Abstract
The avian leukosis virus ΔLR-9 causes a high frequency of B-cell lymphomas within weeks after injection into 10-day-old chicken embryos. These lymphomas result from proviral integrations into the oncogene c-myb. In contrast, LR-9, which lacks the 42-nucleotide gag gene deletion of ΔLR-9, does not cause a high frequency of c-myb-associated short-latency lymphomas. Although viral replication rates and spliced env mRNA levels were found to be similar for both viruses, ΔLR-9 exhibited an increase in readthrough transcription compared to LR-9. The ΔLR-9 deletion is located in the region of the gag gene corresponding to the matrix (MA) protein as well as in the negative regulator of splicing (NRS) element. To test whether disruption of the NRS or of the MA protein was responsible for inducing short-latency lymphomas, we generated viruses with NRS point mutations that maintained the wild-type Gag amino acid sequence. One of the mutant viruses induced an even higher incidence than ΔLR-9 of short-latency lymphomas with viral integrations into c-myb. Thus, we propose that disruption of the NRS sequence promotes readthrough transcription and splicing to the downstream myb gene, causing overexpression of a slightly truncated Myb protein, which induces short-latency tumors.
A defining characteristic of retroviruses is that they randomly integrate their reverse-transcribed genome into the genome of the host cell. Selection for a specific integration site can occur when a retrovirus integrates into a proto-oncogene, resulting in a clonal tumor. When an avian leukosis virus (ALV) is injected into 1-day-old chicks, it typically induces clonal B-cell lymphomas exhibiting proviral DNA integrated into the oncogene c-myc (13). These B-cell lymphomas result from the deregulated expression of c-myc and take several months to develop. However, if an ALV is injected into 10- to 12-day-old chicken embryos, approximately 14% of the hatched chickens develop short-latency B-cell lymphomas that cause death within weeks (29). Moreover, injection of chicken embryos with the recombinant ALV EU-8 (which has a 42-nucleotide [nt] deletion in the gag gene) leads to short-latency B-cell lymphomas in 40 to 80% of the chickens (16, 32).
Most of these short-latency lymphomas contain proviral DNA integrated into the first intron of c-myb (15, 29). Transcription initiates at the ALV promoter in the 5′ long terminal repeat (LTR) and reads through the polyadenylation sequence in the 3′ LTR to transcribe the downstream c-myb gene (see Fig. 1). Splicing then occurs from the 5′ splice site in the 5′ end of the viral gag gene to the 3′ splice site of the c-myb second exon (Fig. 1) (15). Translation of the ALV-myb hybrid RNA yields a Myb protein that is truncated at its N terminus by 20 amino acids (15). Truncated Myb, in contrast to full-length Myb, is highly oncogenic when expressed from retroviral vectors introduced into 12-day-old chicken embryos (15). Further, the transcriptional profile of the short-latency, c-myb-associated tumors is distinct from that of c-myc-associated lymphomas (25).
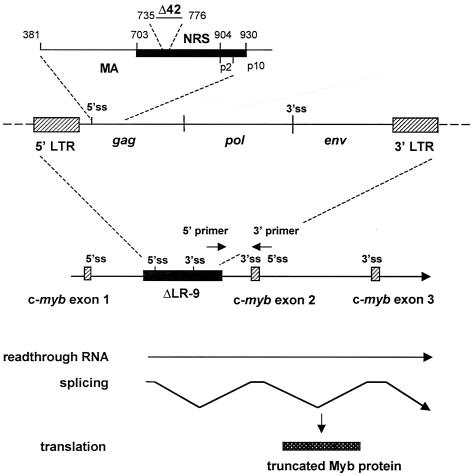
FIG. 1.
Integration of the ΔLR-9 ALV provirus into c-myb results in the expression of a truncated Myb protein. The NRS element spans nt 703 to 930 within the ALV gag gene. ΔLR-9 has a deletion of 42 nt (nt 735 to 776), which lies within both the NRS and the gag region corresponding to the MA protein (nt 381 to 904). When ΔLR-9 integrates into the first intron of c-myb, transcription initiates normally in the 5′ LTR but occasionally reads through the polyadenylation sequence in the 3′ LTR to transcribe c-myb. Splicing then occurs from the viral 5′ splice site (5′ss) to the 3′ splice site (3′ss) of c-myb exon 2. Translation produces a truncated and highly oncogenic Myb protein that is missing its first 20 N-terminal amino acids (15). Viral integrations into c-myb were detected by nested PCR with 5′ primers complementary to the viral LTR and 3′ primers complementary to c-myb exon 2.
The higher incidence of short-latency tumors obtained with EU-8 than with other ALVs has been attributed to a 42-nt deletion in the gag gene of EU-8, which was derived from ring-necked pheasant virus (33). This same region (nt 735 to 776) was deleted from the recombinant ALV LR-9 to produce ΔLR-9. When injected into 10-day-old chicken embryos, ΔLR-9 also causes mortality from short-latency B-cell lymphomas at a higher frequency than LR-9 (33).
The ΔLR-9 genomic deletion is located within the gag gene, and it coincides with the 5′ portion of a viral RNA element termed the negative regulator of splicing (NRS) (Fig. 1) (1, 23). When the NRS of ΔLR-9 was inserted into the intron of a heterologous splicing reporter construct, splicing was increased relative to that in a construct containing the wild-type (WT) LR-9 NRS (33). The NRS has previously been characterized in Rous sarcoma virus (RSV), and its sequence is 95% identical to that of the NRS of the ALV LR-9 (33). The RSV NRS is a 230-nt-long cis-acting element, which limits both RNA splicing and readthrough transcription (1, 10, 24, 28). Both of these processes are critical to c-myb-associated short-latency tumorigenesis. In RSV, the NRS suppresses splicing of the src mRNA but not of env mRNA (28). The NRS has two regions necessary for function. A purine-rich region at its 5′ end binds arginine/serine-rich (SR) proteins and hnRNP H (9, 20, 21). The 3′ end of the NRS contains a sequence that is similar to a 5′ splice site and binds U1 snRNP (14, 22). U1 snRNP binding is thought to be important for NRS activity; mutations in its binding site inhibit NRS activity, and a complementary U1 snRNA mutation partially rescues activity (14).
