Abstract
The subcellular location at which genomic RNA is packaged by Gag proteins during retrovirus assembly remains unknown. Since the membrane-binding (M) domain is most critical for targeting Gag to the plasma membrane, changes to this determinant might alter the path taken through the cell and reduce the efficiency of genome packaging. In this report, a Rous sarcoma virus (RSV) mutant having two acidic-to-basic substitutions in the M domain is described. This mutant, designated Super M, produced particles much faster than the wild type, but the mutant virions were noninfectious and contained only 1/10 the amount of genomic RNA found in wild-type particles. To identify the cause(s) of these defects, we considered data that suggest that RSV Gag traffics through the nucleus to package the viral genome. Although inhibition of the CRM-1 pathway of nuclear export caused the accumulation of wild-type Gag in the nucleus, nuclear accumulation did not occur with Super M. The importance of the nucleocapsid (NC) domain in membrane targeting was also determined, and, importantly, deletion of the NC sequence prevented plasma membrane localization by wild-type Gag but not by Super M Gag. Based on these results, we reasoned that the enhanced membrane-targeting properties of Super M inhibit genome packaging. Consistent with this interpretation, substitutions that reestablished the wild-type number of basic and acidic residues in the Super M Gag M domain reduced the budding efficiency and restored genome packaging and infectivity. Therefore, these data suggest that Gag targeting and genome packaging are normally linked to ensure that RSV particles contain viral RNA.
Gag polyproteins form retrovirus-like particles that bud from the plasma membrane. Gag is synthesized by soluble ribosomes and is targeted to the site of budding by the membrane-binding (M) domain located in the N-terminal region (45). In most retroviruses (e.g., human immunodeficiency virus type 1 [HIV-1]), the M domain contains both myristate and a cluster of basic residues, which form hydrophobic and electrostatic interactions with membrane phospholipids, respectively (2, 10, 12, 48, 49). In Rous sarcoma virus (RSV), an avian retrovirus, the M domain maps to the first 86 residues in Gag (Fig. 1A) (43). Although it is not myristylated, the RSV M domain contains 11 basic residues, and according to the nuclear magnetic resonance structure of the M domain, these basic residues are present on the surface of the molecule (24). Because substitutions of two or more basic residues in the M domain prevent targeting and budding, RSV Gag may bind to the plasma membrane by forming electrostatic interactions with acidic phospholipids (4).
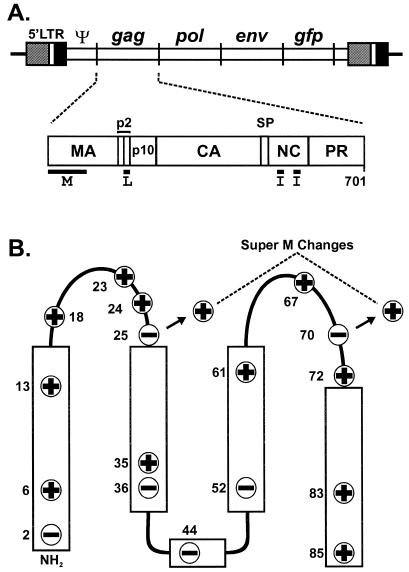
FIG. 1.
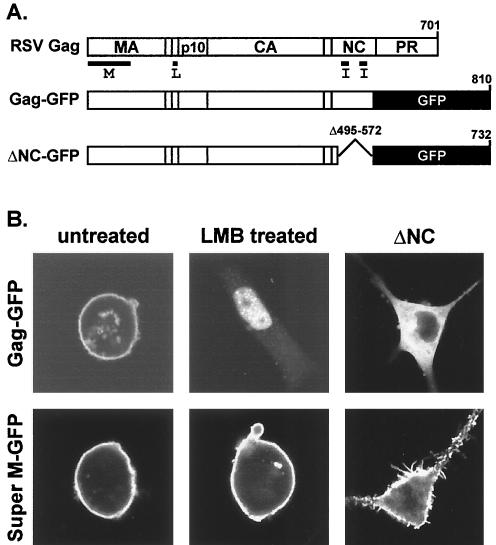
Location of the Super M substitutions in RSV Gag. (A) Diagram of the RSV proviral DNA used in these experiments (top) and the RSV Gag polyprotein (bottom). The 5′ long terminal repeat (LTR) promotes transcription of genome-length mRNA, which can be encapsidated or used for the synthesis of Gag and Gag-Pol proteins. In addition, full-length vRNA can be spliced for the synthesis of Env glycoproteins and, in the recombinant virus used here, for the synthesis of GFP. Intact Gag polyproteins drive budding and are subsequently cleaved by the viral PR into the mature products: MA, CA, NC, etc. The locations of the domains required for budding are indicated below Gag. The M domain is essential for plasma membrane targeting. The I domains promote Gag-Gag interactions and the assembly of dense particles. The L domain is required for a late step in budding. (B) The secondary structure of the RSV M domain consists of five helices (rectangles) connected by flexible loops. The locations of the 11 basic and 6 acidic residues in the M domain are depicted according to their charge. The two acidic-to-basic substitutions in Super M Gag, E25K and E70K, are indicated.
In addition to budding, Gag is responsible for packaging two copies of the retroviral genome. Efficient packaging requires the presence of basic residues and one or two copies of a zinc finger motif (Cys-X2-Cys-X2-His-X4-Cys) in the nucleocapsid (NC) domain of Gag (42). These elements enable specific interactions between Gag and the packaging signal (Ψ), which is located in the 5′ end of the viral RNA (vRNA) (23).
Aside from the interaction between NC and Ψ, many aspects of genome packaging remain unknown. Of particular interest is the subcellular location(s) at which Gag first binds vRNA. Recently, RNA targeting motifs known as A2 response elements have been identified in the genomes of several retroviruses (26). In addition, the RNA-targeting protein Staufen has been implicated in genome packaging by HIV-1 Gag (25). Although the functional significance of these findings is unclear, these data suggest that RNA-targeting proteins might enhance the efficiency of genome packaging by transporting vRNA to the sites of Gag assembly. Given the importance of the M domain in Gag targeting, it is possible that changes in this domain would alter the pathway by which Gag trafficks through the cell, and such changes might also affect the efficiency of genome packaging. There is evidence for this possibility from RSV, where a twofold reduction in the efficiency of genome packaging is caused by adding the myristylated, nine-amino-acid Src M domain to the N terminus of Gag (29). The mutant, known as Myr1E, is also noninfectious, and the packaged vRNA is monomeric.
Stimulated by the potential role of the M domain in genome packaging, we characterized an RSV mutant with two substitutions, E25K and E70K, in the M domain. Although the acidic-to-basic substitutions enhanced budding by this mutant, designated Super M, the extracellular particles were noninfectious and contained only 1/10 the amount of vRNA found in wild-type (WT) particles. To identify the cause(s) of these defects, we considered recent data that suggest that RSV Gag must transit the nucleus to package vRNA (38). In contrast to WT, Super M Gag did not accumulate in the nucleus when cells were treated with leptomycin B (LMB), an inhibitor of nuclear export. In addition, the importance of Gag-Gag interactions in RSV targeting was tested by deleting the NC domain and examining the subcellular distribution of mutant and WT proteins. Interestingly, deletion of the NC sequences prevented WT Gag from accumulating on the plasma membrane, but Super M Gag targeting was unaffected. Together, these results suggest that the Super M substitutions alter the targeting properties of RSV Gag. Because the altered targeting phenotypes of Super M are likely caused by an enhanced affinity for the plasma membrane, it was possible that packaging and infectivity would also be disrupted. To test this possibility, additional substitutions that reestablished the WT number of basic and acidic residues were introduced to the Super M Gag domain, and these changes reduced the budding rate and restored genome packaging and infectivity. Hence, these observations suggest a relationship between genome packaging and membrane targeting by RSV Gag.
