Abstract
This work describes analyses of the function of the murid herpesvirus 4 strain 68 (MHV-68) M2 gene. A frameshift mutation was made in the M2 open reading frame that caused premature termination of translation of M2 after amino acid residue 90. The M2 mutant showed no defect in productive replication in vitro or in lungs after infection of mice. Likewise, the characteristic transient increase in spleen cell number, Vβ4 T-cell-receptor-positive CD8+ T-cell mononucleosis, and establishment of latency were unaffected. However, the M2 mutant virus was defective in its ability to cause the transient sharp rise in latently infected cells normally seen in the spleen after infection of mice. We also demonstrate that expression of M2 is restricted to B cells in the spleen and that M2 encodes a 30-kDa protein localizing predominantly in the cytoplasm and plasma membrane of B cells.
Murid herpesvirus 4 is an endogenous pathogen of free-living rodents of the Apodemus genus, e.g., wood mice (2). Infection of laboratory mice by murid herpesvirus 4 strain 68 (MHV-68; also called γHV-68) is an amenable model system for the study of gammaherpesvirus pathogenesis and for the development of therapeutic strategies against these viruses (1, 8, 21, 23, 37). Following intranasal inoculation of mice with MHV-68, a productive infection occurs in the lung (26). This is cleared around day 10 postinfection (p.i.) by CD8+ T cells (9), though the virus persists in a latent form in epithelial cells at this site (25). MHV-68 spreads to the spleen, where it also becomes latent, predominantly within B lymphocytes, but also in macrophages and dendritic cells (10, 27, 32, 39).
Establishment of latency in the spleen is associated with a marked splenomegaly, increase in lymphocytes in the spleen (splenic lymphocytosis) (29), and a subsequent peripheral mononucleosis that resembles that caused by primary infection of humans by Epstein-Barr virus (EBV) (28). Splenic lymphocytosis, which peaks at day 14 p.i., is driven by CD4+ T cells (9, 29) and is dependent on MHV-68-infected B cells in the spleen (32, 38). Concomitant with the lymphocytosis is a sharp rise in the number of latently infected B cells, the resolution of which to a relatively constant baseline level is achieved by CD8+ T cells (9, 38). CD8+ T cells along with antibody (17, 25) are important in the long-term control of persistent infection (6, 25, 38). Mononucleosis peaks at around day 35 p.i. and is characterized by the massive expansion of a diverse CD8+ T-cell population expressing the Vβ4 T-cell receptor (TCR) chain in conjunction with a variety of TCR α chains (13, 28). This differs from both EBV-associated mononucleosis and classical superantigen reactions in that the majority of T cells are not largely virus specific, nor is their expansion major histocompatibility complex (MHC) dependent (7, 8). However, the Vβ4+ T-cell expansion is dependent upon the presence of MHV-68-infected B cells and CD40 ligand-dependent CD4+ T-cell help (5, 12).
The left end of the unique region of the MHV-68 genome has attracted considerable interest due to the presence of four open reading frames (ORFs) (M1, M2, M3, and M4) and eight viral tRNA-like genes (vtRNAs), none of which have a homologue in other gammaherpesvirus genomes (3, 21, 35). M2 has attracted particular interest because its expression is restricted to latent infection in vitro and in vivo (15, 36), where the peak of expression occurs around day 14 p.i. in the spleen (i.e., during splenomegaly), consistent with a role in the establishment of latency (30). The 193-amino-acid M2 protein, furthermore, contains an actively recognized CD8+ T-cell epitope, suggesting that immune regulation of M2-expressing cells is critical for the resolution of splenomegaly and maintenance of the host-virus equilibrium that underlies viral persistence (15, 30).
Construction of murine gammaherpesvirus with a mutation in M2.
All molecular, cellular, biological, and virological techniques were performed exactly as described in detail by Macrae et al. (20) unless otherwise specified. To assess the function of M2, we generated a recombinant virus with a 4-bp deletion in the M2 ORF. To generate the recombinant, we used a modification of a technique that we had developed for rescue of wild-type (MHV-68) virus from the defective MHV-76 strain (20). Here, the 9.5-kbp deletion at the left end of the unique region of the MHV-76 genome (relative to MHV-68) was repaired by homologous recombination with a cosmid (cA8) that contains the relevant region from the wild-type MHV-68 genome (nucleotide sequence 115165 to 26842). To construct such a recombinant MHV-68 (rescuant) with a lesion in the M2 gene, we utilized a SalI restriction site at genome coordinate 4337 that is unique within cA8 to generate a 4-bp deletion within the M2 ORF (Fig. 1A). This deletion caused a frameshift that would result in premature termination of translation after amino acid residue 90 and mutated the known cytotoxic T-cell epitope in M2 (Fig. 1B) (15). Specifically, following restriction with SalI (Invitrogen), cA8 was digested with mung bean nuclease (Invitrogen) to remove the 5′ overhang and then religated to generate cA8ΔM2. To ensure that the predicted frameshift mutation had been introduced, cosmids that lacked a HincII restriction site within the targeted SalI site were identified and subjected to DNA sequence analysis.
