholar]
- 9.Krafft A E, Duncan B W, Bijwaard K E, Taubenberger J K, Lichy J H. Mol Diagn. 1997;2:217–230. doi: 10.1054/MODI00200217. [DOI] [PubMed] [Google Scholar]
- 10.Swofford D L. paup: Phylogenetic Analysis Using Parsimony, Version 3.1.1. Champaign: Illinois Natural History Survey; 1991. [Google Scholar]
- 11.Kumar S, Tamura K, Nei M. Molecular Evolutionary Genetics Analysis, Version 1.01. University Park: Pennsylvania State Univ.; 1993. [Google Scholar]
- 12.Maddison W P, Maddison D R. macclade: Analysis of Phylogeny and Character Evolution, Version 3. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 13.Shope R E. J Exp Med. 1936;63:669–684. doi: 10.1084/jem.63.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze I T. J Infect Dis. 1997;176, Suppl. 1:S24–S28. doi: 10.1086/514170. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Schulman J, Itamura S, Palese P. J Virol. 1993;67:6667–6673. doi: 10.1128/jvi.67.11.6667-6673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blok J, Air G M. Virology. 1980;107:50–60. doi: 10.1016/0042-6822(80)90271-8. [DOI] [PubMed] [Google Scholar]
- 17.Reid A H, Taubenberger J K. Lab Invest. 1999;79:95–101. [PubMed] [Google Scholar]
- 18.Blok J, Air G M. Virology. 1982;119:229–234. doi: 10.1016/0042-6822(82)90337-3. [DOI] [PubMed] [Google Scholar]
- 19.Castrucci M R, Kawaoka Y. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matrosovich M, Zhou N, Kawaoka Y, Webster R. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou N N, Shortridge K F, Claas E C J, Krauss S L, Webster R G. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 23.Francis T, Moore A. J Exp Med. 1940;72:717–728. doi: 10.1084/jem.72.6.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taubenberger J. Proc Natl Acad Sci USA. 1998;95:9713–9715. doi: 10.1073/pnas.95.17.9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gething M J, Bye J, Skehel J, Waterfield M. Nature (London) 1980;287:301–306. doi: 10.1038/287301a0. [DOI] [PubMed] [Google Scholar]
- 26.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 27.Kawaoka Y, Krauss S, Webster R G. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buonagurio D, Nakada S, Parvin J, Krystal M, Palese P, Fitch W. Science. 1986;232:980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- 29.Sayle R. Greenford, Middlesex, U.K.: Glaxo Research and Development; 1994. [Google Scholar]