Abstract
Multiple cell surface molecules (herpesvirus entry mediator [HVEM], nectin-1, nectin-2, and 3-O-sulfated heparan sulfate) can serve as entry receptors for herpes simplex virus type 1 (HSV-1) or HSV-2 and also as receptors for virus-induced cell fusion. Viral glycoprotein D (gD) is the ligand for these receptors. A previous study showed that HVEM makes contact with HSV-1 gD at regions within amino acids 7 to 15 and 24 to 32 at the N terminus of gD. In the present study, amino acid substitutions and deletions were introduced into the N termini of HSV-1 and HSV-2 gDs to determine the effects on interactions with all of the known human and mouse entry/fusion receptors, including mouse HVEM, for which data on HSV entry or cell fusion were not previously reported. A cell fusion assay was used to assess functional activity of the gD mutants with each entry/fusion receptor. Soluble gD:Fc hybrids carrying each mutation were tested for the ability to bind to cells expressing the entry/fusion receptors. We found that deletions overlapping either or both of the HVEM contact regions, in either HSV-1 or HSV-2 gD, severely reduced cell fusion and binding activity with all of the human and mouse receptors except nectin-1. Amino acid substitutions described previously for HSV-1 (L25P, Q27P, and Q27R) were individually introduced into HSV-2 gD and, for both serotypes, were found to be without effect on cell fusion and the binding activity for nectin-1. Each of these three substitutions in HSV-1 gD enhanced fusion with cells expressing human nectin-2 (ordinarily low for wild-type HSV-1 gD), but the same substitutions in HSV-2 gD were without effect on the already high level of cell fusion observed with the wild-type protein. The Q27P or Q27R substitution in either HSV-1 and HSV-2 gD, but not the L25P substitution, significantly reduced cell fusion and binding activity for both human and mouse HVEM. Each of the three substitutions in HSV-1 gD, as well as the deletions mentioned above, reduced fusion with cells bearing 3-O-sulfated heparan sulfate. Thus, the N terminus of HSV-1 or HSV-2 gD is not necessary for functional interactions with nectin-1 but is necessary for all of the other receptors tested here. The sequence of the N terminus determines whether nectin-2 or 3-O-sulfated heparan sulfate, as well as HVEM, can serve as entry/fusion receptors.
Human herpes simplex virus type 1 (HSV-1) and HSV-2, porcine pseudorabies virus (PRV), and bovine herpesvirus type 1 (BHV-1) are members of the alphaherpesvirus subfamily and have similar viral and cellular requirements for entry into cells (29). Initial attachment is usually mediated by interaction of virion envelope glycoprotein C (gC) and/or gB with cell surface heparan sulfate. The subsequent interaction of virion gD with one of its receptors triggers the penetration of virus, which occurs by fusion of the viral envelope with a cell membrane, and requires four viral glycoproteins, gB, gD, gH, and gL. HSV-induced cell fusion requires the same four viral glycoproteins—gB, gD, gH, and gL—as well as a gD receptor (28, 32). Transfection of cells with plasmids expressing gB, gD, gH, and gL is sufficient, in the absence of virus infection, to induce fusion with target cells expressing HSV entry receptors (2, 6, 21, 23).
Three classes of cell surface molecules can serve as entry/fusion receptors for HSV-1 or HSV-2 (29). These include herpesvirus entry mediator (HVEM), a member of the tumor necrosis factor receptor family; nectin-1 and nectin-2, two members of the immunoglobulin superfamily; and specific sites in heparan sulfate generated by the action of certain isoforms of 3-O-sulfotransferase (3-OST). Human and, as shown here, mouse forms of HVEM can mediate entry of both HSV-1 and HSV-2, but not PRV or BHV-1 (20). Human and mouse forms of nectin-1 can serve as entry receptors for HSV-1, HSV-2, PRV, and BHV-1 (4, 12, 25), whereas human and mouse forms of nectin-2 have limited entry activity. Human nectin-2 can mediate the entry of HSV-2 and PRV but has considerably less activity with HSV-1, unless there is an amino acid substitution at position 25 or 27 of gD (16, 33). Mouse nectin-2 can mediate the entry of PRV but not BHV-1 or HSV strains (27). Specific sites in heparan sulfate generated by human or mouse 3-O-sulfotranferases, 3-OST-3A or 3-OST-3B, can serve as entry receptors for HSV-1 (26).
X-ray structures of a truncated form of HSV-1 gD, alone and in complex with truncated HVEM, were recently determined (3). A portion of the gD ectodomain forms an immunoglobulin fold in both structures, with N-terminal and C-terminal extensions. The N-terminal extension forms a hairpin in the complex with HVEM but is disordered in the crystal of gD alone. The interface between gD and HVEM involves only two short segments of gD (amino acids 7 to 15 and 24 to 32) in the N-terminal hairpin. These observations suggest that the N terminus of gD is conformationally flexible and that gD may have different conformations upon binding different receptors. A single amino acid substitution at position 27 (Q27P or Q27R) in HSV-1 gD prevents binding to HVEM and entry via HVEM (20, 34), a finding consistent with the contact sites identified in the X-ray structure, but has no negative effect on binding to nectin-1 or entry via nectin-1 (12, 14). Intriguingly, these substitutions confer the ability to use nectin-2 as an entry/fusion receptor (23, 33). The inability of PRV and BHV-1 to use HVEM as an entry/fusion receptor can be explained by significant divergence of PRV and BHV-1 gD sequences from HSV-1 and HSV-2 gD in the N-terminal domain that has the contact sites for HVEM. However, there must be structural conservation elsewhere in the proteins to explain the fact that all four viruses can use nectin-1 as an entry/fusion receptor. These observations imply that the interfaces of gD with different receptors are different.
