Abstract
The Moloney murine leukemia virus (MLV) repressor binding site (RBS) is a major determinant of restricted expression of MLV in undifferentiated mouse embryonic stem (ES) cells and mouse embryonal carcinoma (EC) lines. We show here that the RBS repressed expression when placed outside of its normal MLV genome context in a self-inactivating (SIN) lentiviral vector. In the lentiviral vector genome context, the RBS repressed expression of a modified MLV long terminal repeat (MNDU3) promoter, a simian virus 40 promoter, and three cellular promoters: ubiquitin C, mPGK, and hEF-1a. In addition to repressing expression in undifferentiated ES and EC cell lines, we show that the RBS substantially repressed expression in primary mouse embryonic fibroblasts, primary mouse bone marrow stromal cells, whole mouse bone marrow and its differentiated progeny after bone marrow transplant, and several mouse hematopoietic cell lines. Using an electrophoretic mobility shift assay, we show that binding factor A, the trans-acting factor proposed to convey repression by its interaction with the RBS, is present in the nuclear extracts of all mouse cells we analyzed where expression was repressed by the RBS. In addition, we show that the RBS partially repressed expression in the human hematopoietic cell line DU.528 and primary human CD34+ CD38− hematopoietic cells isolated from umbilical cord blood. These findings suggest that retroviral vectors carrying the RBS are subjected to high rates of repression in murine and human cells and that MLV vectors with primer binding site substitutions that remove the RBS may yield more-effective gene expression.
Retroviral vectors based upon the Moloney murine leukemia virus (MLV) have been used widely due to their high efficiency of stable gene transfer. Transcription from the MLV long terminal repeat (LTR) is severely repressed in mouse embryonal carcinoma (EC) cells (7, 23, 42, 45) and mouse embryonal stem (ES) cells (13, 21). Characteristics of MLV that have been shown to mediate poor expression in undifferentiated ES and EC cell lines include the inadequate function of the enhancer in the 5′LTR due to the lack of transcriptional activator binding sites (9, 20, 21, 27, 41, 47), the negative control region (NCR) located in the U3 region of the LTR (1, 5, 10, 12, 47), and the repressor binding site (RBS) (2, 9).
The RBS is an 18-bp DNA element located downstream of the 5′LTR which directly overlaps the MLV primer binding site (PBS) by 17 of its 18 bp (25). The MLV PBS functions by binding a proline tRNA molecule that primes reverse transcription. The existence of the overlapping RBS was first determined due to a spontaneously occurring single-base G-to-A mutation that occurred within the PBS sequence that allowed expression by MLV in F9 EC cells (2). This single base mutation was called the B2 mutation (2). Like the MLV PBS, the B2 PBS binds to a proline tRNA molecule, but with a single base-pair mismatch (4). Several other mutations that have been made within the MLV PBS have also been shown to relieve the repression conveyed by the RBS element (21, 26, 35).
Due to the requirement that vectors derived from MLV must contain a functional PBS to initiate reverse transcription, the overlapping RBS sequence cannot be removed simply by removing the MLV PBS. In order to eliminate this repressive element and still have a functional vector, several groups have replaced the MLV PBS with the PBS from dl587rev, a recombinant between MLV and an endogenous mouse retrovirus (5, 6, 13, 19). The dl587rev PBS sequence contains the B2 mutation plus five additional base pairs different from the MLV PBS and is a perfect match for the glutamine tRNA (6).
The RBS has been shown to repress transcription from the MLV LTR within the context of an MLV genome when located away from its normal position, in either orientation, upstream of the MLV transcription start site (25) or when positioned downstream within an intron (35). In addition to repressing transcription from the MLV LTR, the RBS was shown to substantially repress transcription from two internal heterologous viral promoters, simian virus 40 (SV40) and adenovirus major late promoter, which were inserted downstream of the RBS into an MLV vector with an enhancerless LTR (31, 35).
The mechanism by which the RBS represses transcription is not known. The RBS is thought to function by interacting with an unknown trans-acting factor. In support of this hypothesis, the repressive activity of the RBS was demonstrated to be saturable by transfection of increasing amounts of DNA containing the RBS sequence, suggesting that the repressive activity is mediated by a trans-acting factor or factors (26). With an exonuclease III protection assay, a factor from PC13 EC cell extracts was shown to bind a MLV PBS probe, but not a B2 PBS probe (25). Using an electrophoretic mobility shift assay (EMSA), a predominantly nuclear protein in F9 EC cell nuclear extracts was identified that bound to an MLV PBS probe, but not a B2 PBS probe (35, 48).
Several studies have described the repressive activity of the RBS to be stem cell specific (21, 25, 35, 48), because it has been shown to have repressive activity in several murine EC cell lines (F9, PC13, and PCC4) and D3 ES cells, but not in 3T3 embryonic fibroblasts. Retroviral vectors with multiple modifications, including the dl587rev PBS, have been demonstrated to be superior to vectors containing only MLV components (16), but the specific role of the PBS change has not been analyzed. We evaluated whether the RBS alone was sufficient to cause repression by incorporating this element into a lentiviral vector derived from human immunodeficiency virus type 1 (HIV-1). We also wanted to determine if the repressive activity mediated by the RBS present in ES and EC cell lines was also present in murine hematopoietic stem cells and more differentiated cells and if the repressive activity mediated by the RBS was present in human cells. Because some of these cell types are inefficiently transduced by MLV-based retroviral vectors, we used a series of HIV-1-based lentiviral vectors into which the MLV, B2, or the dl587 PBS sequences were inserted downstream from various internal promoters. We observed that the PBS sequences acted in the context of the lentiviral vector genome as they do within the MLV vector genome context in that the MLV PBS sequence, containing the overlapping RBS, repressed expression from several internal promoters, while the B2 and dl587 PBS sequences did not. Repression specific to the RBS was also documented in a variety of murine and human cells, including hematopoietic stem and progenitor cells.
MATERIALS AND METHODS
Cell culture.
The following cell lines were obtained from the American Type Culture Collection (ATCC) and cultured according to their recommendations: K562 (ATCC CCL-243), U937 (ATCC CRL-1593.2), PA-1 (ATCC CRL-1572), Tera-2 (ATCC HTB-106), NCCIT (ATCC CRL-2073), 70Z/3 (ATCC TIB-158), STO-SNL/2 (ATCC CRL-2225), ES-D3 (ATCC CRL-1934), Jurkat (ATCC TIB-152), KG1a (ATCC CCL-246.1), AMJ2-C11 (ATCC CRL-2456), F9 EC (ATCC CRL-1720), NIH/3T3 (ATCC CRL-1658), and CCRF-CEM (ATCC CCL-119). DU.528 (22) were obtained from Joanne Kurtzberg (Duke University Medical Center, Durham, N.C.) and cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS), 10% horse serum (HS), 2 mM l-glutamine, 0.1 mM sodium pyruvate, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. BM185 cells have been previously described (43) and were cultured in RPMI medium supplemented with 5% FBS, 0.01 mM 2-mercaptoethanol (Sigma, St. Louis, Mo.), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. WTc.F cells (ES cells derived from C57Bl/6 mice) (37), a gift of Andrew Kung (Harvard Medical School, Boston, Mass.), were cultured on irradiated STO-SNL/2 feeder layers in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% FBS, 4 mM l-glutamine, 0.1 mM nonessential amino acids, 0.1 mM 2-mercaptoethanol, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. FDCP-Mix cl.A4 was obtained from Lez Fairbairn (Paterson Institute for Cancer Research, Manchester, United Kingdom) and cultured in Iscove's modification of Dulbecco's medium (IMDM) supplemented with 20% HS and 10 ng of murine interleukin-3 (IL-3; Biosource International, Camarillo, Calif.)/ml.
Mouse embryonic fibroblasts (MEFs) were isolated by trypsinization of embryos dissected at 13.5 days of gestation from outbred CF-1 mice (Charles River Laboratories, Wilmington, Mass.). Each embryo was harvested separately, the brain and internal organs were removed, and the carcasses were minced and incubated in 0.05% trypsin for 30 to 45 min at 37°C. Single-cell suspensions obtained after trypsinization were plated in 10-cm dishes in DMEM supplemented with 10% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM l-glutamine. Experiments with MEFs were performed on early passages (less than five).