The deletion in the ΔLR-9 genome is also within the region of gag corresponding to the matrix (MA) protein, thus generating a truncated MA protein (see Fig. 1) (15). To determine whether the mutation in MA or the NRS was important for short-latency tumorigenesis, we tested viruses with point mutations that code for a WT Gag protein but alter the NRS. In addition, the binding of various splicing factors to the NRS RNA was studied with the NRS mutants. Finally, to further study how the ΔLR-9 deletion promotes tumorigenesis, its effects on viral replication, env mRNA splicing, and readthrough transcription were examined.
MATERIALS AND METHODS
Cells and viruses.
Chicken embryo fibroblasts (CEFs) were cultured in medium 199 with the addition of 2% tryptose phosphate, 1% calf serum, 1% heat-inactivated chick serum, and 1% antibiotic-antimycotic (Life Technologies). The permuted LR-9 (32) and ΔLR-9 (33) ALV constructs are both inserted into the vector pUC13 and were a gift from Bill Hayward. U916A, G919A, and WT NRS ALVs were constructed by inserting RSV fragments from the BspEI site (nt 700) to the KpnI site (nt 4,995) into the corresponding sites in LR-9. U916A and G919A are both silent mutations, which were originally described in RSV by O'Sullivan et al. (28). Ten micrograms of SacI-linearized viral constructs was transfected into CEFs as previously described (1).
Reverse transcriptase assay.
After harvesting, the virion-containing medium was assayed for reverse transcriptase activity as described by Telesnitsky et al. (35). Quantitation was performed on an Instant Imager (Packard).
RNase protection assay.
The readthrough probe contains the entire LR-9 LTR and flanking sequences. The permuted, single-LTR LR-9 genomic plasmid was digested with SacI (nt 255) and HindIII (nt 7,040), and the LTR fragment was inserted into the pGEM-4Z cloning vector (Stratagene). The construct was linearized with BanI prior to in vitro transcription with Sp6 RNA polymerase. Total cellular RNA was harvested with RNA Bee (Tel-Test). Readthrough RNase protections were carried out as described previously (23) by using a hybridization temperature of 42°C and an RNase digestion temperature of 37°C. The bands were quantified on an Instant Imager (Packard), and the values were adjusted for size differences. The fraction of readthrough RNA was determined relative to the sum of normally polyadenylated and readthrough RNA bands.
mRNA selection and Northern blotting.
Polyadenylated RNA was selected from total cellular RNA by binding to oligo(dT) cellulose, and the selected RNA was used for Northern blotting as described by Sambrook and Russell (31). Viral RNA was hybridized to the readthrough (LTR) probe described above, which is complementary to both spliced and unspliced viral RNAs. The bands were quantified on an Instant Imager (Packard).
Chicken injection and tumor detection.
Virus was collected at day 11 after transfection of CEFs, and 105 to 106 infectious U was injected into the chorioallantoic veins of 48 10-day-old chicken embryos from the inbred SC White Leghorn line (Hyline International, Dallas, Iowa). Thirty-seven of the injected embryos hatched, and 34 were monitored for tumor formation. Chickens infected with different viruses were kept in separate cages, and they were sacrificed when ill or at 10 weeks after hatching. Each chicken was examined post mortem for tumors by gross examination and by tissue section staining as described by Neiman et al. (25). Tissue samples were collected for DNA extraction. All chicken manipulations were performed without knowledge of the experimental hypothesis.
DNA extraction and nested PCR.
DNA from chicken bursa and liver tissue was extracted as described by Sambrook and Russell (31). Nested PCR was carried out to detect integrations into c-myb by using primers specific for the proviral LTR and for myb exon 2, as described by Neiman et al. (25). The upstream c-myb integration was detected with a 3′ primer (5′-CGGCACGGAGCGCGCTTTGCGTGCGTG-3′) upstream of exon 1 of c-myb in conjunction with the same LTR primers (25).
Affinity selections.
The viral constructs were used as templates for PCRs with a T7 promoter (underlined)-containing 5′ primer (5′-TAATACGACTCACTATAGGGAGGTCCGGAGTGCATCGAGAAACC-3′) hybridizing to NRS nt 703 to 727 and a 3′ primer (5′-AGGGAAGGATACAAACCACTCCCCACA-3′ for LR-9, ΔLR-9, and the WT; 5′-AGGGAAGGATACAATCCACTCCCCACA-3′ for U916A; and 5′-AGGGAAGGATATAAACCACTCCCCACA-3′ for G919A) hybridizing to nt 930 to 907. RNA synthesis, linkage to adipic acid dihydrazide-agarose beads, and affinity selections were performed as described previously (4, 6). Both affinity-selected proteins and RNAs were examined. For RNA analysis, the washed pellet was digested with 50 μg of proteinase K (Roche) for 30 min at 37°C in 300 μl of proteinase K buffer. The samples were then boiled for 30 s and extracted with phenol and with phenol-chloroform-isoamyl alcohol (25:24:1). After ethanol precipitation, the pellet was resuspended in 20 μl of 8 M urea, electrophoresed on a 6% polyacrylamide-8 M urea gel, and transferred to a Zeta-Probe GT blotting membrane (Bio-Rad) with a transblot semidry transfer apparatus (Bio-Rad). The blot was probed with a U1 snRNA-specific probe (11), which was linearized with EcoRI prior to transcription with T3 polymerase. Affinity-selected proteins were separated on a sodium dodecyl sulfate-13% polyacrylamide gel, transferred, and probed as described by Sambrook and Russell (31). Antibodies against ASF/SF2 were obtained from A. Krainer, and antibodies against hnRNP H were obtained from I. Mattaj. A Coomassie-stained gel (GelCode Blue staining reagent; Perbio) was used to normalize the signals.
RESULTS
LR-9 and ΔLR-9 exhibit similar replication rates and env mRNA levels.