In the present study, we investigated whether the N-terminal domains of HSV-1 and HSV-2 gD were critical for functional interactions with each of the known human and mouse entry/fusion receptors. We identified amino acid substitutions or deletions that significantly altered or reduced physical and functional interactions with HVEM, nectin-2 and 3-O-sulfated heparan sulfate but were without effect on interactions with nectin-1.
MATERIALS AND METHODS
Cells.
Chinese hamster ovary (CHO-K1) cells were provided by J. Esko (University of California, San Diego). CHO-IEβ8 cells, obtained by the stable transfection of CHO-K1 cells with pMLP01, express the Escherichia coli lacZ gene under control of the HSV-1 ICP4 promoter (20). CHO-K1 cells stably expressing human HVEM, nectin-1, or nectin-2 were originally designated CHO-HveA35 (20), CHO-Prr1 (12), or CHO-Prr2 (33), respectively. Here they are called CHO-HVEM, CHO-nectin-1, and CHO-nectin-2, respectively. All cells were grown in Ham F12 medium with 10% fetal bovine serum.
Plasmids.
Plasmids encoding HSV entry receptors included pcDNA3-based constructs as follows: pBEC10 expressing human HVEM (20), pBG38 expressing human nectin-1 (12), human 3-OST-3B whose expression generates HSV-1 entry receptors (26), and pDS106 expressing mouse HVEM and pCR13 expressing mouse nectin-1.
HSV-1 and HSV-2 gD mutants were generated by using the QuickChange site-directed mutagenesis kit (Strategene). HSV-1 gD mutants were generated by using pAZT3 (36), which contains the open reading frame (ORF) of HSV-1(KOS) gD in pUC19 (nucleotides 138419 to 139603 cloned between the EcoRI and SphI restriction sites). HSV-2 gD mutants were generated by using pMY1, constructed by subcloning the EcoRI-KpnI fragment, containing the HSV-2(333) ORF, from pAZD2 (36) into pUC19. pMY1 contains nucleotides 141016 to 142197 cloned between the EcoRI and KpnI sites in pUC19. Nucleotide numbers are from the published sequences of the HSV-1 (17) genome (NC_001806) and the HSV-2(HG52) genome (NC_001798).
Plasmids expressing the various forms of gD and mutants were generated by subcloning the EcoRI-SphI fragment (for HSV-1 gD) or the EcoRI-KpnI fragment (for HSV-2 gD) from the pUC19-based plasmids into pCAGGS/MCS (13). Plasmids expressing soluble gD:Fc hybrids were constructed to contain the first 320 codons of HSV-1(KOS) gD (excluding the signal sequence) or the first 319 codons of HSV-2(333) gD (wild-type or mutant forms) fused to the C terminus of the rabbit immunoglobulin G (IgG) heavy chain. The appropriate region of each gD gene was amplified by PCR with primers that added an upstream HindIII site and a downstream KpnI site and the pUC19-based plasmids as templates. The PCR products were cloned into pDM19, which is a pcDNA3-based plasmid and contains the cytomegalovirus promoter upstream of a HindIII site and 231 codons of the rabbit IgG heavy-chain gene downstream of a KpnI site. For HSV-1 gD:Fc plasmids, the primer pairs consisted of 5′-GGACCAAAGCTTGAATTCCTTTTGTGTGGT-3′ and 5′-TCGAGCTCGGTACCCGGCGATCAGGCCCAT-3′. For HSV-2 gD:Fc plasmids, the primer pairs consisted of 5′-GGACCAAAGCTTGAATTCGTGTGCATCGCG-3′ and 5′-TCGAGCTCGGTACCCGATGATCAGGCCCGG-3′. The introduced restriction endonuclease recognition sequences are shown in boldface. All plasmids used in the present study were verified by DNA sequencing through the gD genes and are listed in Table 1.
TABLE 1.
Summary of gD mutations and plasmids encoding the wild-type and mutant genes
| Mutation | Plasmida associated with:
|
|||||
|---|---|---|---|---|---|---|
| HSV-1(KOS) gD
|
HSV-2(333) gD
|
|||||
| pUC19 | pCAGGS | pcDNA3 | pUC19 | pCAGGS | pcDNA3 | |
| None | AZT3 | MY75 | MY80 | MY1 | AZD2 | MY12 |
| L25P | MY70 | MY76 | MY81 | MY2 | MY7 | MY13 |
| Q27P | MY71 | MY77 | MY82 | MY3 | MY8 | MY14 |
| Q27R | MY72 | MY78 | MY83 | MY4 | MY9 | MY15 |
| L25P/Q27P | ND | ND | ND | MY5 | MY10 | MY16 |
| L25P/Q27R | MY73 | ND | ND | MY6 | MY11 | MY17 |
| L25P/Q27R/T230I | MY74 | MY79 | MY84 | MY18 | MY19 | MY20 |
| Δ7-21 | MY85 | MY89 | MY93 | MY24 | MY29 | MY34 |
| Δ7-15 | MY86 | MY90 | MY94 | MY25 | MY30 | MY35 |
| Δ24-32 | MY87 | MY91 | MY95 | MY26 | MY31 | MY36 |
| Δ7-15/24-32 | MY88 | MY92 | MY96 | MY27 | MY32 | MY37 |
| Δ7-32 | ND | ND | ND | MY28 | MY33 | MY38 |
pUC19, ORF indicated cloned into pUC19 for site-directed mutagenesis; pCAGGS, ORF in the eukaryotic expression plasmid pCAGGS; pcDNA3, hybrid ORF containing the gD ectodomain fused to the Fc domain of rabbit IgG in the eukaryotic expression plasmid pcDNA3; ND, not done.