Mouse bone marrow (BM) cells were isolated by flushing bone marrow from adult mouse femurs with a 27[1/2]-gauge needle. Bone marrow stromal cell populations were isolated by breaking the femurs into small pieces and culturing them with isolated marrow cells. Cells were allowed to adhere to tissue culture plates for 4 to 6 days in IMDM containing 20% HS, 20% FBS, 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 0.1 mM 2-mercaptoethanol, and 6 μM hydrocortisone. Adherent cells were rinsed 5 to 7 times with phosphate-buffered saline (Irvine Scientific, Santa Ana, Calif.) every day for 10 to 15 days to remove nonadherent cells. Adherent cells were allowed to expand to confluency for an additional 2 to 3 weeks. Remaining CD45+ hematopoietic cells were removed with a magnetic column after staining with rat anti-mouse CD45 antibody and magnetic bead-conjugated goat anti-rat immunoglobulin G. CD45− cells were placed back into culture and later verified to be CD45− by flow cytometry.
CD34+ cells were isolated from human umbilical cord blood obtained from normal deliveries, using Miltenyi MiniMACS magnetic separation columns (Miltenyi Biotech, Sunnyvale, Calif.) after Ficoll-Hypaque (Amersham Pharmacia Biotech, Piscataway, N.J.) density gradient centrifugation. Use of these cord blood samples was approved by the Committee on Clinical Investigations at Childrens Hospital, Los Angeles, Calif. To isolate CD34+ CD38− cells, CD34+ cells were washed in phosphate-buffered saline and incubated for 30 min at 4°C in fluorescein isothiocyanate-CD34 (HPCA2; Becton Dickinson Immunocytometry Systems, San Jose, Calif.) and phycoerythrin-CD38 (leu 17; Becton Dickinson Immunocytometry Systems). CD34+ CD38− cells were then isolated by fluorescence-activated cell sorting (FACS), using the gating previously described (17), on a FACSVantage flow cytometer (Becton Dickinson Immunocytometry Systems) and using LysysII software (Becton Dickinson Immunocytometry Systems).
Vector construction.
The pL-eGFP-SN vector was constructed by inserting a BglII-NotI fragment containing the enhanced green fluorescent protein (eGFP) gene (Clontech Laboratories, Palo Alto, Calif.) into the HpaI site of pL-X-SN as previously described (38). Modifications made to pL-eGFP-SN to generate pLD-eGFP-SN, pM-eGFP-SN, pMD-eGFP-SN, and pMND-eGFP-SN were described previously (5, 38).
Self-inactivating (SIN) lentiviral vectors containing the U3 region of the MND LTR (MNDU3; the U3 region from the myeloproliferative sarcoma virus [MPSV] LTR with the NCR removed) (5) as an internal promoter with each of the PBS sequences were constructed as follows. pCCL-hCMV-eGFP (8) (kindly provided by Luigi Naldini, Cell Genesys, Foster City, Calif.) was digested with ClaI and SalI to remove a fragment containing hCMV-eGFP to generate pCCL-X. pMND-Neo (5) was digested with ClaI and Asc1 to isolate a fragment containing the MNDU3 promoter. The MNDU3 enhancer-promoter fragment was blunted and ligated into the EcoR5 site of pIC-20H to generate pMNDU3-20H. pMNDU3-20H was digested with BamHI and BglII to isolate a fragment containing the MNDU3 enhancer-promoter. The MNDU3 enhancer-promoter fragment was inserted into the BamHI site of pCCL-X to generate pCCL-MNDU3-X. pMND-eGFP-SN was digested with XhoI to release a fragment containing eGFP. The eGFP fragment was ligated into the XhoI site of pCCL-MNDU3-X to generate pCCL-MNDU3-eGFP. The MLV PBS was inserted into the SalI site downstream of the MNDU3 promoter using 5′-phosphorylated oligonucleotides which anneal to create the PBS (underlined) flanked by SalI sites and a unique MluI site used to verify insertion: 5′-TCGACACGCGTGGGGGCTCGTCCGGGATCGGGAGACCCCG-3′ and 5′-TCGACGGGGGCTCCCGATCCCGGACGAGCCCCCACGCGTG-3′. The B2 PBS was inserted into the SalI site downstream of the MNDU3 promoter using 5′-phosphorylated oligonucleotides which anneal to create the PBS flanked by SalI sites and a unique MluI site used to verify insertion: 5′-TCGACACGCGTGGGGGCTCGTCCGAGATCGGGAGACCCCG-3′ and 5′-TCGACGGGGGCTCCCGATCTCGGACGAGCCCCCACGCGTG-3′. The dl587 PBS was inserted into the SalI site downstream of the MNDU3 promoter using the following 5′-phosphorylated oligonucleotides which anneal to create the PBS flanked by SalI sites and a unique MluI site used to verify insertion: 5′-TCGACACGCGTGGAGGTTCCACCGAGATTTGGAGACCCCG-3′ and 5′-TCGACGGGGTCTCCAAATCTCGGTGGAACCTCCACGCGTG-3′.
SIN lentiviral vectors containing different internal promoters were constructed using pCCL-hCMV-eGFP. The human cytomegalovirus (hCMV) internal promoter in this vector was removed with ClaI and BamHI and replaced with the human elongation factor 1a (hEF-1a) promoter, which was acquired from pV4.1e-hF.IX (kindly provided by Hiroyuki Nakai, Stanford University, Stanford, Calif.) by PCR with Pfu Turbo polymerase (Stratagene, La Jolla, Calif.) using the primers 5′-GAAGATCGATCGTGAGGCTCCGGTG-3′ and 5′-GGTAGGATCCACGACACCTGAAATG-3′, followed by digestion with ClaI and BamHI. To replace the hCMV promoter in pCCL-hCMV-eGFP with the human ubiquitin C (hUbiqC) promoter, the plasmid was first digested with ClaI, blunt ended with Pfu Turbo polymerase, and then digested with BamHI. The hUbiqC promoter fragment was prepared from pFUGW (kindly provided by Carlos Lois, California Institute of Technology, Pasadena, Calif.) by digesting with PacI, then blunt ending with T4 polymerase (Invitrogen, Carlsbad, Calif.), followed by digestion with BamHI. To replace the hCMV promoter in pCCL-hCMV-eGFP with the mouse phosphoglycerate kinase (mPGK) promoter, the hCMV promoter was removed by digestion with ClaI and AgeI, and an oligonucleotide containing restriction sites ClaI-EcoRV-BamHI-AgeI was inserted. The mPGK promoter was excised from pic20H-mPGK with HincII and BglII and inserted into EcoR5 and BamHI. To replace the hCMV promoter in pCCL-hCMV-eGFP with the SV40 promoter, the hCMV-eGFP cassette was removed by digestion with ClaI and SalI and an oligonucleotide containing restriction sites ClaI-XhoI-EcoRV-BamHI-SmaI-SalI was inserted. An eGFP fragment was excised from pMND-eGFP-SN with BamHI and inserted into this multicloning site. The SV40 promoter was excised from pL-X-SN with XhoI and StuI and inserted into this multicloning site after digestion with XhoI and EcoRV. The MLV PBS was inserted into a BamHI site downstream of the hEF-1a and hUbiqC promoters and upstream of the mPGK promoter using 5′-phosphorylated oligonucleotides that anneal to create the PBS flanked by BamHI sites and a unique NheI site used to verify insertion: 5′-GATCCGCTAGCGGGGGCTCGTCCGGGATCGGGAGACG-3′ and 5′-GATCCGTCTCCCGATCCCGGACGAGCCCCCGCTAGCG-3′. The MLV PBS was inserted into an XhoI site upstream of the SV40 promoter using 5′-phosphorylated oligonucleotides that anneal to create the PBS flanked by XhoI sites and a unique NheI site used to verify insertion: 5′-TCGAGGCTAGCGGGGGCTCGTCCGGGATCGGGAGAC-3′ and 5′-TCGAGTCTCCCGATCCCGGACGAGCCCCCGCTAGCC-3′. The MLV PBS was inserted into an AgeI site downstream of the SV40 promoter using 5′-phosphorylated oligonucleotides that anneal to create the PBS flanked by AgeI sites and a unique NheI site used to verify insertion: 5′-CCGGTGCTAGCGGGGGCTCGTCCGGGATCGGGAGAA-3′ and 5′-CCGGTTCTCCCGATCCCGGACGAGCCCCCGCTAGCA-3′.