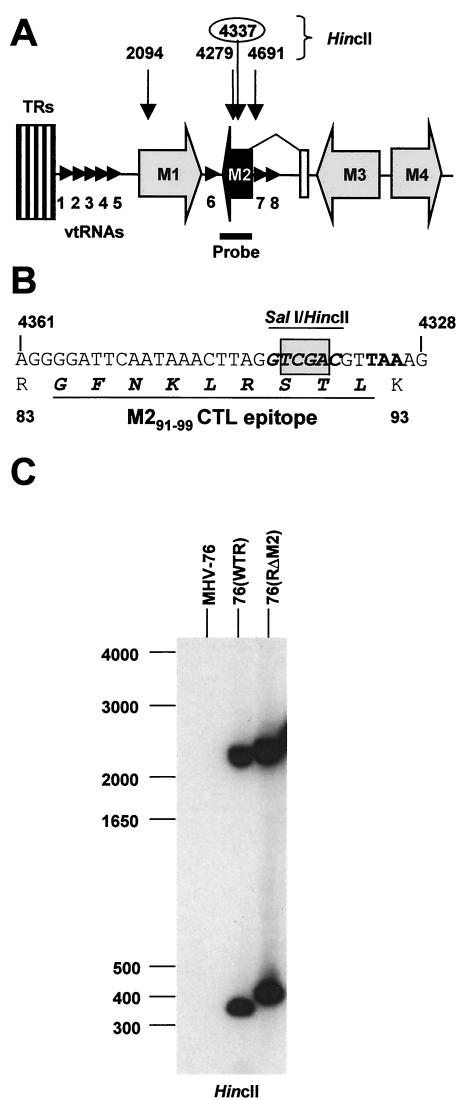
FIG. 1.
Targeted mutation of the M2 gene in MHV. (A) Schematic representation of the left end of the MHV-68 genome. ORFs are shown as shaded arrows, and the eight vtRNAs are shown as small arrowheads. TRs, terminal repeat elements. Note that the M2 ORF is within the second exon of the M2 mRNA. The positions of the HincII restriction sites (genomic coordinates given) used to evaluate recombinant virus DNA are shown above. The position of the M2 probe used in Southern analysis of viral DNA is shown below. (B) Sequence of the M2 gene and protein surrounding the site of the introduced 4-bp deletion (shaded box) at the unique SalI restriction site. The amino acid sequence of the known cytotoxic T-lymphocyte epitope within M2 is shown in underlined italic boldface. (C) Southern blot analysis of DNA isolated from BHK-21 cells infected with MHV-76, 76(WTR), or 76(RΔM2) and digested with HincII. The blot was hybridized to a 32P-labeled M2 DNA probe (nucleotides 4000 to 4606). The positions of molecular size markers (in base pairs) within the gel are shown to the left of the autoradiograph.
To generate recombinant virus, BHK-21 cells were cotransfected with cA8ΔM2 targeting vector and MHV-76 DNA as described previously (20), and the resulting plaques containing MHV with the reinserted 9.5-kbp DNA fragment containing the M2 mutation [76(RΔM2)] were identified by PCR with primers that amplified M2 (20). A wild-type rescuant [76(WTR)] containing the intact M2 gene was similarly generated with cA8 instead of cA8ΔM2. After five rounds of plaque purification by limiting dilution, recombinant virus was characterized by Southern blot hybridization and PCR analysis to ensure that it was free of contaminating MHV-76. The presence and integrity of the M2 and mutated M2 ORF were first tested by restriction digestion of infected-cell DNA with HincII followed by Southern blot hybridization with the M2 ORF as a probe. The results (Fig. 1C) showed the presence of the expected 2.2-kbp and 354-bp HincII fragments in the wild-type rescuant 76(WTR) (the expected 58-bp fragment was too small to be visualized on the blot). In 76(RΔM2) DNA the expected 2.2-kbp fragment was present, and the 354-bp fragment had been replaced by the predicted fragment of 412 bp due to the deletion of the HincII site at position 4337 and amalgamation of the 354- and 58-bp fragments. The integrity of the rest of the leftmost unique portion of the genome was confirmed by Southern analysis of HindIII-digested DNA with cA8 as a probe and by PCR analysis for M1 to M4 ORFs as described by Macrae et al. (20). Further, the gross integrity of the genomes of the two rescuant viruses was confirmed by restriction analysis of viral DNA with BamHI, HindIII, and EcoRI (data not shown). Finally, the integrity of the 4-bp mutation in 76(RΔM2) was confirmed by sequencing the DNA obtained by PCR amplification of the M2 ORF.
M2 is not required for virus replication in vitro.
To assess potential effects of the mutation in M2 on MHV-68 replication, we performed single-step virus growth curves (multiplicity of infection [MOI] of 5) and multistep growth curves (MOI of 0.01) for MHV-76, 76(WTR), and 76(RΔM2) in BHK-21 cells. As shown in Fig. 2A and B, all three viruses replicated with similar efficiency and in a similar fashion as that reported previously for MHV-68 (20). This result was not totally unexpected, since M2 is not expressed to any great extent during productive MHV-68 infection (15, 36), and MHV-76 replicates normally in tissue culture (20). Recent experiments by Jacoby et al. (16), furthermore, have yielded similar results with respect to lack of a contribution by M2 to virus replication.
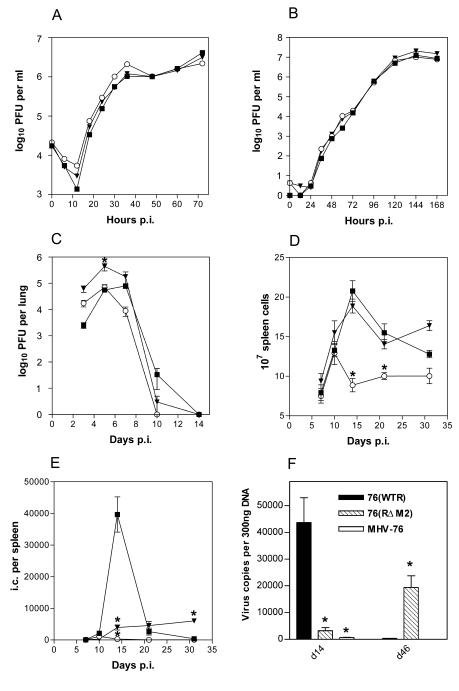
FIG. 2.