Viruses and viral entry assay.
HSV-1(F), HSV-1(U10), HSV-1(KOS), HSV-1(KOS)Rid1, HSV-1(KOS)tk12, and HSV-1(KOS)Rid1/tk12 were described previously (1, 7, 20, 33). Entry assays were based on quantitation of β-galactosidase expressed from the viral genome or expressed in CHO-IEβ8 cells as a result of viral entry. CHO-IEβ8 cells are induced to express β-galactosidase upon viral entry and delivery of the HSV-1 transactivator VP16 into cells (20). At low-input multiplicities of infection, the β-galactosidase activity detected from the reporter gene in virus or cell is proportional to the number of cells infected; when all cells are infected, the enzyme activity reaches a plateau (L. Kwon and P. G. Spear, unpublished data). Parental CHO-K1, CHO-HVEM, or CHO-nectin-1 cells were plated in 96-well plates (ca. 5 × 104 cells per well). The next day, the cells were incubated with serial dilutions of the β-galactosidase reporter viruses, HSV-1(KOS)tk12 or HSV-1(KOS)Rid1/tk12, at 37°C for 6 h. Alternatively, CHO-IEβ8 cells were transfected with plasmids expressing human HVEM (pBEC10), human nectin-1 (pBG38), mouse HVEM (pDS106), or mouse nectin-1 (pCR13) for 6 h and then plated in 96-well plates overnight. Cells were incubated with serial dilutions of HSV-1(KOS), HSV-1(KOS)Rid1, HSV-1(F), or HSV-1(U10) at 37°C for 6 h. The cells were then washed, permeabilized, and incubated with the β-galactosidase substrate, O-nitrophenyl-β-d-galactopyranoside (ONPG; Sigma). The reaction was monitored at 410 nm in a Spectra Max 250 enzyme-linked immunosorbent assay (ELISA) reader.
Preparation of soluble gD:Fc fusion proteins.
Soluble gD:Fc fusion proteins were produced as described previously (10, 11) with modification. Briefly subconfluent CHO-K1 cells in six-well plates were transfected with gD:Fc-expressing plasmids for 6 h by using Lipofectamine (Invitrogen) in serum-free medium and changed into medium containing 10% fetal bovine serum overnight. Cells were then washed and incubated with serum-free medium, and supernatants were collected 48 h later. The culture supernatants were clarified by low-speed centrifugation, and the concentrations of gD:Fc proteins were determined by ELISA by using an anti-rabbit Fc detection system and rabbit IgG for the standard curve.
Receptor-binding assay.
The receptor-binding assay was described previously (10, 11). CHO cells expressing HSV-1 or HSV-2 entry/fusion receptors were grown on coverslips and incubated with gD:Fc at 10 ng/ml and 37°C for 30 min. Cells were washed, fixed with methanol at −20°C for 5 min, and incubated with Alexa 488-conjugated goat anti-rabbit IgG (Molecular Probes). Binding was visualized by confocal microscopy. Alternatively, cell ELISA (CELISA) was performed. Cells grown on 96-well plates were incubated with serial dilutions of gD:Fc for 30 to 60 min at 37°C. Cells were washed and fixed with 2% formaldehyde and 0.2% glutaraldehyde for 10 min. The cells were then sequentially incubated with biotinylated anti-rabbit IgG (Sigma), Amdex streptavidin-conjugated horseradish peroxidase (HRP; Amersham), and HRP substrates (BioFx Lab). Binding was monitored at 370 nm.
Assay for cell surface expression of gD.
CELISA was carried out to detect cell surface expression of gD, as described previously (10, 11). Briefly, subconfluent CHO-K1 cells in six-well plates were transfected with pCAGGS-based plasmids expressing various forms of gD, along with gB, gH, and gL for 6 h and replated on 96-well plates overnight. The cells were then incubated with polyclonal anti-gD rabbit serum (R7) at 37°C for 30 min, fixed, and sequentially incubated with biotinylated anti-rabbit IgG, streptavidin-conjugated HRP, and HRP substrates. Binding was monitored at 370 nm.
Cell fusion assay.
Details of the cell fusion assay were described previously (23). Effector (CHO-K1) cells were transfected with a mixture of plasmids expressing HSV glycoproteins (gB, gD, gH, and gL) and T7 RNA polymerase (pCAGT7). Plasmids expressing the HSV-1(KOS) glycoproteins were described elsewhere (23): pPEP98 (gB), pPEP100 (gH), and pPEP101 (gL). Plasmids expressing the HSV-2(333) glycoproteins have also been described (36): pAZB2 (gB), pAZD2 (gD), pAZH2 (gH), and pAZL2 (gL). The target cells were CHO-HVEM, CHO-nectin-1, CHO-nectin-2, or CHO-K1 cells transiently transfected with human 3-OST-3B, mouse HVEM or mouse nectin-1. All target cells were transfected or cotransfected with a plasmid expressing luciferase under control of the T7 promoter (pT7EMLuc). Effector and target cells were subsequently detached and mixed in a 1:1 ratio and replated for 18 h. Luciferase activity was quantitated by a luciferase reporter assay system (Promega) by using a TD-20/20 luminometer (Turner Designs).