Vector supernatant production.
MLV vector supernatants were generated by a stably transduced GP+E-86 packaging cell line (28). Cells were plated close to confluency in T75 flasks, grown at 37°C for 48 h in 10 ml of DMEM supplemented with 10% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine. Vector supernatants were filtered through a 0.45-μm filter and frozen at −80°C until used. Vesicular stomatitis virus glycoprotein-pseudotyped lentiviral vector supernatants were generated by transient transfection of 293T cells (ATCC CRL-1268) as previously described (40) using 10 μg of vector plasmid, 10 μg of pRΔ8.9 packaging plasmid (49), and 2 μg of pMD.G(VSV) envelope plasmid (32). Twelve hours after transfection, cells were treated with 10 mM sodium butyrate (Sigma Scientific, Inc., Brighton, Mich.) for 12 h as previously described (40). After 12 h of exposure to sodium butyrate, the cells were washed twice with phosphate-buffered saline and refed with fresh medium. Thereafter, supernatants were collected every 12 h for 3 to 5 days, filtered through a 0.2-μm filter flask (Nalgene, Rochester, N.Y.), and concentrated by ultracentrifugation at 50,000 × g for 140 min as previously described (24). Pellets were resuspended in serum-free DMEM and stored at −80°C until used.
Vector supernatant titer determination.
Vector supernatant titers were determined by endpoint dilution. 293 cells (ATCC CRL-1573) were seeded at 105 cells/well in six-well cell culture plates (Corning Inc., Miami, Fla.) in DMEM supplemented with 10% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine and placed in a 37°C incubator for 12 h. Cells were then transduced with 1-ml serial dilutions (i.e., 10−1, 10−2, 10−3) of vector supernatant and analyzed by flow cytometry for eGFP expression 48 h later. Titers were calculated by multiplying the number of cells at the time of vector supernatant addition by the percentage of eGFP-positive cells determined by flow cytometry divided by 100, multiplied by the dilution factor to yield the number of infectious units (IU) per milliliter. Titers ranged between 0.5 × 106 and 10 × 106 IU/ml before ultracentrifugation and 0.5 × 108 and 5 × 108 IU/ml after ultracentrifugation.
Lentiviral vector transductions.
Target cells that were transduced to determine whether they possessed RBS-mediated repressive activity were transduced in parallel and under the same conditions as F9 EC and 293 cells in order to control for small differences in vector titer that could contribute to differences in transduction efficiency. Most cell types were transduced for 12 h using a final vector concentration of 1 × 106 to 2 × 106 IU/ml and a multiplicity of infection of 5 to 10 in their normal growth medium. CD34+, CD34+ CD38−, and primary mouse BM cells required higher concentrations of vector for efficient transduction and were transduced with a final vector concentration of 5 × 107 to 10 × 107 IU/ml (15) plus Polybrene at 8 μg/ml. After transduction, cells were passaged in culture for 6 to 10 days and then analyzed by flow cytometry for eGFP expression.
PCR for relative copy number.
Genomic DNA was isolated using a DNeasy tissue kit (Qiagen, Valencia, Calif.) for use as template in a semiquantitative PCR to determine the relative vector copy number in transduced cells. To generate the standard curve, we used a GP+E-86 cell clone containing five copies of the MND-eGFP-SN vector, as previously described (16). The standard curve was generated using dilutions representing 4, 2, 1, 0.5, 0.25, 0.13, 0.06, and 0.03 copies/cell. DNA of the five-copy clone was mixed with DNA of nontransduced GP+E-86 cells, so that the total input template DNA was maintained constant. Template DNA was diluted to 100 ng in 44 μl of double-distilled (ddH2O) in a PCR tube. Three microliters of this solution, containing 7 ng of template DNA, was removed and placed into a second tube, leaving 41 μl containing 93 ng of template DNA in the first tube. Master mix solutions were then added to make a final volume of 50 μl containing 2.5 U of Pfu Turbo polymerase, 1× Pfu PCR buffer (Stratagene), each primer at a 0.2 μM concentration, and each deoxynucleoside triphosphate at a 0.2 mM concentration. The tube containing 93 ng of template DNA was used for eGFP PCR, and the tube containing 7 ng of template DNA was used for β-actin PCR to control for varying input template content. eGFP primer sequences used were sense, 5′-ATGGTGAGCAAGGGCGAGGAGCTG-3′, and antisense, 5′-GCCGTCGTCCTTGAAGAAGATGGTG-3′, yielding a product of 314 bp. β-Actin primer sequences used were sense, 5′-GTACCACAGGCATTGTGATG-3′, and antisense, 5′-GCAACATAGCACAGCTTCTC-3′, yielding a product of 219 bp, as previously described (44). Reactions were performed on a Perkin-Elmer GeneAmp 9600 PCR system. Both the eGFP and β-actin PCRs were conducted for 28 cycles with denaturation at 94°C for 30 s, annealing at 65°C for 1 min, and extension at 72°C for 1 min. Reaction products were separated by electrophoresis on 0.8% agarose-EtBr gels and visualized using an Eagle Eye charge-coupled device camera (Stratagene).
Nuclear extracts and EMSAs.
Nuclear extracts were prepared essentially as described elsewhere (48). Nuclear extracts were prepared without dialysis against modified Dignam buffer D and were left in Dignam buffer C (20 mM HEPES [pH 7.9], 25% [vol/vol] glycerol, 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol) as previously described (48). EMSAs were performed using previously described binding conditions (21, 48). Probes were prepared by annealing complementary single-stranded oligonucleotides and then end labeling with [γ-32P]ATP (Amersham Biosciences) using T4 polynucleotide kinase (Invitrogen). Extended 28-bp PBS probes were used due to reported increased binding in EMSA compared to the 18-bp core PBS sequence (underlined below) (48). Oligonucleotide probes were as follows: MLV PBS, 5′-GGGGGCTCGTCCGGGATCGGGAGACCCC-3′; B2 PBS, 5′-GGGGGCTCGTCCGAGATCGGGAGACCCC-3′; dl587 PBS, 5′-GGAGGTTCCACCGAGATTTGGAGACCCC-3′ (only one strand for each is shown). Radioactively labeled double-stranded DNA oligonucleotides were purified using G-25 columns (Amersham Biosciences) according to the manufacturer's instructions. Radioactive labeling ranged from 200,000 to 400,000 cpm/ng. Each binding reaction was performed with 5 to 15 μl of nuclear extract in a total volume of 30 μl of Thornell binding buffer (25 mM HEPES [pH 7.9], 1 mM EDTA, 10% [vol/vol] glycerol, 5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) containing 25 ng of poly(dI-dC)/ml, 5 mM NaCl, 5 mM KCl, 3 mM MgCl2, and 0.1 mM ZnCl2, as previously described (48). Prior to the addition of probe, binding reaction mixtures were preincubated at 30°C for 20 min, and then 0.5 ng of double-stranded radioactively labeled probe was added in a 1-μl volume and reactions were incubated for an additional 20 min at 30°C. Binding reactions were run on a 6% native acrylamide gel. The gel was loaded at room temperature, run for 20 min at 130 V, and then transferred to 4°C and run at 175 V for approximately 4 h in Tris-glycine buffer (5 mM Tris [pH 8.5], 38 mM glycine, 0.2 mM EDTA). After electrophoresis, gels were dried and opposed to film for 1 to 4 days at −80°C with intensifying screens.
Mouse bone marrow harvest, transduction, and transplantation.