Biological characterization of viruses. In each panel data obtained from infection with MHV-76 (○), 76(RΔM2) (▾), and 76(WTR) (▪) are shown. All infections of mice (BALB/c) were intranasal with 2 × 105 PFU of virus. (A) Single-step growth curve comparing virus replication in BHK-21 cells infected at an MOI of 5. Data are representative of two independent experiments, each done in duplicate. (B) Multistep growth curve comparing replication of viruses in BHK-21 cells at an MOI of 0.01. Data are representative of two independent experiments, each done in duplicate. (C) Virus replication in the lung; data shown are the mean virus titers obtained from four mice per group at each time point. (D) Total spleen cell number within infected mice; shown are the mean cell numbers obtained from four infected mice per group at each time point. (E) Latently infected cells in the spleens of infected mice as determined by IC assay; data from four mice per group are shown at each time point. No preformed infectious virus was detected in this assay (data not shown). (F) Quantification of viral DNA in the spleen. Shown are the viral genome copy numbers per 300 ng of DNA as determined by QF-PCR at the indicated days p.i. The results are the mean values obtained from four mice per time point. In all panels, where indicated, bars represent standard errors of the means and asterisks over points represent a statistically significant difference from the wild-type value.
Effect of M2 on productive replication in the lung.
The influence of M2 on productive infection in vivo was assessed by infecting groups of BALB/c mice with MHV-76, 76(WTR), and 76(RΔM2) and monitoring infectious virus in the lungs at various times p.i. by plaque assay. The data in Fig. 2C demonstrated that there was no significant quantitative difference between the maximal titers achieved by infectious wild-type rescuant 76(WTR) (the equivalent of MHV-68) and those achieved by MHV-76; however, MHV-76 was cleared slightly more quickly. By contrast, the yield of 76(RΔM2) was minimally but significantly higher than that of either MHV-76 or 76(WTR) (day 5, P < 0.001), and infectious virus remained detectable through day 10 p.i. The results obtained for MHV-76 and 76(WTR) are similar to those reported previously (20). Unexpectedly, the mutation in M2 appeared to slightly enhance virus production in vivo. The reason for this is not clear, and, although the difference was statistically significant, it is unlikely that this relative increase in titer by only 1 log over 76(WTR) in the context of a 4.5-log titer is of biological significance, particularly given that the rate of clearance of 76(RΔM2) from the lungs was similar to that of 76(WTR). Recent experiments by Jacoby et al. (16) also showed the lack of a contribution of M2 to virus replication. Thus, M2 does not negatively influence virus replication upon primary infection.
Splenic lymphocytosis is not affected by mutation of M2.
The total number of splenic lymphocytes was determined in mice at various time points after infection with MHV-76, 76(WTR), and 76(RΔM2). As illustrated in Fig. 2D, 76(WTR) induced a characteristic transient increase in splenic lymphocyte number that peaked at day 14 p.i. MHV-76 infection also caused an increase in cellularity, but cell numbers were significantly lower than those detected after 76(WTR) infection (day 14, P < 0.001; day 21, P < 0.01). In contrast, 76(RΔM2) induced a transient increase in spleen cell number of similar intensity and duration as that induced by 76(WTR). Thus, the mutation in M2, unlike the deletion present in MHV-76, did not affect the ability of MHV-68 to cause splenic lymphocytosis.
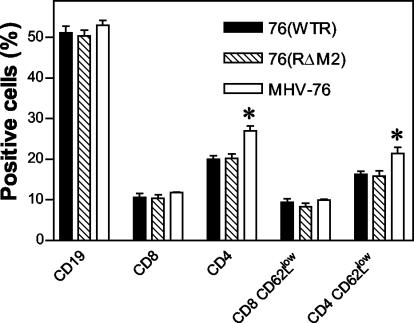
It was possible, however, that even though the mutation in M2 did not influence the overall number of cells in the spleen, specific cell subpopulations could have been affected. Therefore, we determined the relative numbers of CD19+ B cells and CD8+ and CD4+ T cells within spleens at day 14 p.i. by fluorescence-activated cell sorting (FACS) analysis. The results (Fig. 3) indicated that similar relative levels of CD19+ and CD8+ cell populations were observed in mice infected with 76(WTR), 76(RΔM2), and MHV-76. However, while there was a similar level of CD4+ cells in mice infected with 76(WTR) and 76(RΔM2), the number of this subset was significantly elevated by comparison in MHV-76-infected animals (P = 0.007 by t test). In a similar fashion, the activation status of CD8+ and CD4+ T cells was determined by coanalysis of the levels of CD62 ligand (CD62L) expression, as cells expressing low levels of CD62L (CD62Llow) are more activated. As illustrated in Fig. 3, there was no difference in the numbers of activated CD8+ T cells in mice infected with any of the three viruses. However, there was a significantly increased number of activated CD4+ cells in MHV-76-infected mice compared to those infected with 76(WTR) and 76(RΔM2) (P = 0.007 by t test). The relative increase in CD4+ CD62Llow cells in MHV-76-infected mice was similar to the relative increase in CD4+ cells, indicating that there was no difference in the proportion of activated CD4+ cells in these mice. Thus, mutation in M2 did not affect the relative proportions or activation of the major spleen cell subsets that contribute to MHV-68-induced lymphocytosis.
FIG. 3.
FACS analysis of spleen cell subsets. Spleen cells from infected mice (as indicated) were stained with monoclonal antibodies to the indicated cell surface markers and analyzed by FACS. In each case the percentage of positive cells was calculated, and the means with standard errors of four individual animals are shown. Asterisks represent a statistically significant difference from the wild-type value.
Disruption of M2 affects the normal pattern of latency.