Immunofluorescence and microscopy.
Indirect immunofluorescence was carried out as described previously (35). The primary antibodies used were mouse monoclonal anti-nectin-1 antibody R1.302 (17) and rabbit polyclonal anti-HVEM antibody R11874 (20). The secondary antibodies used were Alexa 488-conjuated goat anti-mouse IgG and Alexa 488-labeled anti-rabbit IgG. Immunofluroscence observations were made with a Zeiss LSM 510 confocal microscope equipped with a ×100, 1.4 numerical aperture oil immersion objective lens. Orthogonal sections were made in a z stack of 30 images (0.6-μm interval) according to the LSM 510 operating manual.
RESULTS
Effect of substitutions at position 25 in HSV-1 gD on HVEM-mediated entry.
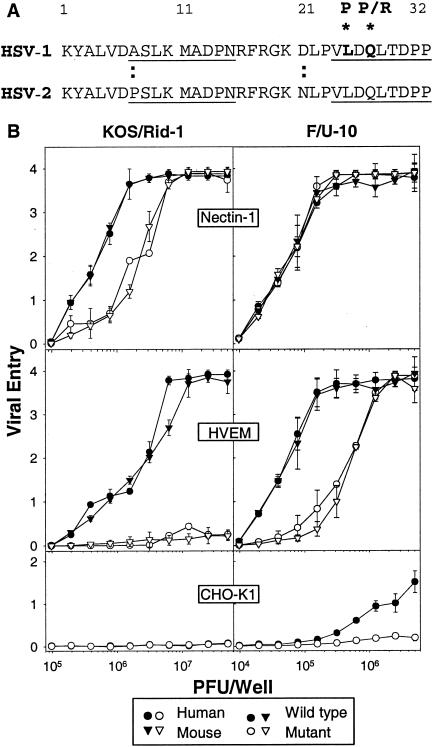
Viral mutants with amino acid substitutions in the N-terminal region of HSV-1 gD have been described previously. As mentioned above, Rid mutants of HSV-1(KOS) have substitutions at position 27 (Q27P or Q27R) that significantly reduce their ability to use human HVEM as an entry receptor and enhance their ability to use human nectin-2 (7, 20, 33). The U10 mutant of HSV-1(F) has a substitution at position 25 (L25P) and has enhanced ability to use human nectin-2 as an entry receptor (16). Results have not previously been reported for effects of the U10 mutation on viral entry via human HVEM or for effects of any of these mutations on entry via mouse HVEM. Therefore, we inoculated CHO cells expressing human or mouse HVEM with the Rid and U10 mutants and HSV-1 parental strains and assessed the efficiency of viral entry by quantitation of β-galactosidase, which was expressed from a reporter gene under control of the immediate-early HSV-1 ICP4 promoter, either in the viral genome or in the cell. Because the inoculated cells were permeabilized for β-galactosidase quantitation at 6 h after the addition of virus, viruses that failed to enter the cells or entered at a very slow rate would score as impaired for entry. As positive controls, CHO cells expressing human or mouse nectin-1 were also inoculated. As negative controls, CHO-K1 cells were inoculated.
The results presented in Fig. 1 show, first, that the human and mouse forms of nectin-1 or HVEM were similar in their ability, or failure, to mediate entry of the mutant and wild-type forms of HSV-1. Second, whereas the Rid mutation (Q27P) eliminated the ability of the mutant to infect cells expressing human or mouse HVEM, the U10 mutation (L25P) had a lesser effect but reduced the efficiency of infection for cells expressing either form of HVEM. At least 10 times more of the U10 mutant than the wild-type parental strain F was required to achieve similar levels of entry. Third, the Rid mutant appeared to infect nectin-1-expressing cells less efficiently than did HSV-1(KOS), whereas there was little difference between the U10 mutant and HSV-1(F) in this regard. Finally, some differences between the parental strains are evident. Lower input multiplicities of HSV-1(F) than HSV-1(KOS) achieved similar levels of viral entry on the nectin-1-expressing and HVEM-expressing cells. Also, infection of the control CHO cells could be detected at high input concentrations of HSV-1(F) but not at all for HSV-1(KOS). This latter difference with respect to CHO cells was reported previously (24) and indicates that there is a weak endogenous strain-dependent CHO entry receptor for HSV-1. It should be noted that the amino acid sequences of the gD genes of these two strains are identical in the ectodomain and differ by only two amino acids in the membrane-proximal region of the cytoplasmic tail (8). The explanation for the observed differences between HSV-1(KOS) and HSV-1(F) in the efficiency of viral entry or early events leading to gene expression could lie in these differences or in differences in other genes.
FIG. 1.