Mice were purchased from Jackson Laboratories (Bar Harbor, Maine) and maintained at the Childrens Hospital Los Angeles Central Animal Facility. All studies were approved by the Institutional Animal Care and Use Committee at Childrens Hospital Los Angeles. Donor marrow was harvested from 8- to 12-week-old male B6/SJL mice 2 days after 5-fluorouracil intravenous injection (150 mg/kg of body weight; American Pharmaceutical Partners, Los Angeles, Calif.). The marrow was cultured at a density of 2 × 106 cells/ml in IMDM supplemented with 20% FBS, 10 ng of murine IL-3 (Biosource International)/ml, 2.5 ng of murine stem cell factor (Biosource International)/ml, 50 ng of human IL-6 (Biosource International)/ml, 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 0.01 mM 2-mercaptoethanol. Harvested cells were prestimulated in cytokine-containing medium for 12 h prior to transduction. Lentiviral supernatant was added to a final concentration of 2 × 107 IU/ml. Polybrene was added to a final concentration of 6 μg/ml. Recipient 8- to 12-week-old female C57Bl/6 mice were exposed to two doses of 600 cGy of gamma irradiation (128 cGy/min from a cesium-137 source) on two consecutive days (total of 1,200 cGy). One to two hours following the second dose of radiation, 2 × 106 to 4 × 106 transduced donor BM cells were injected into the tail vein of each recipient in 200 μl of phosphate-buffered saline containing 50 U of heparin/ml. Antibiotics were added to the drinking water for 3 weeks posttransplantation (200 μg of Maxim-200/ml; Phoenix Pharmaceuticals, St. Joseph, Mo.). The donor-recipient pairs were congenic at the CD45 locus, with donors expressing CD45.1 and recipients expressing CD45.2 isoforms of CD45, which are easily distinguishable by flow cytometry using commercially available antibodies (BD Biosciences, Palo Alto, Calif.).
RESULTS
Modifications made to the MLV vector increase its frequency of expression in F9 EC cells.
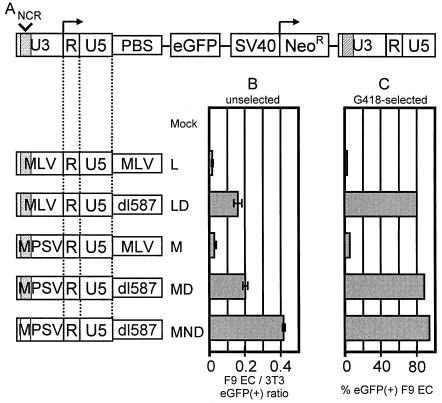
We made a series of retroviral vectors containing from one to three modifications to the MLV vector (5) to remove cis-acting elements reported to restrict expression of MLV in F9 EC cells. Figure 1A shows the arrangement of elements contained in the full-length MLV vector constructs (not drawn to scale). The vector expresses eGFP from the 5′LTR and neomycin resistance (Neor) from an internal SV40 promoter.
FIG. 1.
Modifications made to the MLV vector increase its frequency of expression in F9 EC cells. (A) MLV vector provirus diagram showing the arrangement of elements contained in the full-length MLV vector construct (not drawn to scale). The vector expresses eGFP from the 5′LTR and neomycin resistance from an internal SV40 promoter. Modifications made to the vector are depicted on the left side of the figure. (B) Frequency (± standard deviation) of eGFP expression in unselected F9 EC cells relative to 3T3 cells transduced in parallel. (C) Frequency of eGFP expression in F9 EC cell pools after selection in G418 for expression of Neor from a downstream SV40 promoter.
Figure 1B shows the contribution each of the three modifications, both alone and in combination, made to the frequency of expression in F9 EC cells. To control for differences in titer between vector supernatants, 3T3 cells, which are permissive for MLV expression, were transduced in parallel and under the same conditions as F9 EC cells. Values are expressed as the percentage of eGFP-positive F9 EC cells relative to the percentage of eGFP-positive 3T3 cells 7 days after transduction.
Figure 1B shows that the unmodified MLV vector, L, expresses rarely in F9 EC cells. Replacing the MLV PBS with the dl587 PBS in vector LD, thereby removing the RBS, alleviates repression in a substantial fraction of F9 EC cells. Replacing the MLV U3 with the MPSV U3 alone in vector M, while leaving the MLV PBS and therefore the RBS in place, alleviates very little repression in F9 EC cells. MPSV is a variant of the Moloney sarcoma virus that had greater expression in myeloid cells (34), and the MPSV LTR was isolated and shown to express better than the MLV LTR in EC cells (11, 39). Among the seven single-base differences between the MLV and the MPSV enhancer repeats, one of these single-base differences has been shown to introduce a functional Sp1 transcription factor binding site in the MPSV enhancer repeat that is not present in the MLV enhancer repeat. This Sp1 site accounts for much of the increase in activity of this enhancer in F9 EC cells (14, 36).
Replacing the MLV U3 with the MPSV U3 and replacing the MLV PBS with the dl587 PBS in combination in vector MD, thereby removing the RBS, alleviates repression in a substantial fraction of F9 EC. Removing the NCR from the U3 of MD to make the triply modified vector MND further alleviates repression in F9 EC cells. In each case where the MLV PBS, and therefore the RBS, is present expression is nearly completely repressed.
Figure 1C shows the contribution that each of these three modifications made to expression in F9 EC cells after selection in G418 for expression of Neor from a downstream internal SV40 promoter. Each of these F9 EC pools was selected with 0.75 μg of G418/ml for 10 days from a pool of cells that was transduced with a dilution of vector supernatant that transduced 3T3 cells in parallel to <10% eGFP positivity, so that most cells within these pools should contain only a single copy of the vector. In 0.75 μg of G418/ml, all nontransduced F9 EC cells in control wells were dead within 7 days. Following G418 selection, the cells were passaged in culture for an additional 2 weeks before analysis of eGFP expression by flow cytometry. Figure 1C demonstrates the same pattern of increased expression of eGFP from the modified vectors as in unselected F9 EC cells. Again, in each case, removing the RBS by replacing the MLV PBS with the dl587 PBS made the largest contribution to increased expression, while changing the enhancer or deleting the NCR made only more modest improvements. Vectors containing the MLV PBS, and therefore the RBS (L and M), expressed in less than 5% of the G418-selected F9 cells, whereas the vectors lacking the RBS (LD, MD, and MND) expressed in 80 to 90% of selected cells.
The RBS repressed expression from an internal MNDU3 promoter in a SIN lentiviral vector.
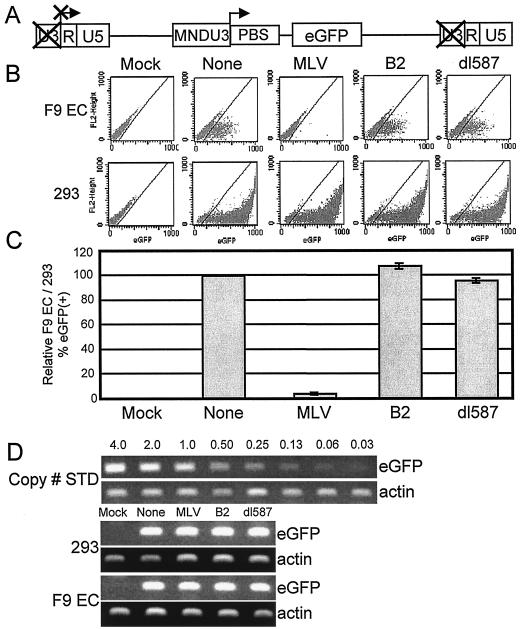
To determine if the RBS was capable of repressing expression outside of its normal retroviral genome context, a series of SIN lentiviral vectors were constructed having an internal MNDU3 promoter driving eGFP expression with one of the three PBS sequences under investigation inserted between the promoter and the eGFP transgene (Fig. 2A). SIN lentiviral vectors do not efficiently express transcript from their own LTR after reverse transcription, and thus eGFP expression reflects the activity of the internal promoter (30).
FIG. 2.