Latent virus in the spleen was first measured at various times p.i. by an infective center (IC) assay (Fig. 2E). In mice infected with 76(WTR), measurable latent (i.e., reactivated) virus could be detected at day 7 p.i. and peaked at day 14 p.i.; by day 31 p.i. a level of approximately 400 ICs per spleen had been established. By contrast, in mice infected with MHV-76, the day 14 spike in the number of latently infected cells characteristic of wild-type MHV-68/76(WTR) was not observed, and overall the amount of latently infected cells detected was significantly less (day 14, P < 0.001). MHV-76 ICs were still detectable at day 31 p.i., at levels of approximately 50 per spleen, i.e., one-eighth of that obtained with wild-type MHV-68 infection. ICs in 76(RΔM2)-infected mice were detectable at day 7 p.i., and by day 14 latency had been established at a relatively constant level of approximately 5,000 ICs per spleen through day 31 p.i. At day 14 p.i. this was a significantly lower level than that observed in 76(WTR)-infected mice (P < 0.001) and significantly higher than that in MHV-76 infection (P < 0.001). The number of latently infected cells did not decrease from day 14 through 21 p.i. and was 1 log higher than that in 76(WTR) infection at day 31 p.i. (P < 0.01). These results indicated that mutation of M2 severely affected the characteristic acute rise in ICs (latently infected cells) within the spleen during primary infection and ultimately resulted in an increased level of latently infected cells. It should be noted that we did not detect preformed infectious virus in the spleen at any of the time points analyzed, which would have indicated a significant presence of productively infected cells. Thus, we conclude that the results obtained with the IC assay represented reactivation from latent infection.
Because the mutation in M2 could be negatively affecting the ability of the virus to reactivate from latency, also resulting in lower estimates of latently infected cells, we determined the viral genome copy number in the spleens of infected mice by quantitative fluorescent PCR (QF-PCR). We have previously shown that genome copy number correlates directly with the number of latently infected cells in MHV-68-infected mice as determined by a limiting dilution reactivation assay (33). DNA was therefore isolated from the spleens of mice at days 14 (peak of lymphocytosis) and 46 p.i. and subjected to QF-PCR. The results (Fig. 2F) indicated that, at day 14 p.i. with all three viruses, viral DNA copy number was in good agreement with the IC assay, i.e., DNA copy number and ICs obtained from mice infected with MHV-76 and 76(RΔM2) were approximately 10-fold lower than in 76(WTR)-infected mice (compare Fig. 2E and F). A positive correlation between copy number and ICs was also observed at day 46 p.i. However, the number of viral genomes was 50-fold higher in 76(RΔM2)-infected mice than in 76(WTR)-infected mice, a difference that was statistically significant (P < 0.0001) as determined by the Student t test. By contrast, in our IC assay, the number of latently infected cells in the spleens of 76(RΔM2)-infected mice was only 10-fold higher than that in the 76(WTR)-infected mice. This suggests that the actual level of latency established by the M2 mutant virus is higher than reactivable latency, indicating a possible defect in the ability of the mutant virus to reactivate from latency.
To confirm that the effects on latency were due to the lesion in M2, we generated a second recombinant virus with a 256-bp deletion within the ORF (coordinates 4239 to 4494). This virus had a similar phenotype in vivo as 76(RΔM2) (data not shown). Thus, the effects seen were due to the mutation of M2 and not other secondary mutations in the viral genome. Finally, to confirm that the observed properties of the 76(RΔM2) virus were attributable to the frameshift mutation within the M2 gene, we determined the sequence of the M2 ORF amplified from DNA extracted from three separate viral plaques resulting from reactivation from three different mice at day 14 p.i. with 76(RΔM2). The results indicated that, in all cases, the only mutation in the M2 ORF was the 4-bp deletion that we had originally introduced (data not shown). Thus, we found no evidence for the selection of M2 revertant or pseudorevertant viruses in vivo and concluded that the striking reduction in the number of latently infected cells observed at day 14 p.i. in mice infected with 76(RΔM2) was indeed a consequence of the mutation within M2.
A decrease in the peak of cells latently infected with M2 mutant viruses was also observed by Jacoby et al. (16). This fits well with previous observations that M2 is expressed during latency (15, 36) and that the peak of M2 expression in the spleen appears to occur around day 14 p.i (30). There was, however, a significantly greater number of latently infected cells present in the spleens of mice infected with the M2 mutant virus than in spleens of mice infected with MHV-76 at day 14 p.i. Because MHV-76 lacks M1 to M4 and the vtRNAs, one or more of these genes in addition to M2 likely contributes to the transient spike or rise in latently infected cells characteristically observed at this point following primary infection. At least part of this is due to the M3 protein, a soluble chemokine binding protein (22, 34), since viruses with M3 deleted have a defect in their ability to cause this transient rise in latently infected splenic lymphocytes (4).
A transient rise in latently infected cells shortly after infection is seen with other gammaherpesviruses such as EBV. The biological function of this is not known, but it is generally believed to promote viral persistence by ensuring that a critical pool of latently infected cells is established prior to maturation of the host antiviral immunosurveillance. Our results suggest that this may not be the case, at least for MHV-68, since both 76(RΔM2) and MHV-76 are capable of establishing long-term latency. However, it is possible that the transient rise in latently infected cells may play a more critical role, such as that proposed in EBV infection, in the context of a naturally acquired MHV-68 infection as opposed to one that is laboratory acquired.