The 42-nt deletion in the gag gene of ΔLR-9 ALV disrupts the NRS sequence, and it also deletes residues 119 to 132 of the MA protein (Fig. 1). In an effort to determine how the deletion in ΔLR-9 promotes tumorigenesis, we first assayed its effect on viral replication. CEFs were infected with equivalent reverse transcriptase units of either LR-9 or ΔLR-9 virus. To monitor growth rates, medium was collected every other day starting with day 3 and assayed for viral reverse transcriptase activity. Based on reverse transcriptase levels, the replication rates of LR-9 and ΔLR-9 were found to be virtually identical (Fig. 2A). Thus, we conclude that although ΔLR-9 has an extensive deletion in MA and in the NRS sequence, this does not affect the replication rate of the virus in CEFs.
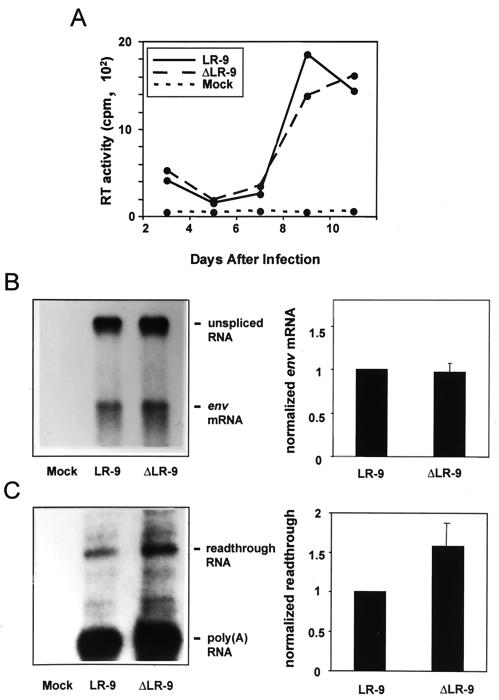
FIG. 2.
Replication rates and env mRNA levels are identical for LR-9 and ΔLR-9, but ΔLR-9 shows an increase in readthrough transcription. (A) To study replication rates, reverse transcriptase (RT) activity was measured every 2 days after infection of CEFs with LR-9 and ΔLR-9. Results representative of those from four experiments are shown. (B) Polyadenylated viral RNA was detected by Northern blotting with an LTR probe. Both LR-9 and ΔLR-9 generated ∼35% spliced env mRNA. A representative gel and the averages of results from three normalized experiments are shown. (C) Readthrough transcription was measured by RNase protections with a readthrough probe, which includes the LTR and flanking sequences and is similar to the one described by O'Sullivan et al. (28). Briefly, the 653-nt-long probe protects 397 nt of correctly terminated 3′ RNA and 498 nt of readthrough RNA. The 255-nt fragment that represents the 5′ RNA initiated in the 5′ LTR is not shown. The percentage of readthrough transcription relative to the total amount of 3′ RNA [readthrough and normally terminated 3′ poly(A)] was found to be ∼10% for LR-9. The figure shows a representative gel and the averages of results from four normalized experiments.
Next, we wanted to determine whether the deletion affected levels of spliced env mRNA in ALV. In RSV, NRS mutations increase src but not env mRNA levels (28). LR-9 and ΔLR-9 constructs were transfected into CEFs, RNA was harvested after 6 days, and the relative amount of spliced env mRNA was determined by Northern blotting with an LTR-specific probe. In this assay, approximately 35% of LR-9 RNA was found to be spliced, and ΔLR-9 RNA was spliced to a similar level (Fig. 2B). Therefore, the deletion in the NRS did not result in an increase in splicing to the viral env gene, which is consistent with the finding that replication was not affected by the deletion.
ALV with an NRS deletion has increased readthrough transcription.
The short-latency tumors induced by ΔLR-9 are likely due to overexpression of truncated Myb, which is translated from a readthrough transcript as shown previously for the related EU-8 ALV (16). Since the NRS has been implicated in promoting polyadenylation of RSV transcripts (10, 24, 28), we next wanted to determine whether the rate of readthrough transcription is different for ΔLR-9 than for the LR-9 virus. To this end, plasmids containing LR-9 and ΔLR-9 DNAs were transfected into CEFs, and total cellular RNA was harvested after 11 days. The levels of readthrough transcription were compared to those of correctly terminated transcripts by RNase protection with a readthrough probe that was complementary to the LR-9 LTR and flanking sequences (Fig. 2C). In these experiments, it was not important where the virus was integrated, because only readthrough into viral sequences was assayed. LR-9 was determined to have an average of 10% readthrough transcripts (relative to the total amount of readthrough and normally terminated polyadenylated RNA), and ΔLR-9 had a 60% higher amount of readthrough (P < 0.005) (Fig. 2C). We conclude that one of the factors that causes ΔLR-9 to induce short-latency lymphomas is that it exhibits a higher level of readthrough transcription into downstream cellular genes than does LR-9. In addition, it was previously shown that the NRS of ΔLR-9 promotes splicing to a downstream reporter gene (33).
Is the deletion in the MA protein or the NRS sequence of ΔLR-9 responsible for the increase in short-latency lymphomas?
The short-latency tumors induced by ΔLR-9 may be due to either an altered MA protein or the disruption of the overlapping NRS sequence (or both). To distinguish between these possibilities, we generated point mutations that changed the NRS sequence but did not affect the amino acid sequence of the MA protein. We constructed two viruses, U916A and G919A, each having a single NRS point mutation but lacking the 42-nt deletion of ΔLR-9. U1 snRNP binding has been shown previously to be important for NRS function (8, 14), and both of these mutations were predicted to disrupt the U1 snRNP binding consensus sequence (Fig. 3A). Nevertheless, only one of these point mutations, G919A, led to an increase in readthrough transcription and splicing to src in RSV (28). Since both readthrough and splicing are thought to be important for the generation of truncated Myb, we predicted that the U916A mutant virus would behave like WT virus. The viruses with point mutations, U916A and G919A, and the WT NRS control virus were constructed by placing the corresponding RSV fragments (nt 700 to 4,995), described previously (28), into the LR-9 background (Fig. 3A). The replication abilities of the constructs were confirmed in CEFs through reverse transcriptase assays (data not shown).