MATERIALS AND METHODS
Proviral expression vectors and mutagenesis.
The WT gag gene used in these studies was obtained from the Prague C strain of RSV (15, 39). To create pRS.V8-EGFP, gag was cloned via the unique SstI and HpaI sites into pRCAS-EGFP, a proviral plasmid that expresses green fluorescent protein (GFP) from the nonessential v-src region of the genome (14, 37).
The substitutions present in Super M Gag and the charge-balanced mutant proviral plasmids were generated by oligonucleotide-directed mutagenesis using previously described methods (20, 46). The sequences of the mutagenic oligonucleotides and the resulting substitutions (underlined) are as follows: for Super M, the E25K oligonucleotide sequence is 5′-CTAAGAAGAAAATAGGGGCCATG and the E70K oligonucleotide sequence is 5′-GAAATCGGGAAAGTTAAAAACC; for K6,18E-Super M, the K6E oligonucleotide sequence is 5′-GCCGTCATAGAGGTGATTTCGTCC and the K18E oligonucleotide sequence is 5′-CTATTGCGGGGAAACTAGTCCTTC; for K13,18E-Super M, the K13E oligonucleotide sequence is 5′-GTCCGCGTGTGAAACCTATTG and the K18E oligonucleotide has the sequence shown above.
Analysis of particle release.
Duplicate cultures of QT6 (quail) cells were transfected with mutant or WT proviral plasmids by the calcium phosphate method. Both cultures were labeled with l-[35S]methionine (0.1 μCi/μl, >1,000 Ci/mmol) in methionine-free Dulbecco's modified Eagle medium (DMEM) at 16 h posttransfection. One set was labeled for only 5 min, while the other was labeled for 2.5 h. After the 5-min labeling, the media were discarded, and the cells were lysed in radioimmunoprecipitation assay buffer. For the 2.5-h labeling, media and cell lysate fractions were prepared. After proteins were immunoprecipitated with antiserum raised against purified RSV, the samples were denatured and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The amounts of labeled Gag in the lysates of the cells labeled for 5 min and the amounts of capsid (CA) protein released into the media after the 2.5-h labeling were quantified by phosphorimagery. The budding efficiency was calculated by dividing the amount of CA released after 2.5 h by the amount of Gag in the 5-min lysate. The data are expressed as percentages of the WT efficiency of release, which was set to 100%.
Pulse-chase analyses were performed by transfecting identical cultures of QT6 cells with WT or mutant proviral (pRS.V8-EGFP) plasmids. After 16 h, the cells were washed and labeled for 10 min with l-[35S]methionine (0.1 μCi/μl, >1,000 Ci/mmol) in methionine-free DMEM. At the end of the pulse (time zero), one culture from each set was placed on ice, and media and cell lysates were prepared with radioimmunoprecipitation assay buffer. For the remaining cultures, the labeling media were removed and serum-free DMEM containing an excess of unlabeled methionine was added. At various times, one culture from each set was placed on ice, and media and cell lysates were prepared. After full-length Gag and the products of Gag proteolysis were immunoprecipitated, the labeled proteins were detected and quantified by phosphorimagery. To determine the rate of Gag disappearance from the cell, the amount present at each time point was divided by the amount detected immediately after the pulse. To determine the rate of Gag accumulation in the medium, the amount of CA present at each time point was quantified, and this number was multiplied by the ratio of the number of methionines in full-length Gag to the number in CA (correction factor = 3). The extrapolated value was then normalized by the amount of Gag detected in the cell lysates immediately after the pulse.
Infectivity assays.
QT6 cells were transfected with mutant or WT proviral DNA. After transfection for 16 h, the cells were washed and fresh media were added. Virus-containing media were collected 48 h later, and the cellular debris was pelleted by centrifugation for 10 min at 2,000 × g. The cell-free supernatants were transferred to new tubes, and reverse transcriptase (RT) assays were performed to determine the concentration of virus. Turkey embryo fibroblasts (TEFs) were then inoculated with equal numbers of particles. After 24 h, the media were changed, and the cells were passaged 1:3 every 3 to 4 days so that infectivity could be determined at days 3, 7, and 14 postinfection. To measure infectivity, the cultures were trypsinized and stored on ice. Approximately 10,000 cells were then examined for the presence of GFP fluorescence with a FACScan (BD Biosciences). As mentioned above, infected cells expressed GFP from the nonessential v-src region of the viral genome. Hence, the percentage of infected cells was determined by dividing the number of green fluorescent cells by the total number examined.
To determine whether infectivity by K6,18E-Super M or K13,18E-Super M resulted from unintended suppressor mutations, virus-containing media were collected from the TEFs at 2 weeks postinfection, the cellular debris was removed by centrifugation (10 min at 2,000 × g), and aliquots of the media were used to determine the virus concentrations by RT assay. Equal numbers of WT and mutant virus were added to fresh, uninfected TEFs, and the percentage of infected (GFP-fluorescent) cells was determined at 1 week postinfection. At this time point, the number of cells infected with the charge-balanced viruses would be similar to the number infected by the WT if the mutant viruses had made additional, compensatory changes (e.g., reversion of the Super M substitutions) during the primary infection. However, cultures inoculated with the charge-balanced mutants contained one-fourth to one-third the number of fluorescent cells found in WT-infected cultures (data not shown).
Analysis of physical properties and protein composition of extracellular virus.
For electron microscopy, QT6 cells were seeded on 60-mm-diameter Permanox dishes and transfected with WT or Super M proviral DNA. After 16 h, the cultures were washed with 0.1 M sodium cacodylate (pH 7.4) and fixed in 4% paraformaldehyde-0.5% glutaraldehyde for 1 h at 4°C. Fixed cells were then incubated for 30 min with 1% osmium tetroxide, dehydrated, and embedded in Epon 812. Thin sections were stained with uranyl acetate and lead citrate, and extracellular particles were detected by electron microscopy.
To determine the density of Super M virus, particle-containing media were collected after labeling transfected QT6 cells for 2.5 h with [35S]methionine. Media were then centrifuged (10 min at 2,000 × g) to remove cellular debris and layered onto a 10 to 50% gradient of sucrose suspended in phosphate-buffered saline (PBS). As an internal control, labeled particles produced by pGag-GFP were added to the same gradient. The samples were then centrifuged at 83,500 × g for 16 h at 4°C. Sixteen fractions of 0.7 ml each were collected from the bottom of the gradient, and the labeled Gag proteins were immunoprecipitated, resolved by SDS-PAGE, and quantified by phosphorimagery. The amount of WT or mutant protein in each fraction is expressed as a percentage of the total detected in the entire gradient. The density of each fraction was determined by examining a 50-μl aliquot by refractometry.
The levels of Env and Pol incorporated into Super M particles were examined by Western blotting. Particles were collected from transfected QT6 cells, and RT assays were performed using exogenous templates (poly[A]) and primers (oligo[dT]). Virus samples corresponding to equal amounts of RT activity were denatured, and the proteins were resolved by SDS-PAGE and blotted to nitrocellulose membranes. To detect the amount of CA in each sample, the membranes were incubated with rabbit polyclonal antisera specific for RSV Gag and the products of Gag proteolysis (anti-Gag). To detect the levels of Env incorporated, membranes were incubated with rabbit antisera raised against the TM subunit of RSV Env (anti-TM). Primary antibody binding was visualized by enhanced chemiluminescence after incubation of the membranes with goat anti-rabbit antibodies conjugated to horseradish peroxidase.
Analysis of genome packaging.