Surprisingly, despite the dramatic reduction in the number of latently infected spleen cells as a consequence of M2 mutation, the splenic lymphocytosis that is concomitant with the peak splenomegaly at day 14 p.i. was not significantly affected. Thus, for the first time we have been able to separate the spike in latently infected cells from the concurrent lymphocytosis, in which all spleen lymphocyte subsets are expanded (29). By contrast, MHV-76 was severely disabled in its ability to induce lymphocytosis, strongly suggesting that one or more of the genes M1, M3, and M4 or the vtRNAs are primarily responsible for this phenomenon. The precise causes of the transient lymphocytosis and rise in latently infected cells are unclear, but it has been assumed that they are interdependent. Specifically, in addition to their temporal overlap during primary infection, both are dependent upon latently infected B cells (32) and CD4+ T cells (29). This led to two alternative hypotheses (24). The first is that the virus drives the expansion of latently infected B cells and that CD4+ and CD8+ T-cell expansion is in reaction to this. The second is that virus infection causes activation and expansion of CD4+ T cells, which in turn drive the expansion of latently infected B cells and CD8+ T cells. In the light of our present results, however, it seems likely that an alternative mechanism is operating, namely, that first the transient expansion of latently infected cells and splenic lymphocytosis are not dependent upon one another; second, transient expansion of the number of latently infected cells is virus driven, with M2 playing a major role; and third, the expansion of noninfected B cells, and CD4+ and CD8+ T cells, appears to be in response to latently infected cells, but this is dependent upon neither high levels of these cells nor M2 expression. Finally, as indicated above, other viral genes not present in MHV-76 are key to the expansion of spleen lymphocyte number.
Given that M2 was required for the expansion of latently infected cells within the spleen during acute infection, it was somewhat surprising that mutation of M2 did not negatively affect the establishment of long-term latency. This is supported by similar observations obtained with other M2 mutants and with MHV-76 (16, 20). Interestingly, the level of both reactivable latency and latent genomes maintained long-term by our M2 mutant in the spleen was significantly higher than that for either wild-type virus or MHV-76 (Fig. 2E and F). An elevation of the level of genome-positive cells in the spleen after peritoneal infection was also observed by Jacoby et al. (16), using a similar mutation in M2 (introduction of a stop codon). It is possible, therefore, in a way analogous to EBV LMP-2A, that M2 functions in part by inhibiting or preventing the overexpansion of virus-infected cells within the host. Since this expansion is not observed in mice infected with MHV-76, this effect is most likely dependent upon other gene products present at the left end of the MHV-68 genome.
M2 does not affect Vβ4-positive mononucleosis.
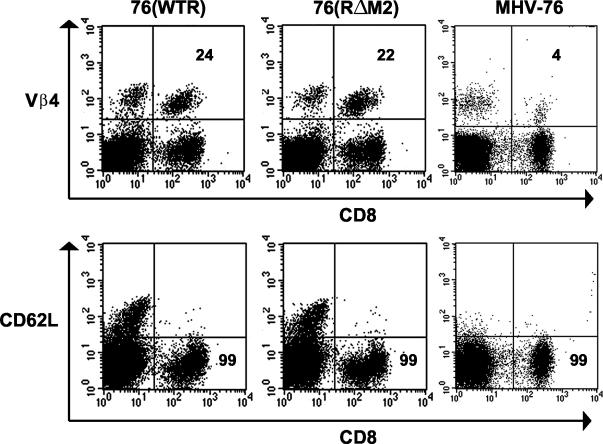
To assess whether the MHV-68-induced mononucleosis is in response to the M2 gene product, C57BL/6 mice were infected with MHV-76, 76(WTR), and 76(RΔM2), and their peripheral blood was assayed by FACS at day 35 p.i. as described previously (12). Two parameters of mononucleosis were measured: the presence of activated CD8+ lymphocytes (CD62Llow) and the levels of Vβ4+ CD8+ T cells. As shown by the representative data presented in Fig. 4, the pattern of CD62L staining was similar regardless of the strain of infecting virus. Specifically, in each case the majority (99%) of CD8+ T cells expressed low levels of CD62L. By contrast, mice infected with either 76(WTR) or 76(RΔM2) had elevated levels of Vβ4+ CD8+ cells (24 and 22%, respectively) relative to mice infected with MHV-76, which as expected had relatively normal levels (4%) of Vβ4+ CD8+ T cells. Thus, the mutation in M2 affected neither the presence of elevated levels of activated CD8+ T cells in the periphery nor the high level of Vβ4+ CD8+ cells in this population. Thus, MHV-68-induced mononucleosis is dependent on a gene other than M2 that is present within the leftmost 9.5 kbp of the unique portion of the MHV-68 genome, i.e., that deleted in MHV-76.
FIG. 4.
FACS analysis of peripheral blood lymphocytes. Peripheral blood cells of mice infected for 35 days with the indicated viruses were stained with labeled with monoclonal antibodies either to CD8 and Vβ4 or to CD8 and CD62L and subjected to FACS. Data are representative of at least three separate experiments.
Like lymphocytosis in the spleen, the viral factors responsible for the generation of either the peripheral CD8+ T-cell mononucleosis or the abnormal expansion of Vβ4 TCR+ cells within this population have not been identified. However, like the transient expansion of spleen cells, Vβ4 TCR+ CD8+ expansion is dependent upon the presence of MHV-68-infected B cells and CD40 ligand-dependent CD4+ T-cell help (5, 12). The mutation in M2 affected neither mononucleosis nor expansion of Vβ4+ T cells (Fig. 4). However, while MHV-76 induced peripheral mononucleosis, it did not promote expansion of Vβ4+ T cells. This indicates that specific viral products are required for Vβ4+ T-cell expansion and that they reside within the deletion present in MHV-76, excluding M2.
M2 expression occurs within B cells in the spleen.