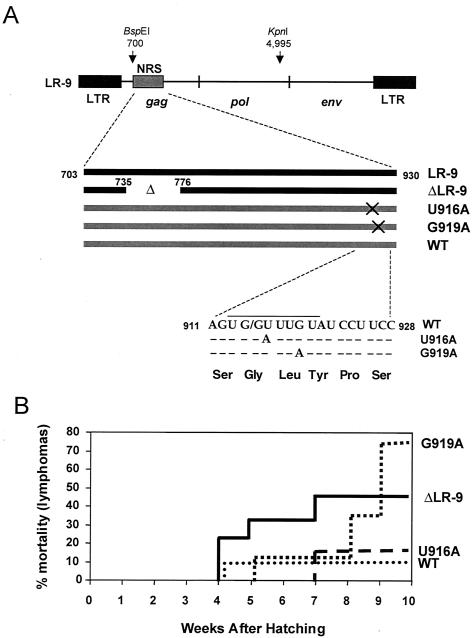
FIG. 3.
Infection of 10-day-old chicken embryos with ΔLR-9 and G919A ALVs causes high mortality from short-latency lymphomas. (A) The mutants with silent point mutations, U916A and G919A, and the WT control were generated by placing a gag-pol fragment (BspEI-KpnI) from the respective RSV constructs (28) into the LR-9 background. The NRS sequences from nt 703 to 930 are shown for LR-9 and ΔLR-9 (black lines) and U916A, G919A, and WT (gray lines; derived from RSV). Point mutations are indicated by X's. The point mutations are in the 3′ region of the NRS, do not overlap with the ΔLR-9 deletion in the 5′ region, and do not affect the Gag amino acid sequence. The line over the WT NRS sequence denotes the NRS region that is similar to the 5′ splice site consensus sequence. (B) Ten-day-old chicken embryos were infected with the viruses shown above, and their mortality from lymphomas was monitored for 10 weeks after hatching (short-latency period).
To determine the tumorigenicity of the viral constructs, we injected 12 10-day-old chicken embryos with one of the following four test viruses: the viruses with point mutations, U916A and G919A, the WT NRS (negative control) virus, and ΔLR-9 (positive control). Out of the total of 48 injected chicken embryos, 37 chicks hatched and 34 were monitored for deaths and the development of tumors for 10 weeks after hatching. Surviving chickens were sacrificed at week 10, and all chickens were examined for tumors at the time of their death. We asked whether short-latency lymphomas (causing death within 10 weeks after hatching) would arise at the same frequency with the viruses carrying NRS point mutations as with the positive control virus, ΔLR-9.
The percentages of chickens that died, or were sacrificed for humane reasons, before 10 weeks after hatching (defined as the short-latency period) and that exhibited lymphomas are plotted in Fig. 3B. Lymphomas were detected both by gross examination and by histological analysis. The group infected with the positive control, ΔLR-9, showed a rate of mortality from lymphomas of 44% (four of nine birds), which is similar to the 54% that Smith et al. (33) observed by week 10 with the same viral construct (Fig. 3B). Remarkably, the G919A viral construct, which has only a single point mutation compared to the WT, induced a 75% (six of eight birds) rate of mortality from short-latency lymphomas in this same time period (Fig. 3B). Moreover, one of these G919A-induced lymphomas was extraordinarily large; the metastatic tumor filled the full length of the chicken's body cavity. The group receiving the negative control WT virus, which has a WT NRS sequence, exhibited a 10% (1 of 10 birds) rate of mortality from lymphomas, similar to the rate that was found by Smith et al. (33) for LR-9 (Fig. 3B). Those infected with the other silent mutant, U916A, showed only a 14% (one of seven birds) mortality rate that was close to that for the negative control (Fig. 3B). U916A did not induce any short-latency tumors observable by gross examination, but one bird, which died 7 weeks after hatching, exhibited a large number of neoplastic follicles (20% of the total) in the bursa (for a detailed discussion of neoplastic follicles, see reference 25). Additional symptoms exhibited by most chickens that died from short-latency lymphomas included anemia, ataxia, cachexia, and hepatomegaly.
Short-latency lymphomas are associated with integrations into c-myb.
We next wanted to determine the integration sites of proviruses in chickens that died from short-latency lymphomas (Fig. 3B). Proviral integrations into the c-myb gene were seen previously in tumors induced by the EU-8 ALV, which has an NRS deletion identical to that of ΔLR-9 (15, 16). DNA was extracted from both bursa and liver tissues of the chickens. Proviral integration into c-myb was assayed by nested PCR by using primers in the viral LTR and either in c-myb exon 2 (Fig. 1B) or upstream of c-myb exon 1 (primer location not shown). A PCR product indicated that there was an integration into c-myb in the same transcriptional orientation as c-myb.
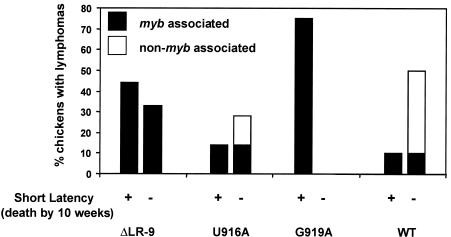
All of the chickens that died with lymphomas during the 10-week period after hatching (defined here as short-latency lymphomas) did indeed show proviral integrations into c-myb (Fig. 4). In addition, a number of birds sacrificed at week 10 were also found to have lymphomas. We asked whether the chickens that developed lymphomas but did not die by week 10 also had proviral integrations into the c-myb gene. The percentage of chickens that developed these lymphomas is shown in Fig. 4. ΔLR-9 induced lethal lymphomas in 44% (four of nine) and nonlethal lymphomas in 33% (three of nine) of the infected chickens; all of these birds exhibited proviral integrations into c-myb (Fig. 4). Nonlethal lymphomas were induced by U916A in 28% (two of seven) of the chickens, but only one of these chickens had a c-myb integration (Fig. 4). Of the chickens infected with the G919A ALV mutant, 75% (six of eight) developed lymphomas and died before week 10; all of these short-latency lymphomas were associated with proviral integrations into c-myb. No nonlethal lymphomas were observed in the remaining two birds infected with G919A. Finally, the negative control (WT) induced nonlethal lymphomas in 50% (5 of 10) of the chickens. Interestingly, only one of these chickens had a proviral integration into c-myb. The non-myb-associated, nonlethal (at week 10) lymphomas induced by either the WT NRS virus or the U916A mutant are likely to have proviral integrations into other proto-oncogenes. There were no morphological differences between the short-latency lymphomas and the nonlethal lymphomas (by 10 weeks) observable by either gross examination or histological staining (data not shown).