(A) Amino acid sequence alignments for the N termini of HSV-1(KOS) and HSV-2(333) gDs. Two segments (amino acids 7 to 15 and 24 to 32, underlined) in HSV-1 gD were shown to be engaged in the interface with HVEM (3). Amino acid substitutions in HSV-1 gD at positions 25 (L25P) and 27 (Q27P or Q27R) have previously been described (1, 7), as discussed in the text. Wild-type forms of HSV-1 and HSV-2 gDs differ at two positions within this region, as marked by the colons. (B) Effects of the Q27P mutation in HSV-1(KOS) and the L25P mutation in HSV-1(F) on entry of virus into CHO cells expressing human or mouse HVEM. In the left panels, CHO-nectin-1 cells, CHO-HVEM cells, CHO-K1 cells transfected with plasmids expressing mouse nectin-1 or mouse HVEM, and control CHO-K1 cells were inoculated with serial dilutions of the β-galactosidase-expressing viruses, HSV-1(KOS)tk12 or HSV-1(KOS)Rid1/tk12. In the right panels, CHO-IEβ8 cells (which can be induced to express β-galactosidase by infection with viruses carrying HSV VP16) were transfected with plasmids expressing the receptors as indicated or empty vector and were then inoculated with serial dilutions of HSV-1(F) or HSV-1(F)U10. After 6 h, the cells were lysed for the quantitation of β-galactosidase activity as a measure of viral entry. Values were optical densities at 410 nm (OD410) of the ONPG reaction product; the means of triplicate determinations with standard deviations for one representative experiment are shown. Similar results were obtained in three additional experiments.
We conclude that mouse and human HVEM are indistinguishable in their entry activities, at least for the viral strains analyzed in Fig. 1, despite our previous mentions of unpublished and preliminary findings that HSV-1(KOS)Rid mutants could infect cells via mouse HVEM (25, 29). We also conclude that the Q27P substitution has a more profound effect on entry via HVEM than does the L25P substitution, although the fact that the mutant viral strains are not isogenic could have influenced the results.
In the present study, we have assessed the effects of both of these mutations, as well as others, in the HSV-1(KOS) and HSV-2(333) genetic backgrounds. Mutations in the N-terminal region of HSV-2 gD have not yet been described. Amino acid substitutions and deletions were introduced into the HSV-1 and HSV-2 gD genes. The altered forms of gD were expressed as soluble hybrid proteins in which the ectodomain of gD was fused to the Fc region of rabbit IgG. The hybrids were used to assess the effects of the mutations on ability of gD to bind to cell surface nectin-1 and HVEM. Binding to the other receptors studied here, nectin-2 and 3-O-sulfated heparan sulfate, could not be assessed in this way, probably because of lower affinities of binding (16, 19). The full-length mutant forms of gD were also expressed in CHO cells, along with gB, gH, and gL, to quantitate effects of the mutations on ability of the glycoprotein-expressing cells (effector cells) to induce fusion with cells expressing the various receptors (target cells). This cell fusion assay provided a convenient way to test the functional activity of multiple gD mutants and yielded results likely to be predictive of the effects of the mutations on viral entry.
Effects of mutations in the N termini of HSV-1 and HSV-2 gDs on binding of gD:Fcs to nectin-1 and HVEM.
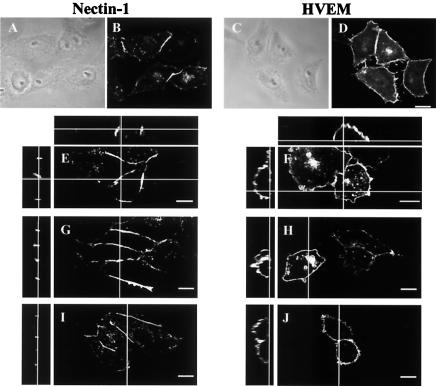
Binding of the mutant forms of gD:Fcs to entry receptors was assessed qualitatively by immunofluorescence and quantitatively by measuring the amounts bound to intact cells in monolayer (CELISA). Figure 2 illustrates differences in the cellular localization of human HVEM and nectin-1 in stably expressing CHO cells, as determined by antibody binding and by the binding of wild-type forms of HSV-1 or HSV-2 gD:Fcs. As previously shown (30, 31, 35), antibodies specific for nectin-1 localized to regions where two cells come in contact (Fig. 2B and E), reflecting the ability of this cell adhesion molecule to engage in homophilic trans-interactions between cells. Nectin-2 exhibits the same cellular localization (30, 31). Antibodies to HVEM, on the other hand, were distributed all over the cell surface (Fig. 2D and F), at least all over the surface not in contact with the substrate. The other panels of Fig. 2 show that HSV-1 gD:Fc (panels G and H) and HSV-2 gD:Fc (panels I and J) bound to the cells in patterns similar to those of the anti-nectin-1 and anti-HVEM antibodies. CHO cells expressing mouse nectin-1 or HVEM bound both HSV-1 and HSV-2 gD:Fcs in patterns indistinguishable from those shown in Fig. 2 for the human receptors (data not shown). Control CHO cells devoid of exogenous receptors failed to bind the anti-nectin-1 antibodies, anti-HVEM antibodies or gD:Fcs (data not shown). We have noted previously the colocalization of anti-nectin-1 antibodies and gD:Fcs at cell contact sites and the enhancement of gD:Fc binding when cell adhesions are disrupted, indicating that homophilic trans-interactions of nectin-1 can reduce, but not prevent, gD:Fc binding (35).
FIG. 2.
Localization of human nectin-1 or HVEM in CHO-nectin-1 or CHO-HVEM cells by immunofluorescence. The cells on coverslips were fixed and stained by incubation with an anti-nectin-1 monoclonal antibody (B and E) or an anti-HVEM rabbit serum (D and F), followed the addition of by Alexa 488-labeled secondary antibodies. Phase-contrast images paired with panels B and D are shown in panels A and C, respectively. Replicate cells were incubated with HSV-1 gD:Fc (G and H) or HSV-2 gD:Fc (I and J), followed by fixation and incubation with Alexa 488-labeled anti-Fc antibodies. Vertical lines across the larger panels in E to J indicate the plane of the z sections shown to the left. Horizontal lines indicate the plane of the z sections shown at the top for the antibody staining only. Bar, 10 μm.