Our previous analyses have shown that M2 mRNA is actively expressed at day 14 p.i. in the spleens of mice infected with MHV-68 and is detectable sporadically in splenic B cells at later time points p.i. (11, 15, 30). In addition to B cells, MHV-68 also infects splenic macrophages and dendritic cells (10, 39). To ascertain in which spleen cell type(s) M2 is expressed, C57BL/6 mice were infected intranasally with MHV-68. B cells (total, activated, and resting), macrophages, and dendritic cells were then isolated by FACS from the spleens at day 14 p.i., and total RNA extracted from these cells was subjected to nested reverse transcription-PCR (RT-PCR) as described previously (11). In addition to M2, the expression of the MHV-68 M3 and cellular β-actin mRNAs were analyzed using primers shown in Table 1. M3 was chosen because it is also characteristically expressed in the spleen at this time point (30, 36); detection of the β-actin mRNA served as an indicator of mRNA integrity and a positive control for amplification. In serial dilution assays, our RT-PCR assay was able to detect M2 mRNA in as few as one S11 cell in 105 MHV-68-negative cells (BJAB B-lymphoma cells; data not shown). S11 is a latently infected murine B-cell tumor line (31) that expresses M2 mRNA in virtually all cells (15), and thus the level of M2 expression in these cells is likely to be an accurate representation of its expression in latently infected cells in vivo. Additionally, upon spiking cDNA synthesized from the total RNA of MHV-68-negative cells with a full-length M2 cDNA, we determined that the sensitivity of our PCR assay was sufficient to detect between one and five copies of the M2 target (data not shown).
TABLE 1.
RT-PCR primers used in this study
| Gene | Round | Sequence | MHV-68 genomic coordinatesa | Product size (bp) |
|---|---|---|---|---|
| M2b | 1st | CTTCCTTAGCCAGTCTCTTC | 5872-5853 | 650 |
| ACCTTCACTGTTACTCCTCG | 4018-4037 | |||
| 2nd | TGAAGCGGTGTTACAGACTC | 5848-5829 | 573 | |
| GGTGGAATAATAGGAAGACG | 4071-4090 | |||
| M3 | 1st | GTGGCACACCAGTGAATGAC | 6916-6897 | 589 |
| GGTGAAGTGAGAGAATCCAG | 6328-6347 | |||
| 2nd | CAGAAGAGACTCCAGAGGAG | 6859-6840 | 505 | |
| AGGTCAGCTAGGATCAAGTG | 6355-6374 | |||
| β-Actin | TGTGATGGTGGGAATGGGTCA | NAc | 514 | |
| TTTGATGTCACGCACGATTTC |
Genomic coordinates are according to the work of Virgin et al. (35).
M2 primer sequences are derived from different exons, and therefore, the product size is shorter than the genomic distance between 5′ and 3′ primers.
NA, not applicable.
To detect gene expression in spleen cell subsets, RNA from a total of at least three independent sorts for each cell type was analyzed. As shown in Table 2, whereas M3 transcripts were detected in all cell subsets analyzed, M2 mRNA was detected only in B cells, both activated and resting. β-Actin mRNA was equivalently amplified in all RNA samples analyzed (data not shown). Our other studies (11) have shown that the frequency of infection within each cell subset (determined by limiting dilution analysis-PCR), though highest in activated B cells, is well within the level of detection of this assay (10−5), as described above. Thus, these data strongly suggest that M2 expression in the spleen is B cell specific.
TABLE 2.
MHV-68 M2 and M3 expression in spleen at day 14 p.i.a
| Cellsb | M2 | M3 |
|---|---|---|
| B cells (total) | 3/3 | 3/3 |
| Resting B cells | 3/5 | 4/5 |
| Activated B cells | 4/5 | 5/5 |
| Mφs | 0/3 | 3/3 |
| DCs | 0/4 | 4/4 |
Data represent positive RT-PCR results per number of individual sorts analyzed for each MHV-68 ORF; β-actin was equivalently detected in all samples. Mφ, macrophage; DC, dendritic cell.
Cells sorted as B220+ CD11b− CD11c− (total B cells; mean purity, 99.64%), B220+ peanut agglutinin (PNA)low (resting B cells; mean purity, 98.15%), B220+ PNAhigh (activated B cells; mean purity, 98.64%), B220− CD11b+ CD11c− (Macrophages; mean purity, 96.34%), and B220− CD11b+/− CD11c+ (dendritic cells; mean purity, 92.86%).
Previous analyses of M2 have shown that it is expressed within the latently infected S11 B-cell line in vitro (15) and during latency in vivo, most abundantly (as determined by RT-PCR) during the peak of lymphocytosis in the spleen at day 14 p.i (30, 36). Here, we extend these analyses by showing that M2 is expressed predominantly, if not solely, within B cells and not in splenic macrophages and dendritic cells that also harbor latent virus. A previous report of M2 expression in spleen and peritoneal exudate cells following intraperitoneal inoculation of μMT knockout mice (36), which fail to develop mature B cells due to an inability to express the immunoglobulin M heavy chain (μm), would seem to be inconsistent with our finding here of B-cell-restricted expression of M2. However, recent findings indicate that μMT mice do indeed generate B cells, through an μm-independent pathway, that specifically produce immunoglobulin A (19). It is possible, therefore, that such B cells were the source of M2 expression in MHV-68-infected μMT mice.
The M2 protein is cytoplasmic and plasma membrane associated in B cells.
The predicted amino acid sequence of the M2 protein lacks sufficient homology to any known protein or functional motif that would suggest its role during MHV-68 infection. Therefore, as a first step in defining M2 function, we transiently expressed it within mouse A20 B cells (18). Because of the lack of an adequate M2-specific antibody, M2 was expressed as either an N- or C-terminal fusion with enhanced green fluorescent protein (EGFP) by using the vectors pEGFP-C1 and pEGFP-N1 (Clontech) to aid in its detection.
Immunoblot analysis of transfected A20 cells revealed the presence of a novel protein of ∼60 kDa in the lysates of those cells transfected with an expression vector encoding either the N- or C-terminal EGFP-M2 fusion protein (data not shown). Accounting for the mass of the EGFPs, the molecular mass of M2 is ∼30 kDa. This was in good agreement with the size of a hemagglutinin-tagged version of M2 stably expressed in A20 cells (data not shown) and previously reported unpublished observations by others (16). Of particular note was the observation that the molecular mass of M2 in the fusion was considerably greater than its predicted molecular mass of 22 kDa. The molecular mass of the protein by in vitro transcription and translation analysis was 26 kDa (data not shown). Thus, the molecular mass of 30 kDa for native M2 is most likely due to a combination of an aberrant migration of the unmodified polypeptide backbone in sodium dodecyl sulfate-polyacrylamide gel electrophoresis and a contribution of approximately 4 kDa by posttranslational modification(s).