QT6 cells were transfected with mutant or WT proviral DNA. After 16 h, the transfection media were removed, the cells were washed, and fresh media were added. Forty-eight hours later, virus-containing media were collected and the cellular debris was removed as described above. To concentrate the extracellular virus, the cell-free media were centrifuged at 126,000 × g for 40 min at 4°C over a solution of 20% sucrose in PBS. The virus pellet was then resuspended in PBS, and a portion was used to determine the concentration of virus by RT assay. Particle-associated RNA was then prepared with the TRI reagent (Sigma) according to the supplier's protocol and linear acrylamide (Ambion; 10 μg/ml) as a coprecipitant. After resuspension in diethyl pyrocarbonate (DEPC)-treated water, RNA sample volumes corresponding to equal numbers of particles were denatured and blotted to nylon membranes with a slot blot apparatus. The amount of genomic RNA packaged by each virus was detected by hybridization with 32P-labeled antisense riboprobes specific for the gag gene, as previously described (6). The amounts of labeled RNA bound to each sample were quantified by phosphorimagery. In a similar manner, the steady-state levels of genomic RNA present in both WT and Super M-producing cells were examined. The levels of genome packaging by the WT and Super M were also examined by RNase protection assay (RPA) and Northern blotting (data not shown). For the Northern blot analyses, particle-associated RNA was purified as described above. RNA sample volumes corresponding to equal numbers of virus were denatured, resolved on 2.2 M formaldehyde-1% agarose gels in MOPS (morpholinepropanesulfonic acid) electrophoresis buffer, and blotted to nylon membranes by standard techniques. Hybridizations with a 32P-labeled gag-specific riboprobe were performed as described above. For analysis by RPA, RNA from equivalent numbers of Super M and WT particles was denatured and hybridized with a 32P-labeled antisense gag riboprobe (532 nucleotides [nt]) and the RPA III kit (Ambion). Protected fragments (502 nt) were separated by electrophoresis on 5% acrylamide-8 M urea gels and quantified by phosphorimagery.
To determine whether Super M contained vRNA dimers, QT6 cells were transfected with WT or mutant proviral DNA and the extracellular particles were collected from the media by ultracentrifugation. RNA from equivalent numbers of particles was purified and resolved on agarose gels under native conditions as described by Fu and Rein (11). After electrophoresis, the RNA was denatured by incubating the gel in 6% formaldehyde for 30 min at 65°C. After several rinses in DEPC-treated water, the gel was soaked in 20× SSC (1× SSC is 0.15 M NaCl plus 0.15 M sodium citrate), and the RNA was blotted to nylon membranes. Blotted RNA was hybridized with the 32P-labeled gag antisense riboprobe and visualized by autoradiography (data not shown).
Packaging of heterologous, Ψ-containing RNA.
The 160-nt RSV minimal packaging sequence, MΨ, was defined by Banks et al. (1). To create pCMV.MΨ.gfp, we used PCR to amplify the DNA sequence between nt 156 and 315, which encodes MΨ in the RSV Prague C proviral plasmid pATV-8. The sequences of the primers used for PCR were as follows: sense, 5′-AATAGATCTGATCCTGCCCTCATCC; antisense, 5′-TTAAGATCTGCGGCCGCCGTCTTCCAACG. These primers created BglII sites (underlined) that flank the MΨ sequence, so the PCR product was digested and inserted into the BglII site (nt 610) of pEGFP-N2 (Clontech), which expresses GFP from the cytomegalovirus immediate-early promoter. Positive clones containing a single insert in the proper orientation were identified by restriction digestion and confirmed by DNA sequencing.
To create the WT gag-only expression vector, the Prague C gag sequences from the proviral plasmid pRC.V8 were isolated by digestion with SstI and HpaI. This fragment was inserted into pEGFP-N2 after the gfp sequences were removed by digestion with SstI and NotI. In this case, the NotI end of the vector DNA was treated with the Klenow fragment of E. coli DNA polymerase I for ligation to the blunt HpaI end of the gag fragment. The resulting plasmid, designated pCMV.GagPR, was transfected into QT6 cells and found to synthesize full-length Gag proteins that were released into the medium and processed by protease (PR) (data not shown). To create pCMV.Super M.GagPR, the SstI/BspEI fragment (453 bp) encoding the Super M changes was exchanged with the corresponding WT sequences in pCMV.GagPR and the resulting clones were screened by DNA sequencing.
The abilities of WT and Super M Gag to package heterologous mRNA were determined as follows. First, both pCMV.MΨ.gfp and pEGFP-N2, designated MΨ(−).gfp, were cotransfected into QT6 cells with either the WT or Super M Gag expression vector. All plasmids were transfected at equimolar amounts, and the total amount of DNA per transfection was 15 μg/35-mm-diameter plate. After 12 h, the transfection media were removed, and the monolayers were washed. The cultures were then trace labeled with [35S]methionine (0.1 μCi/μl, >1,000 Ci/mmol) in complete DMEM containing 1% fetal bovine serum. After approximately 20 h of labeling, the media were collected, the cellular debris was removed by low-speed centrifugation, and a portion of each supernatant was used to determine the concentrations of radiolabeled particles by immunoprecipitation, SDS-PAGE, and phosphorimager analyses. Particles were then pelleted from the remaining media, and the particle-associated RNA was purified with the TRI reagent (Sigma) and linear acrylamide as a coprecipitant. The cell-associated RNA was also purified from the monolayers in a similar manner. Upon resuspension of the RNA in DEPC-treated water, equal volumes were mixed with a 32P-labeled antisense riboprobe (306 nt) specific for the 5′ ends of MΨ.gfp and MΨ(−).gfp mRNA. After hybridization for 16 h at 42°C, RNase digestions were performed with the RPA III kit (Ambion). Protected fragments were resolved on 4% acrylamide-8 M urea gels and quantified by phosphorimagery. Because the labeled riboprobe contained both MΨ and gfp sequences, the presence of the MΨ.gfp mRNA yielded a protected band of 269 nt, whereas MΨ(−).gfp mRNA yielded a protected band of 204 nt. To calculate the packaging efficiency for each RNA species, the amount detected in the medium was divided by the steady-state level detected in the cell. This value was then normalized by the number of radiolabeled particles present in the medium. In each experiment, the efficiency of MΨ.gfp packaging by WT Gag was set to 100%. To determine the abilities of WT and Super M Gag to select for the MΨ-containing mRNA in this assay, the efficiency of MΨ.gfp packaging was divided by the efficiency of MΨ(−).gfp packaging. Hence, this number represents the factor of enrichment for Ψ-containing mRNA in WT and Super M Gag particles.
To compare the abilities of wild-type and Super M Gag to package nonspecific RNA, QT6 cultures were cotransfected with pCMV.MΨ(−).gfp and either pCMV.GagPR or pCMV.Super M.GagPR vectors. After 12 h, the cells were trace labeled with [35S]methionine and particle-associated RNA was purified as described above. Nonspecific packaging of MΨ(−).gfp mRNA was quantified by RPA and normalized to the amount of radiolabeled Gag present in the medium. For comparison, the level of MΨ(−).gfp packaging by the WT was set to 100%.
Subcellular targeting properties of Super M.
Construction of the pGag-GFP expression plasmid was described previously (4). To create pΔNC-GFP, the region between nt 1044 and 1865 in gag was amplified by PCR. The primer sequences were as follows: sense, 5′-GGGCAAGGGTCAGGG; antisense, 5′-GTTTGGGGCCCCCTCTCTCTATTGAC. To prepare the PCR product for cloning, we made use of the endogenous EspI site at nt 1419 in gag and the ApaI site engineered by the antisense PCR primer (underlined). The digested fragment was then exchanged with the EspI/ApaI fragment of pGag-GFP. The resulting sequences encode a chimera in which GFP is fused to the C terminus of the seventh residue of the NC domain in Gag. Hence, the remaining C-terminal residues of Gag, including the I domains and the zinc fingers, are absent in ΔNC-GFP.
To create pSuperM-GFP and pSuperM-ΔNC-GFP, the SstI/BspEI fragment encoding the Super M Gag substitutions was exchanged with the corresponding WT sequences in pGag-GFP and pΔNC-GFP, respectively.