The RBS repressed expression in F9 EC cells when positioned downstream of an internal MNDU3 promoter expressing eGFP in a SIN lentiviral vector. (A) Lentiviral vector provirus diagram showing the arrangement of elements in the SIN lentiviral vector series having an internal MNDU3 promoter driving eGFP expression and one of the three PBS sequences inserted between the promoter and the eGFP transgene. (B) Representative flow cytometric analyses of F9 EC and 293 cells transduced in parallel with vectors containing the indicated PBS sequences. (C) Averages (± standard deviation) of flow cytometry data from 15 experiments performed as described for panel B. The percentage of eGFP-positive F9 EC cells was normalized to the percentage of eGFP-positive 293 cells in the same vector arm to control for small differences in vector titer, relative to the F9 EC/293 transduction ratio achieved with vector with no PBS, multiplied by 100. (D) Semiquantitative PCR demonstrates that gene transfer by the vector containing the MLV PBS occurred at relatively the same frequency as for vectors containing the B2, dl587, or no PBS. Mock-transduced cells served as negative controls for eGFP detection by PCR.
Each vector supernatant in this series was produced and the titer was determined simultaneously on 293 cells. Based on these titer values, F9 EC and 293 cells were transduced in parallel using identical conditions. After 6 to 10 days the cells were harvested and analyzed for eGFP expression by flow cytometry. Flow cytometric analyses from a typical experiment are shown in Fig. 2B. The averages of 15 experiments are plotted in Fig. 2C. In 293 cells, all four vectors were expressed at the same frequency. In contrast, the lentiviral vector containing the MLV PBS shows essentially no expression in the F9 EC cells, whereas the other vectors, with no PBS or with the B2 or dl587 PBS, all expressed at similar frequencies.
An alternative explanation for the results shown in Fig. 2B and C is that the vector containing the MLV PBS was not transferred or integrated at the same frequency as the other vectors in the series. To rule out this possibility, we performed semiquantitative PCR on genomic DNA from transduced 293 and F9 EC cells. Figure 2D shows that the vector containing the MLV PBS was transferred at a similar relative frequency as the vectors containing the B2 and dl587 PBS and the vector containing no PBS. Thus, the difference in expression by the vector carrying the MLV PBS is not due to poor gene transfer and therefore reflects repression of expression.
The RBS repressed expression from internal SV40, hUbiqC, hEF-1α, and mPGK promoters.
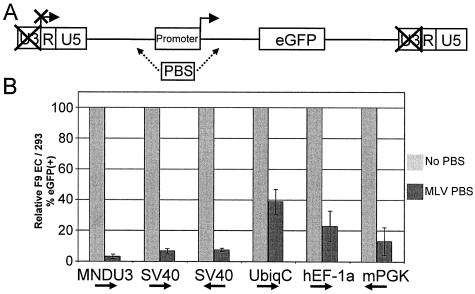
Previous studies have demonstrated that within the context of the retroviral genome, the RBS was able to repress transcription from two downstream internal heterologous viral promoters, SV40 and AdMLP (31, 35). To determine whether the RBS also repressed transcription from cellular promoters, we constructed SIN lentiviral vectors analogous to the vector series introduced in Fig. 2, but having either the SV40, hUbiqC, mPGK, or hEF-1a promoter driving eGFP expression, with and without the MLV PBS sequence inserted either immediately upstream or immediately downstream of the promoter.
F9 EC cells and 293 cells were transduced in parallel with the various vector constructs using identical conditions. Cells were passaged in culture for 6 to 10 days and then analyzed for eGFP expression by flow cytometry. Figure 3 shows the average results from two separate experiments. The data are presented as the percentage of eGFP-positive F9 EC cells normalized to the percentage of eGFP-positive 293 cells in the same vector arm, relative to the F9 EC/293 transduction ratio achieved by vector with no MLV PBS multiplied by 100. Figure 3 shows that the SV40 promoter and the three cellular promoters examined were substantially repressed by the RBS, although to varying degrees. The SV40 promoter was repressed greater than 90% whether the MLV PBS was placed upstream or downstream of the promoter. The hUbiqC promoter was repressed greater than 60%, the hEF-1a promoter was repressed greater than 75%, and the mPGK promoter was repressed greater than 80%. In comparison, the MNDU3 promoter was repressed greater than 95%. These results indicate that although the RBS is capable of substantially repressing heterologous cellular promoters in this context, the repression is not complete when compared to the repression of the MNDU3 promoter.
FIG. 3.
The RBS repressed expression in F9 EC cells by a lentiviral vector with an internal hUbiqC, hEF-1α, mPGK, SV40, or MNDU3 promoter. (A) Lentiviral vector provirus diagram showing arrangement of elements in the vector series having one of five promoters and either no PBS or an MLV PBS inserted either upstream (←) or downstream (→) of the promoter expressing eGFP. (B) Percentage of eGFP-positive F9 EC cells normalized to the percentage of eGFP-positive 293 cells in the MLV PBS vector arm, relative to the F9 EC/293 transduction ratio achieved with vector with no PBS, multiplied by 100 (± standard deviation).
Repression by the RBS is more pronounced in undifferentiated stem cells but is not stem cell specific.
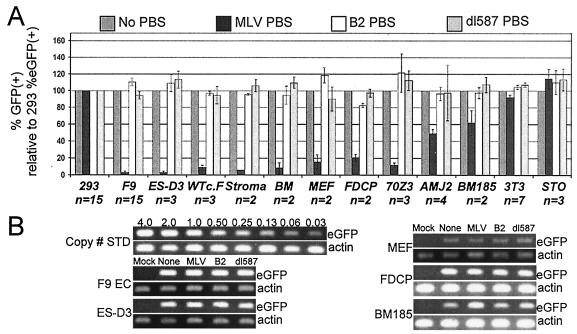
Previous studies have described the repressive activity of the RBS as stem cell specific on the basis that restriction is nearly complete in undifferentiated ES and EC cell lines but not in differentiated 3T3 fibroblasts (21, 25, 35, 48). One study demonstrated that substantial RBS-mediated repressive activity was present in the mouse hematopoietic progenitor cell line FDCP (3). Previous studies have not examined RBS activity in primary mouse cells. To determine the extent of RBS activity in cell types other than EC and ES cell lines, the series of SIN lentiviral vectors containing the different PBS sequences and the MNDU3 promoter driving eGFP expression (Fig. 2A) was used to screen for RBS repressive activity in primary mouse cells and mouse cell lines.
The data presented in Fig. 4 demonstrate that RBS-mediated repressive activity is present to varying degrees in mouse cells other than stem cells. Near-complete repression of the vector containing the MLV PBS was seen in F9 EC and ES-D3 cells, as previously reported, and also in WTc.F cells, an ES cell line generated from C57BL/6 mice (37). In addition, p300 and CBP knockout versions of WTc.F were tested, and similar repression was observed (data not shown).
FIG. 4.
Repression by the RBS is not stem cell specific. (A) eGFP expression in mouse cells from a SIN lentiviral vector with an internal MNDU3 promoter driving eGFP expression with one of the PBS sequences placed between the promoter and the eGFP transgene. Data are presented as the percentage of eGFP-positive target cells (indicated on the x axis) normalized to the percentage of eGFP-positive 293 cells in the same vector arm, relative to the target cell/293 transduction ratio achieved with vector with no PBS, multiplied by 100 (± standard deviation). (B) Semiquantitative PCR demonstrates that gene transfer by the vector containing the MLV PBS occurred at relatively the same frequency as that with vectors containing the B2, dl587, or no PBS.
The vector with the MLV PBS was heavily repressed in the three heterogeneous primary mouse cell populations we analyzed. Expression was repressed in greater than 90% of mouse bone marrow stromal cells (CD45−, adherent cells isolated from adult mouse bone marrow), greater than 90% of whole mouse BM cells isolated from adult mice, and greater than 80% of MEFs isolated from 13.5-day embryos of outbred CF-1 mice.
In addition, we observed that expression was repressed to varying degrees in four hematopoietic cell lines at different stages of differentiation: FDCP-Mix cl.A4 (hematopoietic progenitor), AMJ2-cll (macrophage), 70Z/3 (pre-B cell), and BM185 (pre-B cell). Both the 3T3 and STO cell lines showed no substantial repression when compared to the human 293 cell line. Semiquantitative PCR was performed on selected cell types to verify that differences in expression were not attributable to differences in the level of gene transfer (Fig. 4B). Although the panel of cells examined is limited, these data demonstrate that RBS-mediated repressive activity is not a stem-cell-specific, cell-line-specific, or mouse-strain-specific activity.