To determine the subcellular localization of the M2 protein, transiently expressed EGFP-M2 was examined in A20 cells at 24 h posttransfection by confocal microscopy with a Leica TCSNT confocal system. In some experiments, cells were fixed in 4% (wt/vol) paraformaldehyde and stained with either propidium iodide (PI; in phosphate-buffered saline containing 10 μg of RNase A [Qiagen] and 1 μg of PI [Molecular Probes] per ml for 30 min at 37°C in the dark) to label DNA or tetramethyl rhodamine isocyanate (TRITC)-labeled reagents to detect specific cellular markers. Cells were observed using a combination of laser light wavelength and band filter specific for EGFP (488 and 525/50 nm) or PI-TRITC (568 and 600/30 nm). Image analysis was then used to form a picture with the resulting two images overlaid. Representative results are shown in Fig. 5. Cells transfected with empty vector (pEGFP-C1) displayed a diffuse pattern of EGFP-specific fluorescence that was present throughout the cytoplasm and nucleus as evidenced by the covisualization of EGFP and PI (Fig. 5A to C). In cells transfected with pEGFP-C1/M2 (Fig. 5D to F) the EGFP-M2-specific fluorescence was primarily localized in patches near the cell margins and in the nucleus, as well as diffusely at a lower intensity throughout the nucleus and cytoplasm. However, nuclear staining was not universal and there appeared to be little crossover between the EGFP-M2 and PI fluorescence. A similar pattern of staining was seen with the C-terminal fusion protein encoded by pEGFP-N1/M2 and when a hemagglutinin epitope-tagged molecule was expressed in the same cells (data not shown). These results were consistent with M2 being principally localized in patches at or near the plasma membrane and within areas in the nucleus that did not contain chromatin. Because the cytoplasm in B cells is small and it is thus often hard to distinguish membrane and cytoplasmic fluorescence, we immunostained pEGFP-C1/M2-transfected cells with TRITC-labeled anti-MHC class II (monoclonal antibody SW 73.2 [14]) (Fig. 5G to I). MHC class II glycoprotein molecules are principally localized to the plasma membrane of B cells. The EGFP-M2-specific fluorescence was as observed previously, and a proportion of the patches of EGFP-M2 clearly colocalized with MHC class II, as evidenced by yellow staining when the two images were overlaid. Similar results were obtained when anti-CD19 (another B-cell surface marker) was used instead of anti-MHC class II (data not shown). Additionally, no colocalization was observed between EGFP-M2 and BODIPY ceramide (a marker for the Golgi apparatus) or LysoTracker Red DND-99 (a marker for lysosomes) (data not shown). Thus, the above results are consistent with a proportion of the M2 protein being cytoplasmic and a proportion being localized in patches either at or extremely close to the plasma membrane.
FIG. 5.
Confocal microscopic analysis of M2 expression. A20 cells were transfected with either pEGFP-C1 (A to C) or pEGFP-C1/M2 (D to I) and after 48 h were either fixed, permeabilized, and then counterstained with PI or stained with TRITC-conjugated antibody to MHC class II antigens and then fixed. Cells were then visualized using a Leica TCNTS confocal microscope with combinations of laser light wavelength and band filter specific for EGFP (488 and 525/50 nm [A, D, and G]) or for PI and TRITC (568 and 600/30 nm [B, E, and H]). Image analysis with Leica software enabled the overlaying of these two images (C, F, and I). All images are shown at the same magnification.
Putative function of M2.
Our M2 expression data suggest that the effects of mutation of M2 as discussed above are B cell specific. Thus, in some respects, M2 function may be analogous to the perceived collective function of the EBV growth program of latency gene expression (which includes several B-cell-restricted genes): principally, to drive the rapid expansion of infected cells to establish a critical pool of latently infected cells. The function of M2, however, remains enigmatic. Preliminary biochemical analyses failed to demonstrate any posttranslational modification of the protein that might have shed light on this problem. While the apparent effects of M2 expression in the spleen and its localization to the cytoplasm and plasma membrane may argue for a role in cell signaling, the precise function of M2 and its specific role in MHV-68 latency remain to be determined.
Acknowledgments
This work was funded in part by Wellcome Trust Biomedical Research Collaboration Grant 054503 (to J.P.S), a Royal Society (London) University Research Fellowship (to J.P.S), United States Public Health Service grants CA90208 (to J.T.S. and J.P.S) and AI42927 (to M.A.B.), the Trudeau Institute, and the American Lebanese Syrian Associated Charities (ALSAC). A.I.M. was supported by a Wellcome Trust Veterinary Clinical Scholarship.