In the first 37 amino acids of HSV-1 gD, which includes the N-terminal hairpin present in the gD-HVEM complex, there are only two amino acid differences from HSV-2 gD (A7P and D21N), as shown in Fig. 1A. Thus, the amino acid substitutions previously characterized in HSV-1 could also be investigated in HSV-2. The mutations introduced into HSV-1(KOS) and HSV-2(333) gDs and gD:Fcs are listed in Table 1, along with the names of the relevant plasmids. These mutations included the U10 (L25P) and Rid (Q27P and Q27R) substitutions and combinations of both. They also included a triple substitution (L25P/Q27R/T230I) found in HSV-1(ANG), which results in a phenotype similar to that of the Rid mutants (7, 20). In addition, various deletions encompassing the regions in gD that make contact with HVEM were made. The wild-type and mutant forms of the gD:Fcs were tested by immunofluorescence for their ability to bind to cells expressing nectin-1 or HVEM. The results (not shown) can be summarized briefly. All of the HSV-1 and HSV-2 mutant gD:Fcs bound to cells expressing either the human or mouse forms of nectin-1, in a pattern similar to that observed for the wild-type gD:Fcs (Fig. 2). On the other hand, all of the mutant gD:Fcs, except for L25P, failed to bind to cells expressing either the human or mouse forms of HVEM. Both the HSV-1 and HSV-2 L25P mutant forms of gD:Fc bound to cells expressing either HVEM or nectin-1 in patterns indistinguishable from wild-type.
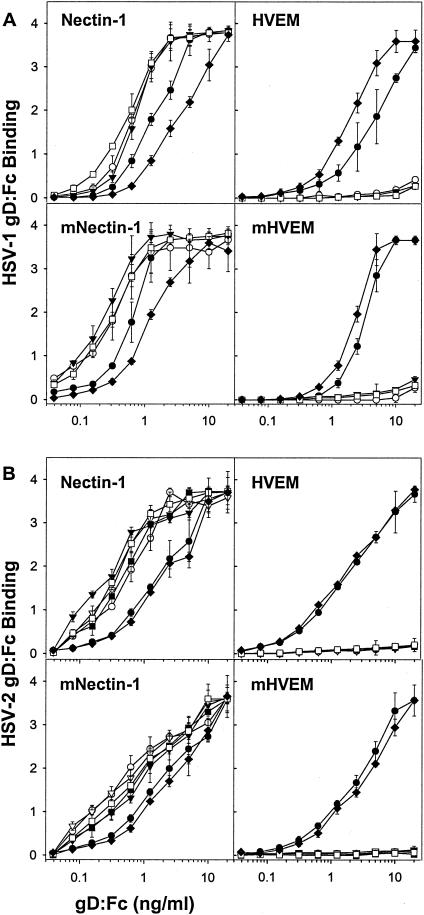
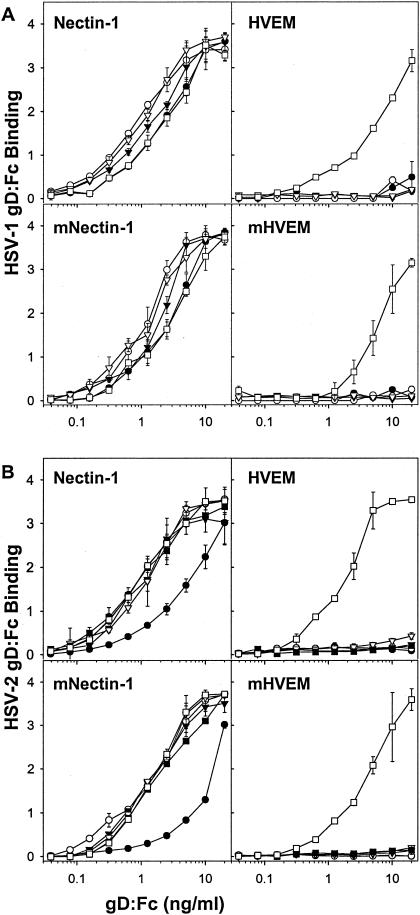
To quantify the binding to nectin-1 and HVEM, serial dilutions of the wild-type and mutant gD:Fcs were incubated with cells expressing human or mouse nectin-1 or HVEM in the CELISA assay. Figure 3 shows the results obtained with the HSV-1 and HSV-2 substitution mutants, and Fig. 4 shows the results obtained with the deletion mutants. All of the mutants bound to human or mouse nectin-1 with certain quantitative differences observed. Specifically, all of the HSV-1 substitution mutants bound more efficiently than wild-type gD:Fc to nectin-1, human or mouse, and all of the HSV-2 substitution mutants, except for L25P, also bound with greater efficiency (based on equivalent binding at input concentrations 1/3 to 1/10 that of wild-type gD:Fc). The HSV-2 L25P mutant was indistinguishable from wild-type HSV-2 gD:Fc in its binding to nectin-1. All of the deletion mutants bound to nectin-1 as efficiently as did wild-type forms of the gD:Fcs, except for HSV-2 deletion 7-21, which required up to 10 times higher concentrations for binding equivalent to that of wild-type gD:Fc. On the other hand, none of the substitution or deletion mutants bound to either form of HVEM, except for HSV-1 and HSV-2 L25P, a finding consistent with the immunofluorescence results (not shown).