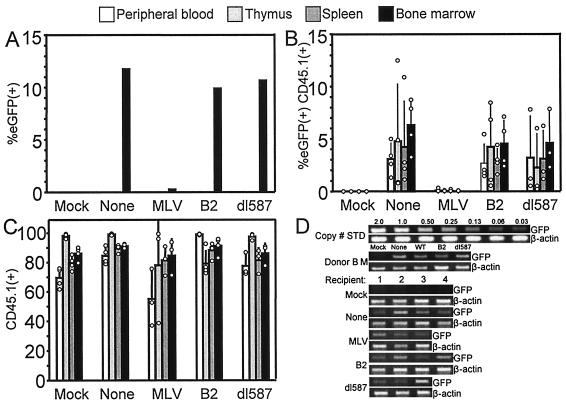
The RBS repressed expression in whole mouse bone marrow and its differentiated progeny after bone marrow transplant.
As described above (Fig. 4), expression was repressed in greater than 90% of whole mouse BM cells transduced with the SIN lentiviral vector containing the MLV PBS. To determine the fate of the progeny of the bone marrow progenitors in this cell population carrying repressed vector, we transplanted transduced murine BM cells into radiation-ablated mice. Figure 5A shows the percentage of cells expressing eGFP in donor mouse bone marrow transduced for transplantation but maintained in culture for 1 week. Approximately 10 to 12% of the cells expressed eGFP from each vector, except <1% expression was seen in cells transduced by the lentiviral vector containing the MLV PBS. After 10 weeks, the transplanted mice were sacrificed and hematopoietic cells were harvested from the bone marrow, peripheral blood, thymus, and spleen. Figure 5B shows the percentage of donor-derived CD45.1+ cells that expressed eGFP from each mouse in each hematopoietic tissue examined. Expression was nearly completely absent from the vector containing the MLV PBS in CD45+ cells from all four hematopoietic compartments examined. This stands in contrast to the B2 and dl587 PBS vector arms that yielded frequencies of eGFP expression comparable to the vector having no PBS. The absence of eGFP expression in these differentiated hematopoietic cell populations may not reflect active RBS-mediated repression but instead could be due to an inherited epigenetic mechanism of silencing, such as DNA methylation or chromatin condensation, that occurred secondary to RBS-mediated repression in a repopulating hematopoietic progenitor.
FIG. 5.
The RBS repressed expression in whole mouse bone marrow and its differentiated progeny after bone marrow transplant. (A) eGFP expression of donor bone marrow kept in culture for 7 days following transduction. (B) Mean eGFP expression (+ standard deviation) in CD45.1+ donor cells harvested from recipient mice 10 weeks after transplant with transduced bone marrow shown in panel A. Circles represent values of individual mice. (C) Percentage of donor CD45.1+ cells recovered from transplanted mice. (D) Semiquantitative PCR demonstrates that gene transfer by the vector containing the MLV PBS occurred at relatively the same frequency as the B2, dl587, and no PBS vectors in donor bone marrow and bone marrow harvested from each recipient mouse.
To rule out the possibility that the lack of eGFP expression observed in the cells transduced with the vector containing the MLV PBS was due to a difference in engraftment efficiency in this arm, harvested cells from all arms were stained with anti-CD45.1 antibody to differentiate donor-derived CD45.1 cells from recipient-derived CD45.2 cells. Figure 5C shows the percentage of harvested cells that were derived from donor cells according to CD45.1 antibody staining. The frequency of CD45.1+ cells recovered from blood, thymus, spleen, and bone marrow of mice transplanted with cells transduced with the MLV PBS-containing vector was in the same range as in the other three vector arms and in the mock-transduced arm. Thus, it can be concluded that the lack of expression seen in the MLV PBS vector arm was not due to inefficient engraftment of donor cells.
To rule out the possibility that the observed lack of eGFP expression seen in the cells transduced by the vector containing the MLV PBS was due to inefficient gene transfer into the donor bone marrow, semiquantitative PCR was performed on genomic DNA isolated from an aliquot of the transduced donor BM cells and bone marrow isolated from each recipient mouse. Donor bone marrow was transduced to the same relative efficiency (Fig. 5D) in each vector arm. All three bone marrow samples from mice transplanted with bone marrow transduced with the vector containing the MLV PBS were positive for eGFP sequence, and the signal intensity was not significantly different from that seen in the 11 other mice in the three other vector arms. Therefore, the lower expression seen from the vector with the MLV PBS was not due to inefficient gene transfer but rather reflects repression of expression.
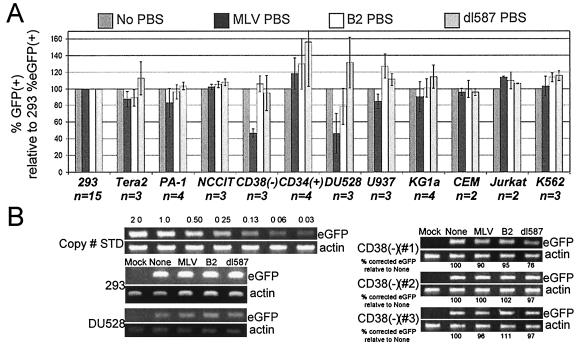
The RBS repressed expression in the human hematopoietic cell line DU.528 and primary human CD34+ CD38− cells isolated from umbilical cord blood.
Previous studies have not examined the effects of the RBS in human cells. The series of SIN lentiviral vectors containing the different PBS sequences and the MNDU3 promoter driving eGFP expression (Fig. 2A) was used to screen for RBS repressive activity in a panel of primary human cells and human cell lines. Of all the cell types we examined, only DU.528 and primary CD34+ CD38− cells demonstrated any repressive activity (Fig. 6). DU.528 is a hematopoietic progenitor cell line capable of generating progeny with characteristics of at least three hematopoietic lineages, in vitro: T-lymphoid, granulocytic/monocytic, and erythroid (21). CD34+ CD38− cells constitute about 3.5% of CD34+ cells present in umbilical cord blood and are enriched for primitive hematopoietic progenitor cells (17, 18). About 50% of the cells in each of these cell populations were repressed for expression of the MLV PBS-containing vector. In contrast, little repression was seen in transduced CD34+ cells, which are a heterogeneous population of cells that are more mature than the CD34+ CD38− cells. In contrast to mouse EC cells (Fig. 4), substantial repression was not observed in the three human EC cells we tested: Tera-2, PA-1, and NCCIT. In addition, we did not observe substantial repression in any of the five human hematopoietic cell lines we tested: U937 (myeloid), KG1a (myeloid), CEM (T cell), Jurkat (T cell), and K562 (myeloid). These findings suggest that expression of a factor(s) that binds to the RBS and represses expression occurs mainly in the more primitive and pluripotent human hematopoietic stem and progenitor cells.
FIG. 6.
The RBS repressed expression in the human hematopoietic cell line DU.528 and primary human CD34+ CD38− cells isolated from umbilical cord blood. (A) eGFP expression in human cells from a SIN lentiviral vector with an internal MNDU3 promoter driving eGFP expression with one of the PBS sequences placed between the promoter and the eGFP transgene. (B) Semiquantitative PCR demonstrates that gene transfer by the vector containing the MLV PBS occurred at relatively the same frequency as the B2, dl587, and no PBS vectors.
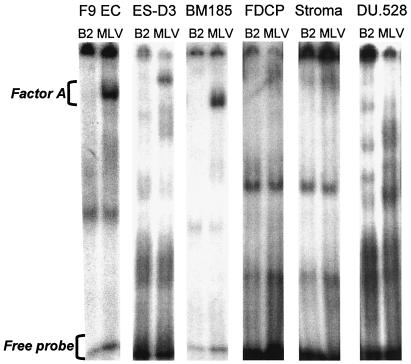
Binding factor A was present in mouse cells that have RBS-mediated repressive activity.
The differential binding of factor A, from EC cell nuclear extracts, to a MLV PBS probe, but not a B2 PBS probe, has been previously demonstrated by an EMSA (35). The EMSA gel pictures shown in Fig. 7 demonstrate that the factor A bandshift was present in nuclear extracts from primary mouse cells and mouse cell lines other than EC cells, in each cell type where the MLV PBS was repressive, including cell types where repression by this element was only partial (i.e., BM185 cells).