REFERENCES
- 1.Blackman, M. A., E. Flano, E. Usherwood, and D. L. Woodland. 2000. Murine gamma-herpesvirus-68: a mouse model for infectious mononucleosis? Mol. Med. Today 6:488-490. [DOI] [PubMed] [Google Scholar]
- 2.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J. Gen. Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 3.Bowden, R. J., J. P. Simas, A. J. Davis, and S. Efstathiou. 1997. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J. Gen. Virol. 78:1675-1687. [DOI] [PubMed] [Google Scholar]
- 4.Bridgeman, A., P. G. Stevenson, J. P. Simas, and S. Efstathiou. 2001. A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J. Exp. Med. 194:301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, J. W., A. M. Hamilton-Easton, J. P. Christensen, R. D. Cardin, C. L. Hardy, and P. C. Doherty. 1999. Requirement for CD40 ligand, CD4+ T cells, and B cells in an infectious mononucleosis-like syndrome. J. Virol. 73:9650-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardin, R. D., J. W. Brooks, S. R. Sarawar, and P. C. Doherty. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J. Exp. Med. 184:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppola, M. A., E. Flaño, P. Nguyen, C. L. Hardy, R. D. Cardin, N. Shastri, D. L. Woodland, and M. A. Blackman. 1999. Apparent MHC-independent stimulation of CD8+ T cells in vivo during latent murine gammaherpesvirus infection. J. Immunol. 163:1481-1489. [PubMed] [Google Scholar]
- 8.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehtisham, S., N. P. Sunil-Chandra, and A. A. Nash. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 67:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 11.Flano, E., I. J. Kim, D. L. Woodland, and M. A. Blackman. 2002. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J. Exp. Med. 196:1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flano, E., D. L. Woodland, and M. A. Blackman. 1999. Requirement for CD4+ T cells in Vβ4+CD8+ T cell activation associated with latent murine gammaherpesvirus infection. J. Immunol. 163:3403-3408. [PubMed] [Google Scholar]
- 13.Hardy, C. L., S. L. Silins, D. L. Woodland, and M. A. Blackman. 2000. Murine gamma-herpesvirus infection causes Vβ4-specific CDR3-restricted clonal expansions within CD8+ peripheral blood T lymphocytes. Int. Immunol. 12:1193-1204. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins, J., B. M. Dutia, and I. McConnell. 1986. Monoclonal antibodies to sheep lymphocytes. I. Identification of MHC class II molecules on lymphoid tissue and changes in the level of class II expression on lymph-borne cells following antigen stimulation in vivo. Immunology 59:433-438. [PMC free article] [PubMed] [Google Scholar]
- 15.Husain, S. M., E. J. Usherwood, H. Dyson, C. Coleclough, M. A. Coppola, D. L. Woodland, M. A. Blackman, J. P. Stewart, and J. T. Sample. 1999. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 96:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby, M. A., H. W. Virgin, and S. H. Speck. 2002. Disruption of the M2 gene of murine gammaherpesvirus 68 alters splenic latency following intranasal, but not intraperitoneal, inoculation. J. Virol. 76:1790-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, I. J., E. Flano, D. L. Woodland, and M. A. Blackman. 2002. Antibody-mediated control of persistent gamma-herpesvirus infection. J. Immunol. 168:3958-3964. [DOI] [PubMed] [Google Scholar]
- 18.Kim, K. J., C. Kanellopoulos-Langevin, R. M. Merwin, D. H. Sachs, and R. Asofsky. 1979. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J. Immunol. 122:549-554. [PubMed] [Google Scholar]
- 19.Macpherson, A. J., A. Lamarre, K. McCoy, G. R. Harriman, B. Odermatt, G. Dougan, H. Hengartner, and R. M. Zinkernagel. 2001. IgA production without mu or delta chain expression in developing B cells. Nat. Immunol. 2:625-631. [DOI] [PubMed] [Google Scholar]
- 20.Macrae, A. I., B. M. Dutia, S. Milligan, D. G. Brownstein, D. J. Allen, J. Mistrikova, A. J. Davison, A. A. Nash, and J. P. Stewart. 2001. Analysis of a novel strain of murine gammaherpesvirus reveals a genomic locus important for acute pathogenesis. J. Virol. 75:5315-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry, C. M., J. P. Simas, V. P. Smith, C. A. Stewart, A. C. Minson, S. Efstathiou, and A. Alcami. 2000. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart, J. P. 1999. Of mice and men: murine gammaherpesvirus 68 as a model. EBV Rep. 6:31-35. [Google Scholar]
- 24.Stewart, J. P., E. J. Usherwood, B. Dutia, and A. A. Nash. 1998. Immunobiology of murine gamma herpesvirus-68, p. 149-163. In P. G. Medveczky, H. Friedman, and M. Bendinelli (ed.), Herpesviruses and immunity. Plenum Press, New York, N.Y.
- 25.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 27.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 28.Tripp, R. A., A. M. Hamilton-Easton, R. D. Cardin, P. Nguyen, F. G. Behm, D. L. Woodland, P. C. Doherty, and M. A. Blackman. 1997. Pathogenesis of an infectious mononucleosis-like disease induced by a murine gamma-herpesvirus: role for a viral superantigen? J. Exp. Med. 185:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Usherwood, E. J., A. J. Ross, D. J. Allen, and A. A. Nash. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J. Gen. Virol. 77:627-630. [DOI] [PubMed] [Google Scholar]
- 30.Usherwood, E. J., D. J. Roy, K. Ward, S. L. Surman, B. M. Dutia, M. A. Blackman, J. P. Stewart, and D. L. Woodland. 2000. Control of gammaherpesvirus latency by latent antigen-specific CD8+ T cells. J. Exp. Med. 192:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Usherwood, E. J., J. P. Stewart, and A. A. Nash. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J. Virol. 70:6516-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Usherwood, E. J., J. P. Stewart, K. Robertson, D. J. Allen, and A. A. Nash. 1996. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J. Gen. Virol. 77:2819-2825. [DOI] [PubMed] [Google Scholar]
- 33.Usherwood, E. J., K. A. Ward, M. A. Blackman, J. P. Stewart, and D. L. Woodland. 2001. Latent antigen vaccination in a model gammaherpesvirus infection. J. Virol. 75:8283-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Berkel, V., J. Barrett, H. L. Tiffany, D. H. Fremont, P. M. Murphy, G. McFadden, S. H. Speck, and H. W. Virgin. 2000. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol. 74:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virgin, H. W., R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 38.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weck, K. E., S. S. Kim, H. W. Virgin, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]