The subcellular distributions of the GFP-tagged chimeras in transfected QT6 cells were examined by confocal microscopy as previously described (4). To determine the effects of LMB treatment on WT and mutant Gag-GFP localization, transfected QT6 cells were treated with the drug at a final concentration of 10 ng/ml for 3 h before examination by confocal microscopy (38).
RESULTS
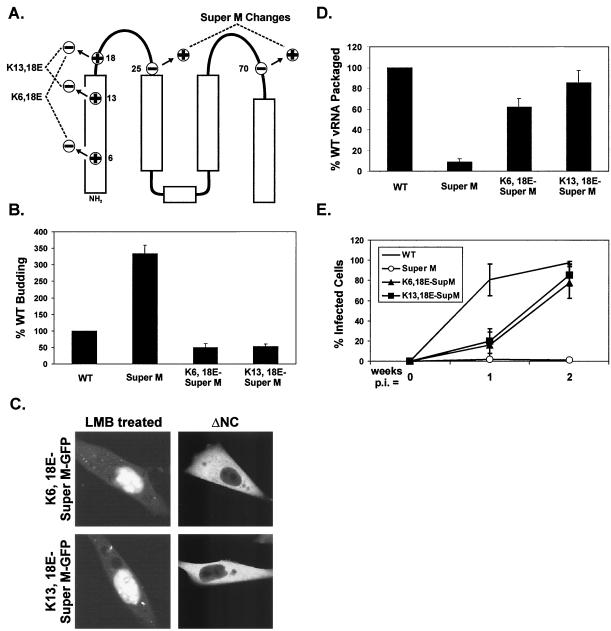
In a previous report, the 11 basic residues in the RSV Gag M domain were shown to be essential for Gag targeting to the plasma membrane (4). To further investigate the possibility that Gag binds the plasma membrane by forming electrostatic interactions with acidic phospholipids, additional basic residues were introduced into the M domain, and these changes were shown to enhance budding from mammalian (COS-1) cells. The most dramatic increase in budding was observed with a mutant, designated Super M, whose Gag contains two substitutions, E25K and E70K (Fig. 1B). These substitutions simultaneously increased the number of basic residues in the M domain from 11 to 13 and decreased the number of acidic residues from 6 to 4. Hence, the Super M substitutions cause a net shift of +4 in the M domain.
Analysis of Super M virus budding and infectivity.
To further our analysis of Super M, the mutant sequence was subcloned into pRS.V8-EGFP, a proviral plasmid derived from pRCAS-EGFP (Fig. 1A). Metabolic labeling of transfected QT6 cells subsequently showed that Super M produced three times more virus than the WT control, whereas the level of the previously described Src chimera, Myr1E, was enhanced by about twofold (Fig. 2A). The enhanced rate of release of these two mutants was confirmed by pulse-chase analyses. As shown in Fig. 2B, the pulse-labeled mutants disappeared from the cells more quickly than the WT, with the levels of Super M and Myr1E being reduced to less than one-half of WT levels after chasing for 30 min. Concurrent with their increased rate of disappearance from the cell, Super M and Myr1E proteins accumulated more rapidly in the media (Fig. 2C). Therefore, Super M and Myr1E bud much faster than WT virus.
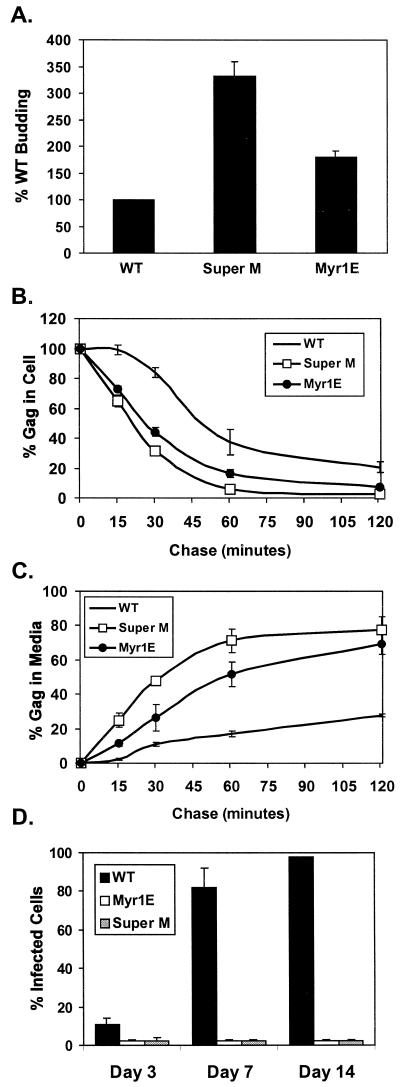
FIG. 2.
Super M budding and infectivity. (A) QT6 (quail) cells were transfected with proviral (pRCAS-derived) plasmids and labeled for 2.5 h with [35S]methionine. Viral proteins were immunoprecipitated from the cell and media fractions, separated by SDS-PAGE, and quantified by phosphorimager analysis. The amounts of viral proteins in the media were normalized to the levels of intracellular gag expression. The budding efficiency of WT virus was set to 100% for comparison to Super M and the previously described RSV mutant Myr1E, which encodes the Src membrane-binding domain as an extension from the N terminus of Gag. The data are the averages of 10 (Super M) or 4 (Myr1E) experiments, and the error bars measure 1 standard deviation (SD) from the mean. Pulse-chase analyses of Gag levels in the cell (B) and medium (C) fractions were performed with QT6 cells transfected with proviral plasmids. The percentage of Gag in the cells at each time point was determined by dividing the amount remaining by the amount detected at the beginning of the chase. Similarly, the amount of Gag present after the pulse was used to normalize the amount of viral antigen detected in the medium at each time point. The data are the averages of four experiments, and the error bars measure 1 SD from the mean. (D) WT and mutant viruses were collected from transfected QT6 cells, normalized by RT assay, and transferred to cultures of TEFs. Infected cells were detected by FACS analysis because the proviruses expressed gfp from the long terminal repeat. The numbers of infected (fluorescent) cells are expressed as percentages of the total population examined at days 3, 7, and 14 postinfection. The results show the averages of six experiments, and the error bars measure 1 SD from the mean.
Interestingly, 50 to 60% of the amount of wild-type Gag synthesized during the pulse was recovered in the cell and media fractions at the end of the chase (120 min), whereas approximately 80% of Super M Gag was accounted for. Given the correlation between increased recovery and enhanced budding, it was possible that rapid escape from the cytoplasm reduced Gag loss due to premature processing by the viral PR or degradation by cellular machinery. To determine whether PR affected recovery, PR− versions of the WT and Super M were expressed from gag-only vectors and analyzed as shown in Fig. 2B and C. In this case, approximately 90% of the pulse-labeled WT and Super M proteins were recovered at the final chase point (data not shown). Thus, the difference in recovery between PR+ versions of the WT and Super M is likely due to differences in the relative budding rates rather than a gross change in protein stability.
As mentioned above, Myr1E was previously shown to be noninfectious (29). Since Super M and Myr1E exhibit similar, enhanced budding rates, we considered the possibility that Super M would be noninfectious as well. To test this, WT and mutant viruses were collected from transfected QT6 cells. Media containing equal numbers of particles were then transferred to primary cultures of TEFs. The kinetics of virus spreading were determined by examining the cultures at days 3, 7, and 14 postinfection. Infected cells were detected by fluorescence-activated cell sorting (FACS) because the proviruses expressed gfp from the nonessential, v-src region of the genome (Fig. 1A). To determine the sensitivity of this method, serial twofold dilutions of infected (GFP-positive) cells were made with uninfected (GFP-negative) cells. Subsequent analyses revealed that FACS reliably detected GFP-positive cells in a linear range from 1 to 100% of the total population examined (10,000 cells) (data not shown). Despite the apparent sensitivity of this assay, fluorescent cells were not detected in the TEF cultures incubated with Super M virus (Fig. 2D). Moreover, no infectivity was detected when 104-fold more Super M virus (relative to the WT control) was incubated with the TEF cultures (data not shown). Therefore, Super M is noninfectious.