FIG. 7.
EMSA for differentially binding factor A. Differential binding of the factor A bandshift, to an MLV PBS probe but not a B2 PBS probe, was observed in nuclear extracts of several mouse cell lines and primary mouse cells, but not from human DU.528 cells.
We observed RBS-mediated repressive activity in the human cell line DU.528 and primary CD34+ CD38− cells in our expression assay. To determine if an orthologue of factor A protein was detectable in human cells that showed repression by the MLV PBS, we generated nuclear extracts from DU.528 cells for use in the EMSA. In two experiments, differential binding of any factor to the MLV PBS and B2 PBS probes was not observed. Due to the number of cells required to generate nuclear extracts, we have not attempted this assay with primary human CD34+ CD38− cells.
DISCUSSION
Repression by the RBS outside of the context of MLV genome.
Previous studies have demonstrated that the RBS is active in either orientation when positioned upstream of the MLV transcription start site or downstream of its normal position within an intron. Although the minimal length of the RBS sequence required for repressive activity was shown to be 18 bp, all previous studies have examined the RBS within the context of the MLV genome, leaving open the possibility that other sequences within the MLV genome might be required to cooperate with the RBS for full repressive activity. The only elements from MLV present in the lentiviral vectors we studied were the U3 region from MPSV (with the NCR deleted) and the MLV PBS. In this context, the internal MNDU3 promoter was fully repressed by the RBS, suggesting that additional elements outside of the PBS sequence and the MPSV U3 are not required for full PBS activity, although our study does not exclude the possibility that the RBS may cooperate with elements present in the lentiviral vector backbone. No effect was seen from the B2 or dl587 PBS.
Repression of the SV40, hUbiqC, mPGK, and hEF-1a promoters by the RBS.
A previous study demonstrated that within the context of the MLV genome, the RBS was able to repress transcription from a position upstream of two internal heterologous viral promoters, SV40 and AdMLP (35). In this study, we showed that in addition to the SV40 viral promoter, three cellular promoters were each substantially repressed in F9 EC cells when the RBS was present in the vector, although each of the heterologous promoters was repressed to differing degrees and none were as fully repressed as the MNDU3 promoter. Nevertheless, these experiments demonstrate that the mechanism of repression of the RBS is able to dominantly repress transcription directed by many different transcription factors.
RBS-mediated repressive activity in mouse cells is not stem cell specific.
Previous studies have referred to RBS-mediated repressive activity as an undifferentiated stem cell-specific activity. Most studies of the RBS have focused on repression of MLV in EC and ES cell lines compared to the lack of repression observed in 3T3 fibroblasts. Our study of RBS-mediated repression in mouse cells demonstrates that the repressive activity of the RBS, although most pronounced in ES and EC cell lines, is not restricted to undifferentiated stem cells. Our findings demonstrate that there is substantial RBS-mediated repressive activity in several mouse progenitor and differentiated hematopoietic cell lines as well as three heterogeneous primary cell populations isolated from embryonic and adult mice. The only mouse cells we tested that did not demonstrate repression of vector expression by the RBS were 3T3 and STO fibroblasts. Both 3T3 and STO cells were originally isolated from primary MEF cultures (29, 46), which in this study showed nearly the same degree of repression as ES and EC cell lines. The lack of RBS-mediated repressive activity in the 3T3 and STO cell lines might be due to either aberrant gene expression following transformation, a founder effect, or the loss of repressor expression during differentiation.
Vectors derived from MLV have been used for a variety of purposes where stable gene transfer and expression is necessary, including preclinical studies leading to gene therapy clinical trials. Often times, these initial expression studies are performed in mouse cell culture or animal models. This study demonstrated that several more-differentiated hematopoietic cell lines possess an incomplete but substantial degree of RBS-mediated repressive activity, compared to ES cells. Primary isolates of MEFs and mouse bone marrow stromal cells isolated from adult mice also demonstrated RBS-mediated repressive activity comparable to ES and EC cells. Although we have not examined fully differentiated cells outside the hematopoietic compartment, it is possible that many differentiated cell types in the mouse may possess substantial RBS-mediated repressive activity that may affect the outcome of gene expression studies that employ MLV-based vectors containing the MLV PBS. Several commercially available retroviral expression systems contain the MLV PBS and so may not be ideal tools for gene expression studies in mice.
RBS-mediated repressive activity in human cells.
Our data using the DU.528 human hematopoietic cell line and primary human CD34+ CD38− hematopoietic stem cells provide the first evidence that RBS-mediated repressive activity is present in human cells. The RBS is hypothesized to effect transcriptional repression by an undefined mechanism through an as-yet-unidentified trans-acting factor or factors identified by EMSA as binding factor A. If mouse binding factor A is the trans-acting factor that conveys repression through its interaction with the RBS, then human cells that repress expression from vectors containing the RBS may express an orthologue. We have been unable to demonstrate using an EMSA that nuclear extracts from the human cell line DU.528 contain an obvious differentially binding factor analogous to the differentially binding factor in the nuclear extracts of mouse cells that repress expression of vectors containing the RBS. This may be because the binding conditions used in the EMSA were optimized for use with nuclear extracts from mouse cells that contain high RBS-mediated activity, and the conditions have not been optimized for human nuclear extracts.
The observation that human cells possess RBS-mediated repressive activity has important implications for human gene therapy studies. Past and ongoing clinical trials have been performed using MLV-based vectors containing the MLV or B2 PBS. Because both the tRNA sequence and the PBS sequence are copied during reverse transcription, the double-stranded DNA product of reverse transcription of a vector having the B2 PBS contains a single base mismatch within the PBS region. The cellular machinery that repairs this single base mismatch in the double-stranded DNA provirus corrects one or the other base so that approximately 50% of the progeny provirus contain a B2 PBS and 50% revert to the MLV PBS sequence (4). Although our studies with human cell lines representing mature hematopoietic lineages did not detect any repressive activity of the RBS, it has not yet been determined whether RBS-mediated repressive activity is present in primary differentiated human hematopoietic lineages or in differentiated nonhematopoietic cell types. We were able to detect RBS-mediated repressive activity in human CD34+ CD38− cells, but it is not yet known whether this repression is maintained or reversed when these cells differentiate in vivo. If the RBS-mediated repressive activity we observed in human CD34+ CD38− cells is sustained through differentiation, as we observed in mouse bone marrow transplant experiments, then integrated proviruses containing the RBS in patients might be repressed.
At present, there are MLV-based vectors readily available with PBS replacements which do not contain the RBS (MND [5], MSCV [19], and HSC1 [33]). Our results suggest that it would be advisable to use the modified vectors instead of those containing the RBS for gene transfer and expression studies in mice and that there is the potential for the RBS present in some clinically approved retroviral vectors to have deleterious effects on vector expression in human gene therapy clinical trial patients.
Acknowledgments
These studies were supported by a grant from the National Cancer Institute, NIH 1P01 CA59318.
We thank Andrew Kung for providing WTc.F cells, Joanne Kurtzberg for providing DU.528 cells, Lez Fairbairn for providing FDCP-Mix cells, Hiroyuki Nakai for providing the hEF-1a promoter, and Carlos Lois for providing the UbiqC promoter.