FIG. 3.
Quantitation of the binding of wild-type and mutant (substitution mutant) gD:Fcs to CHO cells expressing human and mouse forms of nectin-1 and HVEM. CHO-nectin-1 cells, CHO-HVEM cells, or CHO-K1 cells transfected with mouse nectin-1 (mNectin-1) or mouse HVEM (mHVEM) were incubated with serial dilutions of the wild-type or substitution mutant forms of gD:Fc indicated (HSV-1 [A] and HSV-2 [B]). The cells were then washed, fixed, and incubated with an anti-Fc detection system. The horse radish peroxidase reaction product was quantitated by determining the OD370. The values presented are means and standard deviations of triplicate determinations. The results presented for this experiment are representative of two other experiments. Symbols: •, L25P; ○, Q27P; ▾, Q27R; ▿, L25P/Q27P; ▪, L25P/Q27R; □, L25P/Q27R/T230I; ♦, wild type.
FIG. 4.
Quantitation of the binding of wild-type and mutant (deletion mutant) gD:Fcs to CHO cells expressing human and mouse forms of nectin-1 and HVEM. CHO-nectin-1 cells, CHO-HVEM cells or CHO-K1 cells transfected with mouse nectin-1 (mNectin-1) or mouse HVEM (mHVEM) were incubated with serial dilutions of the wild-type or deletion mutant forms of gD:Fc indicated (HSV-1 [A] and HSV-2 [B]). The cells were then washed, fixed, and incubated with an anti-Fc detection system. The HRP reaction product was quantitated by determining the OD370. The values presented are means and standard deviations of triplicate determinations. The results presented for this experiment are representative of two other experiments. Symbols: •, Del 7-21; ○, Del 7-15; ▾, Del 24-32; ▿, Del 7-15/24-32; ▪, Del 7-32; □, wild type.
Effects of mutations in the N termini of HSV-1 and HSV-2 gDs on cell fusion.
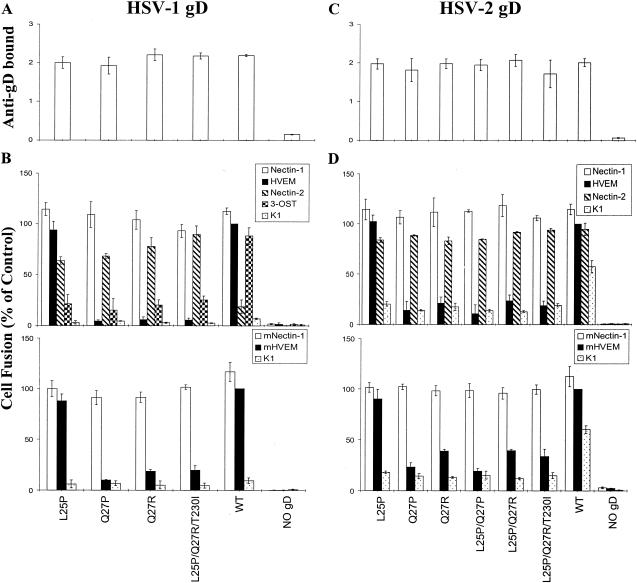
It was previously shown that cells expressing the Q27P mutant of HSV-1 gD, along with wild-type forms of HSV-1 gB, gH, and gL, had significantly reduced ability to induce fusion with target cells expressing HVEM, enhanced ability to induce fusion with cells expressing nectin-2, and unchanged ability to induce fusion with cells expressing nectin-1 (23), a finding consistent with the entry phenotypes of Rid mutant viruses. None of the other gD mutations described here have been tested previously for their effects on cell fusion. CHO cells were transfected with each wild-type or mutant form of HSV-1 or HSV-2 gD, along with wild-type forms of HSV-1 or HSV-2 gB, gH, and gL in homologous combinations. These effector cell populations were then divided, and portions were replated in 96-well dishes for CELISA assays to quantitate the cell surface expression of gD, while the remainder of each population was mixed with target cells expressing each of the known HSV-1 or HSV-2 fusion/entry receptors for cell fusion assays. Cell fusion was quantitated by measuring the activity of luciferase, which can be expressed only after effector and target cells have fused. The results are presented in Fig. 5 for the substitution mutants and in Fig. 6 for the deletion mutants.
FIG. 5.
Expression of wild-type and mutant (substitution mutant) forms of gD on the surfaces of effector cells and fusion of these cells with target cells expressing various human and mouse fusion receptors. CHO-K1 cells were transfected with mixtures of plasmids expressing HSV-1 (A and B) or HSV-2 (C and D) gB, gD (wild-type [WT] or substitution mutant forms as indicated), gH, and gL. In control mixtures, the gD plasmid was left out and replaced with empty vector (NO gD). The cell populations were then divided, and a portion was plated in 96-well plates for CELISA (A and C). The remaining cells were mixed with the various target cells indicated for cell fusion assays (B and D). In panels A and C, anti-gD bound is expressed as HRP product measured at OD370. In panels B and D, cell fusion is expressed as units of luciferase, normalized to the values obtained with wild-type gD on HVEM-expressing cells. The results are expressed as the means and standard deviations of three independent experiments, each done in triplicate.