Physical properties and composition of the Super M virus.
To uncover the cause(s) of the Super M infectivity defect, several features of the mutant virus were examined. Thin sections of Super M-producing cells examined by electron microscopy revealed particles of normal size with electron-dense cores like those of mature, WT virus (Fig. 3A). Analysis of particles by isopycnic sucrose gradient centrifugation showed that the density of Super M virus was like that of WT particles, with the peak of each population banding at a density of 1.16 g/ml in sucrose (Fig. 3B).
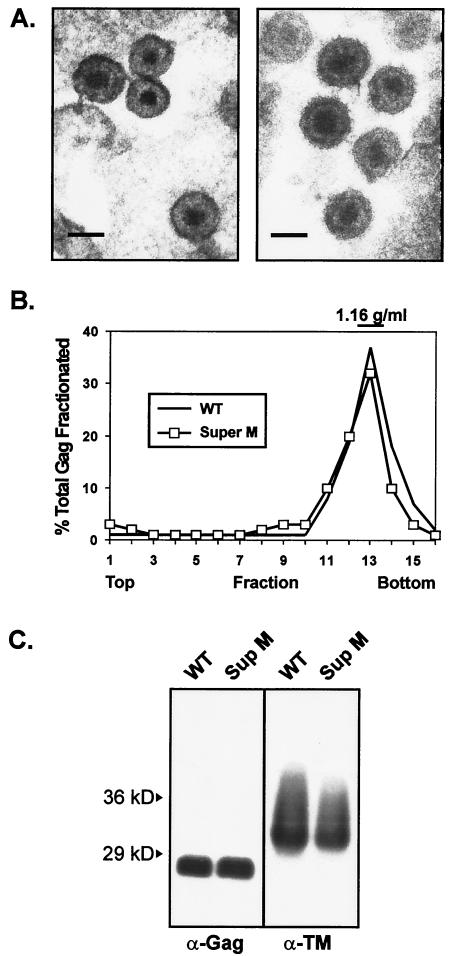
FIG. 3.
Characterization of Super M particles. (A) Electron micrographs of Super M (left) and WT (right) virus released from QT6 cells transfected with proviral plasmids. (Scale bars = 100 nm). (B) Viral particles were produced by metabolically labeled cells and separated by sucrose density gradient centrifugation. Gag proteins were immunoprecipitated from each gradient fraction, resolved by SDS-PAGE, and quantified by phosphorimager analysis. The amount of Gag in each fraction is expressed as a percentage of the total recovered from all of the fractions collected. WT virus was used as a control for normal density. (C) WT and Super M virions were collected from transfected cells, pelleted through 20% sucrose, and resuspended in PBS. Virus samples corresponding to equivalent amounts of RT activity were denatured, separated by SDS-PAGE, and transferred to nitrocellulose membranes for Western blot analyses. CA bands were detected with polyclonal Gag-reactive antiserum (α-Gag). Env incorporation was determined by using an antibody against the TM subunit (α-TM).
To determine the levels of Env and Pol incorporation, particles were collected from transfected QT6 cells and analyzed by RT assays using exogenous templates and primers. Virus sample volumes corresponding to equal amounts of RT activity were denatured, separated by SDS-PAGE, and probed by Western blotting. With a primary antibody specific for RSV Gag and Gag-derived proteins (anti-Gag), equal amounts of CA (27 kDa) were detected in the WT and Super M lanes (Fig. 3C). Since these samples were first normalized by RT activity, this result indicates that the ratios of Gag to Pol for WT and Super M viruses are similar. Likewise, an antibody specific for the TM subunit of Env (anti-TM) yielded bands of equal intensities when the WT and Super M samples were normalized by RT activity. Here, however, the TM bands migrated faster than the published mobility of 37 kDa, a difference probably due to fewer glycosylation sites in the TM subunit of this particular strain of RSV (7, 44).
In contrast to the levels of Env and Pol, the amounts of genomic RNA packaged by Super M were significantly reduced. This defect was first identified by slot blot analyses using RNA purified from equal numbers of WT and mutant particles (Fig. 4A). Specifically, Super M contained only 10% of the amount of vRNA found in WT virus. Northern blotting and RPAs confirmed the severity of this defect (data not shown). To determine whether the Super M packaging deficiency was due to a paucity of vRNA inside the cell, perhaps as a result of vRNA instability due to the Super M mutations, RNA was also purified from the transfected monolayers. As shown in Fig. 4A, however, the Super M-expressing cells contained normal amounts of genomic RNA. Therefore, the Super M packaging defect is not due to a gross reduction in the level of intracellular vRNA.
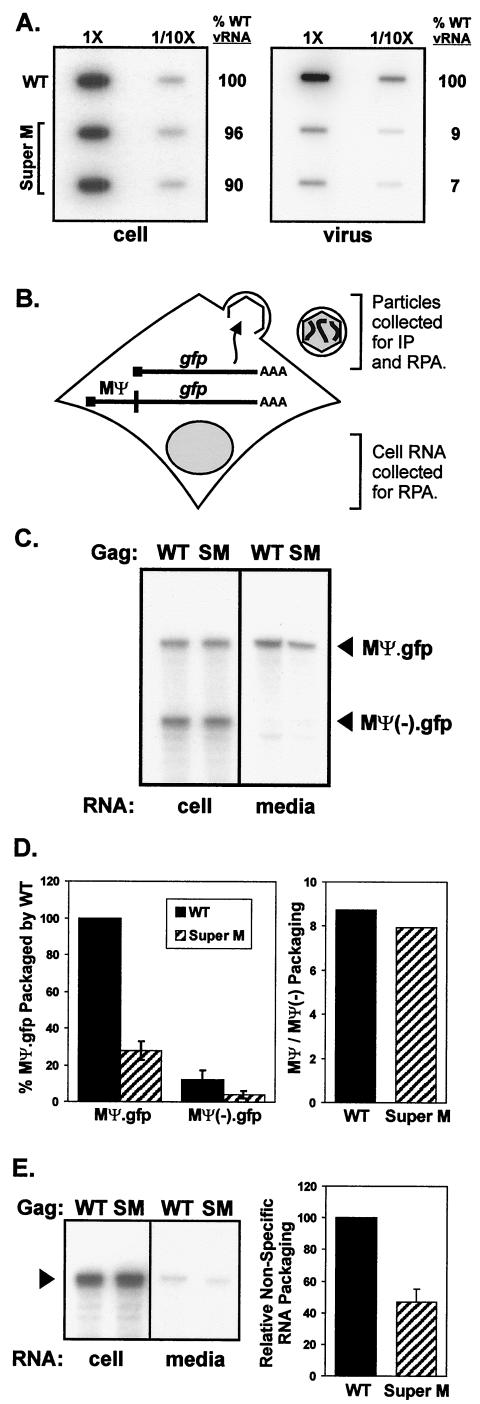
FIG.4.