FIG. 4.
All short-latency tumors were associated with viral integrations into c-myb. All of the chickens that died with short-latency lymphomas during the 10 weeks after hatching (bars on left; also plotted in Fig. 3B) had viral integrations into c-myb as observed by nested PCR. The bursa and liver of each chicken that exhibited nonlethal lymphomas 10 weeks after hatching were also examined for viral integration into c-myb (bars on right). These nonlethal lymphomas were associated with integrations into c-myb in all of the ΔLR-9-induced tumors but to a lesser extent in the tumors induced by U916A and WT viruses. G919A caused only short-latency lymphomas.
Finally, by this PCR assay, c-myb integrations into DNA from both the bursa and liver were observed in the absence of obvious tumor formation in six chickens (three infected with U916A, one with G919A, and two with WT) but not in a control uninfected chicken. Even though observable tumors did not develop in these chickens, there appears to have been some selection for cells with proviral integration into c-myb.
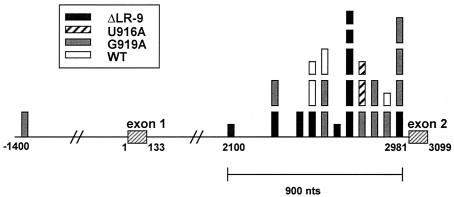
Integration sites cluster within 900 nt upstream of c-myb exon 2.
To determine whether specific integration sites correlated with the induction of short-latency lymphomas, we mapped the approximate sites of proviral insertion into c-myb based on the sizes of the PCR products. Strikingly, most of the integration sites clustered within 900 nt upstream of the start of c-myb exon 2 (Fig. 5). In addition, one integration site was found upstream of c-myb exon 1 in the bursa and the liver of a G919A-infected chicken (by using an alternate upstream myb primer). There was no apparent difference between the integration sites found in lymphomas induced by the different viral constructs. In addition, short-latency lymphomas and lymphomas that did not lead to death in the time course of the experiment exhibited similar proviral integration sites. Most chickens that had an integration into c-myb intron 1 had only one such integration; however, multiple c-myb integrations (two or three) were seen in six chickens.
FIG. 5.
Proviral integration sites cluster within 900 nt upstream of c-myb exon 2. Each bar represents an independent integration site that was determined by nested PCR. Nearly all integration sites were within the 3′ third of the 2.8-kb first intron of c-myb; in addition, one integration site was found about 1.4 kb upstream of myb exon 1. Larger boxes indicate integrations associated with short-latency lymphomas, and smaller boxes indicate integrations that resulted in lymphomas that were not lethal within 10 weeks after hatching.
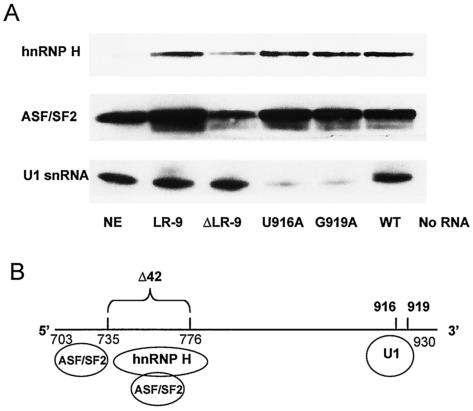
Effects of mutations on the binding of factors to the NRS.
The NRS was shown to be important in preventing tumorigenesis since the virus with the NRS point mutation G919A caused a high frequency of short-latency lymphomas, even though it encoded a WT MA protein. Since the U916A mutant did not cause a high frequency of short-latency lymphomas, we wanted to determine whether the mutant RNAs differed in the binding of U1 snRNP or other factors. Affinity selection experiments were carried out with NRS RNAs from LR-9, ΔLR-9, U916A, G919A, and WT (RSV) viral constructs (Fig. 3A). The NRS RNAs (RSV nt 703 to 930) were transcribed in vitro and covalently linked to agarose beads. After incubation with HeLa nuclear extract, the levels of binding of hnRNP H and ASF/SF2 proteins and U1 snRNA were determined by Western and Northern blot analysis, respectively. These particular factors were selected because they have been shown previously to have a functional interaction with the RSV NRS (9, 14, 20, 21, 22). The levels of binding of both hnRNP H and ASF/SF2 were found to be substantially reduced for ΔLR-9 RNA relative to those for LR-9 RNA and RNAs of all the other constructs (Fig. 6A). This suggests that the primary binding sites for ASF/SF2 and hnRNP H in the NRS are in the region spanning nt 735 to 776. However, ΔLR-9 RNA did show a low level of binding of both ASF/SF2 and hnRNP H, suggesting that there may be additional binding sites outside of the deleted region. McNally and McNally reported that ASF/SF2 binding was greatly decreased by the deletion of nt 715 to 748 of the RSV NRS (20), suggesting that there may be additional binding sites upstream of the deletion in ΔLR-9, as shown in Fig. 6B.
FIG. 6.