REFERENCES
- 1.Akgün, E., M. Ziegler, and M. Grez. 1991. Determinants of retrovirus gene expression in embryonal carcinoma cells. J. Virol. 65:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barklis, E., R. C. Mulligan, and R. Jaenisch. 1986. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47:391-399. [DOI] [PubMed] [Google Scholar]
- 3.Baum, C., S. Hegewisch-Becker, H. Eckert, C. Stocking, and W. Ostertag. 1995. Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J. Virol. 69:7541-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berwin, B., and E. Barklis. 1993. Retrovirus-mediated insertion of expressed and non-expressed genes at identical chromosomal locations. Nucleic Acids Res. 21:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Challita, P. M., D. Skelton, A. el-Khoueiry, X. J. Yu, K. Weinberg, and D. B. Kohn. 1995. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J. Virol. 69:748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colicelli, J., and S. P. Goff. 1987. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J. Virol. 57:37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Auriol, L., W. K. Yang, J. Tobaly, F. Cavalieri, J. Peries, and R. Emanoil-Ravicovitch. 1981. Studies on the restriction of ecotropic murine retrovirus replication in mouse teratocarcinoma cells. J. Gen. Virol. 55:117-122. [DOI] [PubMed] [Google Scholar]
- 8.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feuer, G., M. Taketo, R. C. Hanecak, and H. Fan. 1989. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J. Virol. 63:2317-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan, J. R., A. M. Krieg, E. E. Max, and A. S. Khan. 1989. Negative control region at the 5′ end of murine leukemia virus long terminal repeats. Mol. Cell. Biol. 9:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franz, T., F. Hilberg, B. Seliger, C. Stocking, and W. Ostertag. 1986. Retroviral mutants efficiently expressed in embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 83:3292-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman, C. M., P. W. Rigby, and D. P. Lane. 1985. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell 42:519-526. [DOI] [PubMed] [Google Scholar]
- 13.Grez, M., E. Akgun, F. Hilberg, and W. Ostertag. 1990. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA 87:9202-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grez, M., M. Zornig, J. Nowock, and M. Ziegler. 1991. A single point mutation activates the Moloney murine leukemia virus long terminal repeat in embryonal stem cells. J. Virol. 65:4691-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas, D. L., S. S. Case, G. M. Crooks, and D. B. Kohn. 2000. Critical factors influencing stable transduction of human CD34+ cells with HIV-1-derived lentiviral vectors. Mol. Ther. 2:71-80. [DOI] [PubMed] [Google Scholar]
- 16.Halene, S., L. Wang, R. M. Cooper, D. C. Bockstoce, P. B. Robbins, and D. B. Kohn. 1999. Improved expression in hematopoietic and lymphoid cells in mice after transplantation of bone marrow transduced with a modified retroviral vector. Blood 94:3349-3357. [PMC free article] [PubMed] [Google Scholar]
- 17.Hao, Q. L., A. J. Shah, F. T. Thiemann, E. M. Smogorzewska, and G. M. Crooks. 1995. A functional comparison of CD34+ CD38− cells in cord blood and bone marrow. Blood 86:3745-3753. [PubMed] [Google Scholar]
- 18.Hao, Q. L., F. T. Thiemann, D. Petersen, E. M. Smogorzewska, and G. M. Crooks. 1996. Extended long-term culture reveals a highly quiescent and primitive human hematopoietic progenitor population. Blood 88:3306-3313. [PubMed] [Google Scholar]
- 19.Hawley, R. G., F. H. Lieu, A. Z. Fong, and T. S. Hawley. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1:136-138. [PubMed] [Google Scholar]
- 20.Hilberg, F., C. Stocking, W. Ostertag, and M. Grez. 1987. Functional analysis of a retroviral host-range mutant: altered long terminal repeat sequences allow expression in embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 84:5232-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempler, G., B. Freitag, B. Berwin, O. Nanassy, and E. Barklis. 1993. Characterization of the Moloney murine leukemia virus stem cell-specific repressor binding site. Virology 193:690-699. [DOI] [PubMed] [Google Scholar]
- 22.Kurtzberg, J., S. H. Bigner, and M. S. Hershfield. 1985. Establishment of the DU.528 human lymphohemopoietic stem cell line. J. Exp. Med. 162:1561-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linney, E., B. Davis, J. Overhauser, E. Chao, and H. Fan. 1984. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. Nature 308:470-472. [DOI] [PubMed] [Google Scholar]
- 24.Liu, M. L., B. L. Winther, and M. A. Kay. 1996. Pseudotransduction of hepatocytes by using concentrated pseudotyped vesicular stomatitis virus G glycoprotein (VSV-G)-Moloney murine leukemia virus-derived retrovirus vectors: comparison of VSV-G and amphotropic vectors for hepatic gene transfer. J. Virol. 70:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh, T. P., L. L. Sievert, and R. W. Scott. 1990. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol. Cell. Biol. 10:4045-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh, T. P., L. L. Sievert, and R. W. Scott. 1988. Negative regulation of retrovirus expression in embryonal carcinoma cells mediated by an intragenic domain. J. Virol. 62:4086-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loh, T. P., L. L. Sievert, and R. W. Scott. 1987. Proviral sequences that restrict retroviral expression in mouse embryonal carcinoma cells. Mol. Cell. Biol. 7:3775-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markowitz, D., S. Goff, and A. Bank. 1988. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J. Virol. 62:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, G. R., and M. J. Evans. 1975. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc. Natl. Acad. Sci. USA 72:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi, H., U. Blomer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modin, C., F. S. Pedersen, and M. Duch. 2000. Lack of shielding of primer binding site silencer-mediated repression of an internal promoter in a retrovirus vector by the putative insulators scs, BEAD-1, and HS4. J. Virol. 74:11697-11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldini, L., U. Blömer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 33.Osborne, C. S., P. Pasceri, R. Singal, T. Sukonnik, G. D. Ginder, and J. Ellis. 1999. Amelioration of retroviral vector silencing in locus control region beta-globin-transgenic mice and transduced F9 embryonic cells. J. Virol. 73:5490-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostertag, W., K. Vehmeyer, B. Fagg, I. B. Pragnell, W. Paetz, M. C. Le-Bousse, F. Smadja-Joffe, B. Klein, C. Jasmin, and H. Eisen. 1980. Myeloproliferative virus, a cloned murine sarcoma virus with spleen focus-forming properties in adult mice. J. Virol. 33:573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen, R., G. Kempler, and E. Barklis. 1991. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol. Cell. Biol. 11:1214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prince, V. E., and P. W. Rigby. 1991. Derivatives of Moloney murine sarcoma virus capable of being transcribed in embryonal carcinoma stem cells have gained a functional Sp1 binding site. J. Virol. 65:1803-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rebel, V., A. L. Kung, E. A. Tanner, H. Yang, R. T. Bronson, and D. M. Livingston. 2002. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. USA 99:14789-14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbins, P. B., X. J. Yu, D. M. Skelton, K. A. Pepper, R. M. Wasserman, L. Zhu, and D. B. Kohn. 1997. Increased probability of expression from modified retroviral vectors in embryonal stem cells and embryonal carcinoma cells. J. Virol. 71:9466-9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seliger, B., R. Kollek, C. Stocking, T. Franz, and W. Ostertag. 1986. Viral transfer, transcription, and rescue of a selectable myeloproliferative sarcoma virus in embryonal cell lines: expression of the mos oncogene. Mol. Cell. Biol. 6:286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speck, N. A., and D. Baltimore. 1987. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol. Cell. Biol. 7:1101-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, C. L., H. Stuhlmann, D. Jahner, and R. Jaenisch. 1982. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 79:4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stripecke, R., D. C. Skelton, T. Gruber, D. Afar, P. K. Pattengale, O. N. Witte, and D. B. Kohn. 1998. Immune response to Philadelphia chromosome-positive acute lymphoblastic leukemia induced by expression of CD80, interleukin 2, and granulocyte-macrophage colony-stimulating factor. Hum. Gene Ther. 9:2049-2062. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, T., T. Suda, J. Suda, T. Inoue, Y. Hirabayashi, H. Hirai, F. Takaku, and Y. Miura. 1991. Stimulatory effects of granulocyte colony-stimulating factor on colony-forming units-spleen (CFU-S) differentiation and pre-CFU-S proliferation in mice. Blood 77:2597-2602. [PubMed] [Google Scholar]
- 45.Teich, N. M., R. A. Weiss, G. R. Martin, and D. R. Lowy. 1977. Virus infection of murine teratocarcinoma stem cell lines. Cell 12:973-982. [DOI] [PubMed] [Google Scholar]
- 46.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsukiyama, T., O. Niwa, and K. Yokoro. 1989. Mechanism of suppression of the long terminal repeat of Moloney leukemia virus in mouse embryonal carcinoma cells. Mol. Cell. Biol. 9:4670-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamauchi, M., B. Freitag, C. Khan, B. Berwin, and E. Barklis. 1995. Stem cell factor binding to retrovirus primer binding site silencers. J. Virol. 69:1142-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]