Genome packaging properties of Super M. (A) QT6 cells were transfected with proviral plasmids, and particles were prepared as for Fig. 3C. RNA was purified from equivalent numbers of cells and particles, bound to nylon membranes using a slot blot apparatus, and then probed with radiolabeled, antisense RNA specific for the viral genome. Labeled bands were quantified by phosphorimager analysis, and the amounts of vRNA detected in WT cell and virus samples were defined as 100%. To ensure that the RNAs were analyzed within the linear range of this assay, the samples in the right column of each blot contained 1/10 of the total RNA blotted in the left column. (B) Vectors expressing gfp mRNA with or without the RSV minimal packaging sequence (MΨ) were created. These were cotransfected into QT6 cells with WT or Super M gag-only expression vectors (pCMV.GagPR). The cultures were trace labeled with [35S]methionine in 1% serum-containing media. After the labeling, the media were collected and aliquots of each were used to determine the numbers of radiolabeled VLPs present. The remaining media were used for the purification of particle-associated RNA, and the cell-associated RNA was purified from the transfected monolayers. The amounts of MΨ.gfp and MΨ(−).gfp mRNA in both the cell and media fractions were determined by RPA and quantified by phosphorimager analysis. IP, immunoprecipitation. (C) A representative autoradiograph shows the amounts of MΨ.gfp and MΨ(−).gfp mRNA detected in the cell and media fractions by RPA. The specific gag-only expression vector, WT or Super M (SM), cotransfected is indicated above each lane. (D) The packaging efficiencies of MΨ and MΨ(−).gfp mRNA were calculated by dividing the amount detected in the medium by the amount present in the cell. This ratio was then normalized by the number of particles present in the medium. For comparison, the efficiency of MΨ.gfp packaging by WT Gag was set to 100%. The results are the averages of four experiments, and the error bars measure 1 standard deviation (SD) from the mean. In addition, the abilities of WT and Super M Gag proteins to select for Ψ-containing mRNA were calculated by determining the ratio of MΨ to MΨ(−).gfp packaged into extracellular particles. (E) MΨ(−).gfp RNA was coexpressed with either WT or Super M Gag in QT6 cells. RNA was purified from cell and medium fractions, and the amounts of MΨ(−).gfp (arrowhead) present were determined by RPA (left). The relative abilities of WT and Super M Gag to package nonspecific RNA were calculated by normalizing the amount of MΨ(−).gfp RNA in the medium to the amount of Gag released (right). For comparison, the amount of MΨ(−).gfp packaging by WT Gag was set to 100%. The data are the averages of six experiments, and the error bar measures 1 SD from the mean.
Although the mutations that cause the two acidic-to-basic substitutions in Super M lie downstream of the packaging signal, it was possible that these mutations somehow altered folding in the 5′ end of the vRNA and disrupted stem-loop structures that are critical for Ψ function. Alternatively, the Super M changes might have altered folding in the NC domain and prevented Gag from recognizing the Ψ sequence. To test these possibilities, a plasmid expressing MΨ.gfp, an RNA consisting of the 160-nt RSV minimal packaging sequence, MΨ, at the 5′ end of a nonviral (gfp) mRNA, was generated (Fig. 4B) (1). With this vector, designated pCMV.MΨ.gfp, the abilities of WT and mutant Gag proteins (expressed from pCMV.GagPR, a gag-only expression vector) to package identical, MΨ-containing mRNA were compared. To determine the specificity of packaging in this system, the pCMV.MΨ.gfp plasmid was cotransfected with pCMV.MΨ(−).gfp, a vector that expresses a gfp mRNA lacking the MΨ sequence. Under these conditions, Super M Gag packaged about one-fourth the amount of MΨ.gfp found in WT virus-like particles (VLPs) (Fig. 4C and D). Thus, the genome packaging defect seen in provirus-transfected cells is likely due to the amino acid substitutions in the Super M protein rather than to the mutations made in the genomic RNA. With MΨ.gfp, however, the Super M defect was less pronounced (i.e., genome packaging was 1/10 WT levels, whereas MΨ.gfp packaging was 1/4 WT levels). A possible explanation for this difference is that the number of MΨ.gfp mRNAs (produced by the cytomegalovirus immediate-early promoter) may have exceeded the number of viral genomes that Gag normally encounters in an infected cell. Nevertheless, packaging of MΨ.gfp mRNA by WT and Super M Gag was apparently specific because incorporation of the MΨ− control was only 1/5 to 1/10 as efficient. This result reveals that, although the absolute amounts differed, the ratios of MΨ to MΨ(−).gfp mRNA in WT and Super M VLPs were similar (Fig. 4D). Thus, the ability to select for MΨ-containing RNA suggests that the Super M substitutions do not weaken NC's specificity for vRNA. Instead, the Super M domain may prevent packaging by an indirect mechanism, perhaps by enabling Gag to leave the site of genome packaging more quickly.
Given recent data suggesting that RNA is required to serve a structural role during retrovirus assembly (27), the ability of Super M to incorporate nonspecific, cellular RNA was investigated. In this case, either WT or Super M gag-only (pCMV.GagPR) expression vectors were cotransfected into QT6 cells with pCMV.MΨ(−).gfp, which makes mRNA that can only be packaged nonspecifically. The amounts of MΨ(−).gfp mRNA in the cell and medium fractions were determined by RPA, and the packaging efficiency was calculated by normalizing to the amount of Gag released. Under these conditions, Super M Gag packaged about one-half the amount of MΨ(−).gfp mRNA found in WT VLPs (Fig. 4E). Hence, this result suggests that Super M is able to “compensate” for the vRNA packaging defect by incorporating nonspecific, cellular RNA. However, because Super M Gag exhibited a twofold reduction in the relative level of MΨ(−).gfp incorporation, it seems likely that the rapid rate of budding inhibits nonspecific RNA packaging, albeit to a much lesser degree than vRNA packaging. Despite the reduced amount of RNA incorporation, Super M virus appeared normal by electron microscopy and exhibited WT density in sucrose (Fig. 3). Therefore, the level of total RNA incorporation is apparently sufficient to enable Gag-Gag interactions and efficient release.
Previous studies of Myr1E provide a precedent for the finding that M domain changes can reduce genome packaging in RSV. However, packaging in Myr1E is reduced by only twofold. More strikingly, the vRNA packaged by this mutant is monomeric. To determine whether Super M exhibits a dimerization defect, vRNA was purified and resolved on agarose gels under native conditions. Although the genomes of WT virus were clearly dimeric and those from Myr1E migrated as monomers, we were unable to resolve a clear band of vRNA from the Super M samples (data not shown). Efforts to increase the signal by loading purified RNA from 10-fold-more Super M particles resulted only in higher backgrounds. One possible explanation for these results is that the amount of vRNA packaged by Super M is below the level of detection for this particular assay.
Targeting properties of Super M Gag.
Previous studies of HIV-1 Gag have identified M domain changes that enhance membrane binding but do not stimulate the release of extracellular particles (16, 17, 28, 40). Instead, these mutants bind membranes indiscriminately, and, in some instances, particles have been detected in the endoplasmic reticulum or in intracellular vesicles. To examine the membrane specificity of Super M, GFP was fused in place of a C-terminal region of Gag that is nonessential for budding (Fig. 5A). Confocal microscopy revealed that Super M Gag-GFP, like an analogous construct bearing the WT M domain, was found at the plasma membrane under steady-state conditions (Fig. 5B). In addition, Super M Gag-GFP did not accumulate at intracellular membranes, which is consistent with the enhanced rate of extracellular virus production by this mutant.
FIG. 5.
Subcellular localization of Super M Gag. (A) Gag-GFP was created by replacing nonessential, C-terminal sequences of RSV Gag with GFP. The ΔNC-GFP construct contains a large deletion encompassing the I domains in NC. Super M versions of Gag-GFP and ΔNC-GFP were generated by replacing the WT M domain sequences with those with the E25K and E70K substitutions. (B) QT6 cells transfected with Super M or WT Gag-GFP vectors were left untreated or were treated with an inhibitor of nuclear export (LMB) prior to being examined by confocal microscopy. The untreated cells revealed the steady-state localization patterns of WT and Super M Gag-GFP and also served as controls for cells treated with LMB. The ΔNC panels show the effects of deleting NC on the localization of Gag-GFP proteins with WT or Super M domains.