Reduced binding of hnRNP H and ASF/SF2 proteins, but not U1 snRNA, to the ΔLR-9 NRS RNA. (A) Affinity selections were performed with in vitro transcribed NRS RNA (nt 703 to 930), which was covalently bound to agarose beads and incubated in HeLa nuclear extract. Bound hnRNP H and ASF/SF2 proteins were visualized by Western blotting, and bound U1 snRNA was visualized by Northern blotting. Equal loadings of total selected proteins in each lane were confirmed by a Coomassie-stained gel (data not shown). NE, nuclear extract. (B) The NRS has a bipartite structure, binding ASF/SF2 and hnRNP H at its 5′ end and U1 snRNP at its 3′ end.
Surprisingly, levels of U1 snRNA binding were found to be identical for LR-9 and ΔLR-9 RNAs in this in vitro assay (Fig. 6A). Thus, ASF/SF2 binding to nt 735 to 776 of the NRS does not seem to be necessary for U1 snRNP recruitment to the 3′ end of the NRS. Furthermore, the two viruses with NRS point mutations, U916A and G919A, both showed sharply diminished U1 snRNA binding (Fig. 6A). Thus, the difference in short-latency tumor induction by the U916A and G919A viruses could not be explained by differential U1 snRNP binding in this assay. In summary, the 5′ region of the NRS (specifically nt 735 to 776) binds hnRNP H and ASF/SF2, and the 3′ region of the NRS binds U1 snRNP (Fig. 6B). We were unable to differentiate between U916A and G919A on the basis of the binding of these factors; however, there may be other proteins that bind differentially to these two mutants.
DISCUSSION
In this study, LR-9 and ΔLR-9 ALVs, which differ only by a 42-nt deletion in the gag gene, were found to have the same replication rates and the same amounts of spliced env mRNA. However, readthrough transcription was found to be higher with ΔLR-9 than with LR-9. In addition, the ΔLR-9 NRS was shown previously to increase splicing to a downstream reporter gene (33). When injected into 10-day-old chicken embryos, the G919A, but not the U916A, NRS point-mutation virus showed an even higher rate of tumor formation than ΔLR-9. Thus, the deletion in the viral MA protein was not found to be necessary for short-latency lymphoma formation. We conclude that the high frequency of short-latency tumors with proviral integrations into c-myb is associated with inactivation of the NRS, implicating RNA processing as the prime determinant of short-latency tumorigenesis in this system.
The NRS is not necessary for viral replication in CEFs.
The replication rates of LR-9 and ΔLR-9 were found to be identical in CEFs. Thus, a 42-nt deletion in the 5′ portion of the NRS did not impair replication rates. Similarly, O'Sullivan et al. (28) did not find substantial replication impairment in RSV mutants bearing single point mutations in the 3′ portion of the NRS. However, some replication impairment was observed at early time points with RSV containing three scattered point mutations in the 3′ region of the NRS (14). The ΔLR-9 deletion is also in the MA protein-encoding region (corresponding to residues 119 to 132) of the virus. MA is the adapter protein that anchors the viral core to the cell's Env-studded lipid bilayer so that new virions can bud out of the cell (34). Consistent with our findings, Nelle and Wills (26) found that the entire C-terminal half of RSV MA (residues 87 to 155) is dispensable for budding and infectivity in QT6 cells. Thus, this region of the virus does not seem to be necessary for replication, even though removing it deletes portions of the MA protein and of the NRS. Furthermore, the level of splicing to the viral gene env was not affected by the 42-nt deletion in ΔLR-9 (Fig. 2B) or by NRS point mutations in RSV (28).
The NRS promotes polyadenylation.
Expression of truncated Myb to induce short-latency lymphomas necessitates readthrough transcription followed by splicing to c-myb (15) (Fig. 1). Readthrough transcription was found to be increased in the NRS mutant ΔLR-9 in this study. The level of readthrough transcription was determined to be ∼10% for LR-9. The deletion in the NRS increased the level of readthrough by 60%. The NRS was previously implicated by Miller and Stoltzfus in the regulation of polyadenylation of RSV RNA (24). In that study, large deletions, which overlapped in a 606-nt region including the NRS, led to readthrough of 50% of the viral RNA. More recently, Fogel et al. (10) and O'Sullivan et al. (28) found that more-limited mutations in the NRS also increased the level of readthrough transcription in RSV. Thus, one of the functions of the WT NRS appears to be the promotion of efficient polyadenylation at a site 6.5 kb downstream. Since polyadenylation can be promoted by an upstream 3′ splice site (27) and the ALV env 3′ splice site is weak (17), the NRS may promote polyadenylation either directly or indirectly through interaction with the 3′ splice site.
The NRS regulation of polyadenylation may be modulated through hnRNP H or SR protein binding. This study reports that ΔLR-9, which exhibited reduced hnRNP H and SR protein binding (Fig. 6A), showed an increase in readthrough transcription (Fig. 2C) and a concomitant increase in short-latency tumor formation (Fig. 3B). There are two GGGA sequences in the deleted region (nt 735 to 776); this sequence has been shown to be a binding site for hnRNP H (5). Further, a member of the hnRNP H family, DSEF-1, has been reported to bind to a G-rich region downstream of the simian virus 40 poly(A) sequence, where it helps to recruit cleavage-stimulating factor to aid both in cleavage and in polyadenylation (2). hnRNP H may perform a similar function for the NRS. Binding of hnRNP H to an RSV NRS fragment containing nt 740 to 770 was previously reported by Fogel and McNally (9). However, an RSV mutant bearing extensive point mutations in this region failed to affect the polyadenylation frequency in transient transfection assays with a replication-defective virus (10). SR proteins may also modulate polyadenylation efficiency. For instance, SR proteins SAP75 and U2AF65 can interact with the poly(A) polymerase; however, this interaction has been seen to exert a negative effect on polyadenylation efficiency (18). Nevertheless, different SR proteins may have a positive effect on polyadenylation.