Recent data suggest that at least some RSV Gag passes through the nucleus (38). Since nuclear trafficking is inhibited by the Src sequence present on the Myr1E M domain, we asked whether Super M is similarly deficient by treating pGag-GFP- and pSuperM-GFP-transfected cells with LMB, an inhibitor of the CRM-1 pathway of nuclear export (19). As expected, LMB treatment trapped WT Gag-GFP in the nucleus (Fig. 5B). However, Super M Gag-GFP was found at the plasma membrane in LMB-treated cells and did not appear in the nucleus. Although the functional significance of nuclear trafficking by WT Gag is unclear, it is possible that the acidic-to-basic substitutions in the Super M domain prevent nuclear targeting by enhancing Gag's affinity for the plasma membrane. This interpretation is supported by the role of NC in Super M targeting.
Although NC is best known for packaging vRNA during assembly, studies of HIV-1 Gag have revealed that NC is also required for efficient plasma membrane targeting (35, 36). This is because NC contains the I domains, which enable Gag to bind RNA and use it as a scaffold for assembly (5, 27, 47). Since RNA binding promotes interactions between Gag proteins, the requirement for the I domains in membrane targeting suggests that Gag cannot bind to the plasma membrane as a monomer. Upon oligomerization, however, Gag proteins form a multivalent membrane-binding complex that can stably bind membranes with high avidity. Prompted by the importance of NC and the I domains in membrane targeting by HIV-1 Gag, we investigated the role of NC in the subcellular distribution of RSV Gag by deleting the majority of the NC sequence from our GFP-tagged chimera (Fig. 5A). As shown in Fig. 5B, this mutant adopted a diffuse pattern in the cytoplasm of transfected cells. Thus, the NC domain is required for efficient plasma membrane localization by RSV Gag. In contrast, removal of the NC domain did not prevent plasma membrane targeting by Super M Gag-GFP (Fig. 5B). This result is significant because it suggests that Super M is not dependent on RNA interactions for targeting to the plasma membrane. Because Gag normally binds the viral genome in infected cells, Super M's ability to target in the absence of RNA interactions suggests that genome packaging is inhibited by Super M's enhanced membrane-binding potential. This interpretation implies that, by reducing Super M's membrane-binding potential, its dependence on RNA binding for plasma membrane targeting would be increased and genome packaging would be restored.
Second-site suppressors of the Super M phenotype.
To determine whether Super M's genome packaging and infectivity defects are due to enhanced membrane targeting, pairs of lysine-to-glutamate substitutions were made at residues 6 and 18 or at residues 13 and 18, while maintaining the original E25K and E70K substitutions present in Super M Gag (Fig. 6A). Hence, although the positions of the charged residues were changed, these substitutions restored the balance of 11 basic and 6 acidic residues found in the WT M domain. Consequently, reestablishing the charge balance reduced the budding efficiency of proviral constructs to within 50% of WT levels (Fig. 6B). Moreover, these substitutions restored both the ability to trap mutant Gag-GFP proteins in the nuclei of LMB-treated cells and the dependence on NC for plasma membrane targeting (Fig. 6C). These results suggest that Super M's membrane-binding potential can be weakened by reducing the net positive charge in the M domain. As predicted, the charge-balancing substitutions also restored genome packaging (Fig. 6D) and the ability to infect cultured cells (Fig. 6E). Although the slower replication kinetics exhibited by the charge-balanced mutants could be due to some defect in virus entry, K6,18E-SuperM and K13,18E-SuperM bud only half as well as the WT, so the relative rates of spreading by these mutants are slowed, at least in part, by their reduced budding efficiencies. The observed infectivity was not due to additional, unintended mutations (e.g., reversion) because mutant particles isolated at the end of these experiments (2 weeks postinfection) replicated with the same kinetics as those initially collected from the transfected QT6 cells (data not shown). Thus, the ability to affect genome packaging and infectivity by modulating the net charge in the M domain suggests that Gag targeting and genome packaging are linked in RSV.
FIG. 6.
Creation and analysis of suppressors of Super M. (A) The WT numbers of positively and negatively charged residues were restored in the Super M domain by basic-to-acidic substitutions at positions 6 and 18 or at positions 13 and 18. (B) Analysis of the budding efficiencies of the charge-balanced mutants was performed with proviral vectors as for Fig. 2A. (C) The localization patterns of ΔNC-GFP and Gag-GFP proteins containing charge-balanced M domains in untreated or LMB-treated cells were examined as for Fig. 5B. (D) The genomic RNA content of extracellular virus was determined by using proviral vectors as for Fig. 4A. (E) Infectivity assays were performed at 1 and 2 weeks postinfection as for Fig. 2D. For the results shown in panels B, D, and E, the data are the averages of at least three experiments and the error bars represent 1 standard deviation from the mean.
DISCUSSION
The data presented here reveal that small changes (i.e., two substitutions) in the M domain can have a profound impact on genome packaging and infectivity of RSV. Whether the infectivity block exhibited by Super M is due entirely to the genome packaging defect is unclear. Given the severity of the packaging defect, it is likely that Super M incorporates no more than one vRNA copy per particle. If this is the case, then an inability to form vRNA dimers would also block infectivity. Alternatively, if a few Super M particles contained vRNA dimers, then additional defects (e.g., tRNA primer incorporation) would remain to be identified. However, the correlation between the vRNA content and infectivity of the mutants described here suggests that the genome packaging deficiency is the major reason why Super M is noninfectious. Hence, these data raise the question of how the M domain affects genome packaging by RSV Gag.
Although the severity of the Super M packaging defect is similar in magnitude to that caused by deleting both zinc fingers in NC (both mutants package only 1/10 the amount of vRNA found in WT particles), numerous studies of RSV Gag have shown that the MA region, including the M domain, is not involved in direct vRNA interactions (21, 23). For instance, MA can be replaced by the Src M domain without affecting the levels of vRNA incorporation (21, 34). Moreover, an RSV Gag chimera containing NC sequences from murine leukemia virus (MLV) packages the MLV genome. Hence, RNA specificity is not determined by the MA region in chimeric Gag proteins (9). This result is consistent with the observation that RSV MA does not bind vRNA with any specificity in vitro (41). So, if the M domain is not involved in vRNA binding, then how does it affect genome packaging? Given the well-established role of the M domain in Gag targeting, one possibility is that the M domain directs Gag to the site of genome packaging.
Recent data suggest that the RSV M domain contains a nuclear localization signal (NLS), and because the absence of nuclear import by Myr1E Gag correlates with a genome packaging defect, it was proposed that genome binding occurs in the nucleus (38). However, the dispensability of the RSV M domain in vRNA packaging contradicts this hypothesis. Indeed, vRNA packaging is not affected when the MA region is replaced by the Src M domain, a change that would block nuclear trafficking by removing the putative NLS in the M domain (21, 34). Likewise, deletions or single substitutions in the NLS in HIV-1 MA prevent nuclear import, but these changes do not affect vRNA incorporation (8). Moreover, retroviral vector systems in which Gag specifically packages mRNA that exists exclusively in the cytoplasm have been designed (18, 22). In those studies, recombinant alphaviruses (e.g., Semliki Forest virus) or poxviruses (e.g., vaccinia virus) were used to synthesize transcripts in the cytoplasm of Gag-expressing cells, and the high titers achieved by this approach reveal that Gag can readily package cytoplasmic mRNA. Therefore, these data argue against a role for nuclear trafficking in genome packaging. Consequently, the Super M packaging defect may not be due to a lack of nuclear import per se, but the absence of a nuclear phase suggests that the Super M substitutions alter Gag targeting in some manner. This possibility may be significant because, if genome packaging occurs at a specific location in the cytoplasm, then perhaps the Super M substitutions prevent targeting to this site as well. Alternatively, genome packaging may not require Gag transport to a particular location in the cell but may depend on the accumulation of Gag proteins at the site of synthesis. In this case, packaging would be inhibited if Gag begins targeting from the site of synthesis before binding to the viral genome. Support for this possibility is provided by the role of the NC domain in Gag targeting (35, 36).