Besides hnRNP H and SR proteins, components of the U1 snRNP, which also binds to the NRS, have been shown to recruit polyadenylation factors. The U1 snRNP-associated protein U1A has been found to interact with the cleavage-polyadenylation specificity factor and to aid in its recruitment (19). In addition, the U1 snRNP 70,000-molecular-weight protein has been found to bind to the polyadenylation polymerase and to inhibit polyadenylation of bovine papillomavirus RNA (12). In that case, however, the U1 snRNP binding site is immediately upstream of the AAUAAA polyadenylation signal. Thus, the NRS may promote polyadenylation not only through interactions with hnRNP H and SR proteins but also through binding of U1 snRNP or other factors.
The NRS is a bipartite element: the function of the 5′ end is not limited to recruiting U1 snRNP to the 3′ end.
Our experiments showed that a single point mutation in the NRS can cause short-latency lymphomas that are associated with integrations into the oncogene c-myb, similar to the previously characterized lymphomas induced by ΔLR-9. Thus, we have observed that mutations at either end of the NRS (the 5′ end in ΔLR-9 and the 3′ end in G919A) can increase the frequency of short-latency lymphomas. This confirms the original observation that both 5′ and 3′ NRS regions are important for the function of the NRS (1, 23). However, it has been proposed that the main function of the 5′ region of the NRS is to bind SR proteins in order to recruit U1 snRNP to the 3′ region of the NRS. McNally and McNally (22) found that U1 snRNP binding to the NRS was greatly reduced in an affinity selection assay with a mutant that was missing the 5′ region of the NRS containing the SR protein binding sites. In contrast, affinity selection experiments carried out in our lab demonstrated that the 3′ region of the NRS (nt 903 to 939) was sufficient to bind U1 snRNP (data not shown). In addition, in the present study the ΔLR-9 mutation (nt 735 to 776) did not inhibit U1 snRNP recruitment to the full-length NRS construct (Fig. 6A). Our data indicate that the function of the 5′ region of the NRS is not limited to recruiting U1 snRNP to the 3′ end since the mutation in ΔLR-9 partially inhibits NRS function (suppression of readthrough and splicing) (Fig. 2C) (33) but the ΔLR-9 NRS is nevertheless able to recruit U1 snRNP efficiently in vitro (Fig. 6A).
The G919A NRS mutant causes a high incidence of short-latency lymphomas.
In the short-latency lymphomas studied here, the locations of proviral integration sites in c-myb intron 1 were similar to those in EU-8-induced tumors previously observed (15, 16). Since most of the integrations that produced lymphomas clustered within 900 nt upstream of the start of exon 2, it is possible that the processivity of the transcribing polymerase may be a limiting factor for tumor development (reviewed in reference 30). In addition, all of the selected integrations are downstream of a transcriptional pause site found in intron 1 of c-myb, which has been studied in human and murine c-myb genes (3, 7). Also, integration into c-myb alone may be sufficient to cause short-latency lymphomas, because two of four tumors examined by inverse PCR had no other integrations (data not shown).
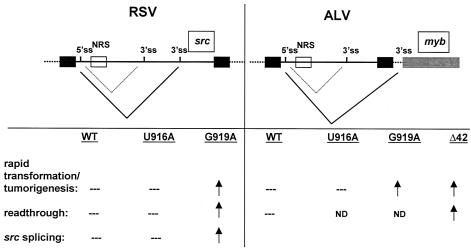
We observed that a silent NRS point mutation (G919A) caused short-latency lymphomas effectively, showing that a defective NRS and not a truncated MA protein is the cause of the high frequency of short-latency tumor induction by ΔLR-9. In the case of ΔLR-9, increased readthrough may contribute to increased expression of truncated Myb, thus explaining, at least partially, why ΔLR-9 causes a higher level of short-latency lymphomas than LR-9. Perhaps the NRS functions in ALVs to limit readthrough transcription into a wide variety of downstream cellular genes. Interestingly, the U916A mutant in RSV shows RNA readthrough levels that are identical to those of WT RSV, while readthrough is increased by 50% with the RSV G919A mutant virus (Fig. 7) (28). Thus, short-latency tumor induction with ALV mutants G919A and ΔLR-9 correlates with increased readthrough of the poly(A) site, as determined in RSV for the mutants with point mutations (Fig. 7).
FIG. 7.
Comparison of the phenotypes of NRS mutants in RSV and ALV. The RSV NRS mutant G919A, but not U916A, was previously found to cause increased readthrough of the viral poly(A) site and increased splicing to v-src compared to WT RSV (28). Accordingly, G919A transformed CEFs more rapidly than WT RSV (28). The RSV U916A mutant virus was indistinguishable from WT virus in these assays. Similarly, in the LR-9 ALV background, the NRS G919A mutant virus and viruses with the 42-nt deletion (Δ42), but not the U916A mutant virus, caused a high frequency of short-latency lymphomas. We propose that the RSV NRS regulation of splicing to v-src is similar to the ALV NRS regulation of splicing to downstream cellular genes like c-myb. 5′ss, 5′ splice site; 3′ss, 3′ splice site; ND, not determined.
In addition, once readthrough has occurred, splicing to c-myb is most likely also increased in NRS mutants, analogous to the increase in splicing to src in RSV (Fig. 7) (28). RSV has two alternate 3′ splice sites (for env and the cell-derived src) (Fig. 7). O'Sullivan et al. (28) found that the WT NRS in RSV limits splicing to the src gene but does not affect env splicing. Similarly, we did not observe an increase in env mRNA levels with an ALV bearing a deletion in the NRS. We propose that in an ALV, which lacks src, the NRS regulates splicing to a downstream cellular 3′ splice site, like that of c-myb, after readthrough transcription (Fig. 7). Furthermore, the RSV U916A mutant virus showed only a minimal increase in splicing to src, while RSV G919A showed a threefold increase in splicing to src compared to WT RSV (28). Considering the increase in both readthrough transcription (1.5-fold) and splicing (threefold) relative to those in the WT NRS virus, we predict that integration of the G919A virus into c-myb should promote approximately five times more expression of truncated Myb protein. U916A, on the other hand, does not exhibit a substantial increase in readthrough transcription or in splicing to src in RSV (28), and it also did not cause a high frequency of short-latency lymphomas in this study. Thus, the U916A mutant's phenotype is similar to that of the WT NRS virus in vivo; however, the U916A NRS differs from the WT NRS in that it does not bind U1 snRNP in vitro (Fig. 6A). Additional work will be necessary to resolve the apparent inconsistency between the effects of this NRS mutation in the viral context, assayed in vivo, and as an isolated RNA element assayed in vitro. In conclusion, the ALV NRS seems to inhibit both readthrough and splicing to downstream cellular genes, such as c-myb. In this manner, the NRS suppresses short-latency tumorigenesis, and it may also limit the occurrence of transducing viruses like RSV.
Acknowledgments
We thank William Hayward for the LR-9 and ΔLR-9 viral constructs and for helpful discussions. We thank Seyung Chung for generating the U916A and G919A ALV constructs. Antibody against ASF/SF2 was obtained from Adrian Krainer, and the antibody against hnRNP H was obtained from Iain Mattaj. Thanks to lab members for discussions concerning this work.
This work was supported by the National Institutes of Health grants RO1CA48746 to K.L.B. and RO1CA20068 to P.E.N. T.S.P. was supported in part by National Institutes of Health predoctoral training grant T32GM07231.
REFERENCES
- 1.Arrigo, S., and K. Beemon. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 8:4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagga, P., G. Arhin, and J. Wilusz. 1998. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 26:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, T., C. Thompson, and W. Kuehl. 1987. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science 237:1473-1476. [DOI] [PubMed] [Google Scholar]
- 4.Caputi, M., A. Mayeda, A. Krainer, and A. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caputi, M., and A. Zahler. 2001. Determination of the RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 276:1706-1709. [DOI] [PubMed] [Google Scholar]
- 6.Caputi, M., and A. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castellano, M., J. Golay, A. Mantovani, and M. Introna. 1992. Detection of a transcriptional block in the first intron of the human c-myb gene. Int. J. Clin. Lab. Res. 22:159-164. [DOI] [PubMed] [Google Scholar]
- 8.Cook, C., and M. McNally. 1999. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J. Virol. 73:2394-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogel, B., and M. McNally. 2000. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 275:32371-32378. [DOI] [PubMed] [Google Scholar]
- 10.Fogel, B., L. McNally, and M. McNally. 2002. Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res. 30:810-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gontarek, R., M. McNally, and K. Beemon. 1993. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 7:1926-1936. [DOI] [PubMed] [Google Scholar]
- 12.Gunderson, S., M. Polucarpou-Schwarz, and I. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly (A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 13.Hayward, W., B. Neel, and S. Astrin. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475-480. [DOI] [PubMed] [Google Scholar]
- 14.Hibbert, C., R. Gontarek, and K. Beemon. 1999. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA 5:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, W., M. Kanter, I. Dunkel, R. Ramsay, K. Beemon, and W. Hayward. 1997. Minimal truncation of the c-myb gene product in rapid-onset B-cell lymphoma. J. Virol. 71:6526-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanter, M., R. Smith, and W. Hayward. 1988. Rapid induction of B-cell lymphomas: insertional activation of c-myb by avian leukosis virus. J. Virol. 62:1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz, R., and A. Skalka. 1990. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol. Cell. Biol. 10:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko, B., and S. Gunderson. 2002. Identification of new poly (A) polymerase-inhibitory proteins capable of regulating pre-mRNA polyadenylation. J. Mol. Biol. 318:1189-1206. [DOI] [PubMed] [Google Scholar]
- 19.Lutz, C., K. Murthy, N. Schek, J. O'Conner, J. Manley, and J. Alwine. 1996. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 10:325-337. [DOI] [PubMed] [Google Scholar]
- 20.McNally, L., and M. McNally. 1996. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 70:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNally, L., and M. McNally. 1998. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol. Cell. Biol. 18:3103-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNally, L., and M. McNally. 1999. U1 small nuclear ribonucleoprotein and splicing inhibition by the Rous sarcoma virus negative regulator of splicing element. J. Virol. 73:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNally, M., R. Gontarek, and K. Beemon. 1991. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology 185:99-108. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J., and C. Stoltzfus. 1992. Two distant upstream regions containing cis-acting signals regulating splicing facilitate 3′-end processing of avian sarcoma virus RNA. J. Virol. 66:4242-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neiman, P., J. Grbiç, T. Polony, R. Kimmel, S. Bowers, J. Delrow, and K. Beemon. 2003. Functional genomic analysis reveals distinct neoplastic phenotypes associated with c-myb mutation in the bursa of Fabricius. Oncogene 22:1073-1086. [DOI] [PubMed] [Google Scholar]
- 26.Nelle, T., and J. Wills. 1996. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 70:2269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa, M., S. Rose, and S. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552-1559. [DOI] [PubMed] [Google Scholar]
- 28.O'Sullivan, C., T. Polony, R. Paca, and K. Beemon. 2002. Rous sarcoma virus splicing suppressor selectively affects src mRNA splicing and promotes polyadenylation. Virology 302:405-412. [DOI] [PubMed] [Google Scholar]
- 29.Pizer, E., T. Baba, and E. Humphries. 1992. Activation of the c-myb locus is insufficient for the rapid induction of disseminated avian B-cell lymphoma. J. Virol. 66:512-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proudfoot, N., A. Furger, and M. Dye. 2002. Integrating mRNA processing and transcription. Cell 108:502-512. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Simon, M., W. Neckameyer, W. Hayward, and R. Smith. 1987. Genetic determinants of neoplastic diseases induced by a subgroup F avian leukosis virus. J. Virol. 61:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, M., R. Smith, I. Dunkel, V. Hou, K. Beemon, and W. Hayward. 1997. Genetic determinant of rapid-onset B-cell lymphoma by avian leukosis virus. J. Virol. 71:6534-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanstrom, R., and J. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 35.Telesnitsky, A., S. Blain, and S. Goff. 1995. Assays for retroviral reverse transcriptase. Methods Enzymol. 262:347-362. [DOI] [PubMed] [Google Scholar]