The importance of NC, and in particular the I domains, in both genome packaging and Gag targeting suggests that these two functions are normally linked. The basis of this connection seems to be the need for Gag to use RNA as a scaffold for assembly. Indeed, the I domains, in conjunction with the zinc fingers in NC, facilitate specific interactions with the Ψ sequence in the viral genome. As additional Gag proteins attach nonspecifically to vRNA via the I domains, they assemble into a multivalent complex that can bind membranes with high avidity. Since the formation of this multivalent complex relies on RNA interactions, genome packaging may normally precede Gag targeting. Consequently, the requirement for RNA interactions in Gag targeting may ensure that the retroviral genome is packaged before nascent particles are released from the cell.
Based on the model described above, the vRNA packaging defects exhibited by Super M and Myr1E may be due to the ability of these mutants to target to the plasma membrane before genome binding and Gag-Gag interactions occur. Support for this interpretation is provided by the enhanced budding properties of these mutants and the maintenance of Super M targeting in the absence of the NC domain. Although Gag-Gag interactions may be unnecessary for Super M targeting, this mutant nevertheless assembles efficiently because the extracellular particles resemble the WT in appearance and density (Fig. 3). Importantly, reestablishing the WT balance of basic and acidic residues in the Super M Gag domain improved genome packaging and restored infectivity (Fig. 6). Hence, these data provide further evidence that the genome packaging defect is caused by the enhanced membrane-binding potential of the Super M Gag domain.
The idea that genome packaging precedes Gag targeting is not new. A well-known example of this mechanism is provided by the Mason-Pfizer monkey virus (MPMV). Unlike those of RSV and HIV-1, MPMV Gag proteins first form complete capsids in the cytoplasm, which then target to the plasma membrane for budding (32, 42). A precedent for M domain changes alleviating the assembly prerequisite for Gag targeting is provided by the effects of substitution R55W in the MA domain of MPMV Gag (33). Specifically, this change prevents the accumulation of intracellular capsids and enables particle assembly at the plasma membrane. Interestingly, the mutant was also shown to bud faster than WT MPMV, so the R55W change may enhance membrane affinity. Like Super M, the R55W mutant is noninfectious, and further studies suggested that the infectivity defect is due to the incorporation of fewer Env glycoproteins. However, the efficiency of genome packaging was not examined in these studies and therefore may be defective as well. This prediction is based on the assumption that MPMV genomes are present at the intracellular sites of capsid assembly, though surprisingly the presence of vRNA in intracellular particles has not been reported.
Despite the likelihood that the extra basic residues in the Super M domain enhance Gag's affinity for the plasma membrane, other factors could explain why this mutant buds so quickly. For instance, we have shown that Super M does not localize to the nucleus in LMB-treated cells. Hence, if WT Gag normally travels through the nucleus before targeting to the plasma membrane, then Super M may bud faster simply by avoiding the nuclear “detour.” In addition, it is possible that the substitutions in Super M promote Gag-Gag interactions. That the MA region could be a site of direct, albeit weak, contact between Gag proteins is suggested by cross-linking studies, yeast two-hybrid data, and the observation that purified HIV-1 MA forms trimers upon crystallization (3, 13, 30, 31). Thus, the Super M substitutions may enhance interactions between Gag proteins and enable the formation of a multivalent membrane-binding complex in the absence of the I domains and RNA binding. Certainly, additional studies will be needed to test these hypotheses, and the insights gained by these efforts should shed more light on the relationship between membrane targeting and genome packaging by Gag.
Acknowledgments
We thank Joshua Loomis for construction of pΔNC-GFP and Roland Meyers for thin-sectioning and electron microscopy. The anti-TM antibody was generously provided by Judy White.
This work was supported by grant CA47482 from the National Institutes of Health to J.W.W.
REFERENCES
- 1.Banks, J. D., A. Yeo, K. Green, F. Cepeda, and M. L. Linial. 1998. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 72:6190-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J. Virol. 74:11222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duesberg, P. H., G. S. Martin, and P. K. Vogt. 1970. Glycoprotein components of avian and murine RNA tumor viruses. Virology 41:631-646. [DOI] [PubMed] [Google Scholar]
- 8.Dupont, S., N. Sharova, C. DeHoratius, C. M. Virbasius, X. Zhu, A. G. Bukrinskaya, M. Stevenson, and M. R. Green. 1999. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402:681-685. [DOI] [PubMed] [Google Scholar]
- 9.Dupraz, P., and P. F. Spahr. 1992. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J. Virol. 66:4662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz, R. A., J. H. Omer, J. H. Weis, S. A. Mitsialis, A. J. Fara, and R. V. Guntaka. 1982. Restriction endonuclease and nucleotide sequence analysis of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J. Virol. 42:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiernan, R. E., A. Ono, and E. O. Freed. 1999. Reversion of a human immunodeficiency virus type 1 matrix mutation affecting Gag membrane binding, endogenous reverse transcriptase activity, and virus infectivity. J. Virol. 73:4728-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konetschny, C., G. W. Holzer, and F. G. Falkner. 2002. Retroviral vectors produced in the cytoplasmic vaccinia virus system transduce intron-containing genes. J. Virol. 76:1236-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 21.Lee, E.-G., A. Yeo, B. Kraemer, M. Wickens, and M. L. Linial. 1999. The Gag domains required for avian retroviral RNA encapsidation determined by using two independent assays. J. Virol. 73:6282-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, K. J., and H. Garoff. 1998. Packaging of intron-containing genes into retrovirus vectors by alphavirus vectors. Proc. Natl. Acad. Sci. USA 95:3650-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 24.McDonnell, J. M., D. Fushman, S. M. Cahill, W. Zhou, A. Wolven, C. B. Wilson, T. D. Nelle, M. D. Resh, J. Wills, and D. Cowburn. 1998. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J. Mol. Biol. 279:921-928. [DOI] [PubMed] [Google Scholar]
- 25.Mouland, A. J., J. Mercier, M. Luo, L. Bernier, L. DesGroseillers, and E. A. Cohen. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol 74:5441-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouland, A. J., H. Xu, H. Cui, W. Krueger, T. P. Munro, M. Prasol, J. Mercier, D. Rekosh, R. Smith, E. Barbarese, E. A. Cohen, and J. H. Carson. 2001. RNA trafficking signals in human immunodeficiency virus type 1. Mol. Cell. Biol. 21:2133-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parent, L. J., T. M. Cairns, J. A. Albert, C. B. Wilson, J. W. Wills, and R. C. Craven. 2000. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 74:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepinsky, R. B. 1983. Localization of lipid-protein and protein-protein interactions within the murine retrovirus gag precursor by a novel peptide-mapping technique. J. Biol. Chem. 258:11229-11235. [PubMed] [Google Scholar]
- 31.Pepinsky, R. B., D. Cappiello, C. Wilkowski, and V. M. Vogt. 1980. Chemical crosslinking of proteins in avian sarcoma and leukemia viruses. Virology 102:205-210. [DOI] [PubMed] [Google Scholar]
- 32.Rhee, S. S., and E. Hunter. 1987. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 61:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee, S. S., and E. Hunter. 1990. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell 63:77-86. [DOI] [PubMed] [Google Scholar]
- 34.Sakalian, M., J. W. Wills, and V. M. Vogt. 1994. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J. Virol. 68:5969-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag). J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 38.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz, D. E., R. Tizard, and W. Gilbert. 1983. Nucleotide sequence of Rous sarcoma virus. Cell 32:853-869. [DOI] [PubMed] [Google Scholar]
- 40.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steeg, C. M., and V. M. Vogt. 1990. RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J. Virol. 64:847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 43.Verderame, M. F., T. D. Nelle, and J. W. Wills. 1996. The membrane-binding domain of the Rous sarcoma virus Gag protein. J. Virol. 70:2664-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral Gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 46.Wills, J. W., R. C. Craven, and J. A. Achacoso. 1989. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J. Virol. 63:4331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan, X., X. Yu, T.-H. Lee, and M. Essex. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 67:6387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]