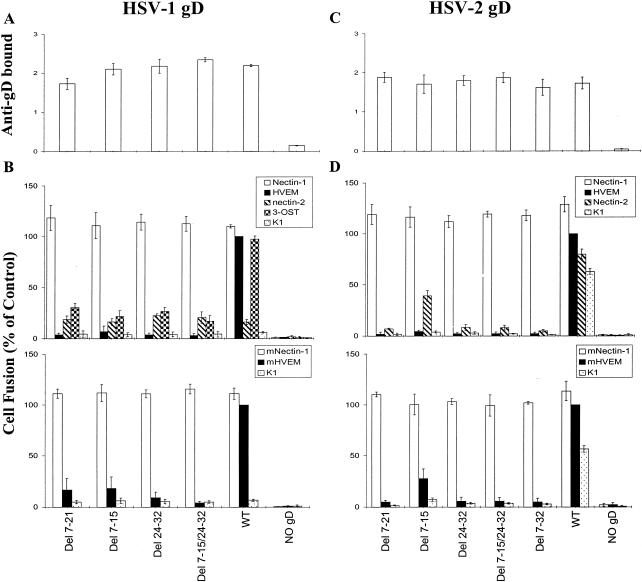
FIG. 6.
Expression of wild-type and mutant (deletion mutant) forms of gD on the surfaces of effector cells and fusion of these cells with target cells expressing various human and mouse fusion receptors. CHO-K1 cells were transfected with mixtures of plasmids expressing HSV-1 (A and B) or HSV-2 (C and D) gB, gD (wild-type [WT] or deletion mutant forms as indicated), gH, and gL. In control mixtures, the gD plasmid was left out and replaced with empty vector (NO gD). The cell populations were then divided, and a portion was plated in 96-well plates for CELISA (A and C). The remaining cells were mixed with the various target cells indicated for cell fusion assays (B and D). In panels A and C, anti-gD bound is expressed as HRP product measured at OD370. In panels B and D, cell fusion is expressed as units of luciferase, normalized to the values obtained with wild-type gD on HVEM-expressing cells. The results are expressed as the means and standard deviations of three independent experiments each done in triplicate.
Panels A and C in both figures show that the mutant and wild-type forms of gD were expressed at equivalent levels on the surfaces of the effector cells. The cell fusion results (panels B and D in Fig. 5 and 6) demonstrate the following. (i) All of the amino acid substitutions in HSV-1 gD, except L25P, reduced cell fusion with human HVEM to ca. 5% of the wild-type level and with mouse HVEM to 10 to 20% of the wild-type level. Similar results were obtained with the HSV-2 substitution mutants except that the reduction in levels of cell fusion was not quite so pronounced. (ii) All amino acid substitutions at positions 25 and/or 27 in HSV-1 gD significantly increased cell fusion activity with target cells expressing human nectin-2 and reduced fusion with cells expressing 3-OST-3B. These mutations in HSV-2 gD were without effect on the already significant level of cell fusion observed with wild-type HSV-2 gD and human nectin-2. (iii) All of the amino acid substitutions in HSV-2 gD reduced cell fusion with parental CHO-K1 cells to 20 to 35% of the wild-type level. (iv) All of the deletions in HSV-1 or HSV-2 gD significantly reduced cell fusion with cells expressing any of the receptors except nectin-1. Values close to the background values found in the absence of gD were noted for some of the deletions, particularly for those missing at least amino acids 24 to 32 and for all target cells except those expressing nectin-1 and those expressing human nectin-2 or 3-OST-3B when tested with the HSV-1 mutants. (v) None of the amino acid substitutions or deletions had any appreciable effect on cell fusion when the target cells expressed either human or mouse nectin-1.
Figure 4 demonstrated that deletion 7-21 in HSV-2 gD, but not in HSV-1 gD, reduced the efficiency of binding of the soluble gD:Fc to nectin-1; however, neither mutation had any discernible effect on fusion with nectin-1-expressing cells (Fig. 6). It is possible that (i) the fusion assay cannot discriminate among different levels of activity above a certain threshold, (ii) the different forms of gD used in the binding and fusion assays were affected differently by the deletion in the case of HSV-2 (both forms were checked for presence of the appropriate deletion and absence of unwanted mutations), or (iii) a wide range of binding affinities can be associated with the same level of fusion activity. As we noted previously, mutations in nectin-1 that reduced gD binding did not necessarily affect HSV entry activity (18, 30). A deletion of 7 to 21 amino acids in HSV-1(F) gD was previously characterized (9). Virus expressing this mutant gD appeared indistinguishable from wild-type HSV-1(F) for plaque formation on Vero cells and replication in a human cell line, suggesting that the receptor used by this mutant might have been nectin-1.
We can conclude that the N-terminal region of either HSV-1 or HSV-2 gD is dispensable for the binding of gD to nectin-1 and for fusion with cells expressing nectin-1, whereas this region is required for full activity with all of the other receptors tested. Moreover, the amino acid sequence within the N terminus governs the level of activity with each of these receptors.
DISCUSSION
We were surprised to find that all but one of the mutations described here had effects on functional interactions of both HSV-1 and HSV-2 gDs with all of the known HSV fusion/entry receptors except nectin-1. The results with HVEM were largely predictable, but the other results were not, for the reasons given below.
The importance of amino acid Q27 in the HSV-1 gD-HVEM interaction was evident from the crystal structure of the HVEM-gD complex (3). This showed that Q27 is part of a short extended strand that forms hydrogen bonds with a β strand in HVEM. The side chain of Q27 points toward an α-helix in gD that is sandwiched between the N-terminal hairpin and the immunoglobulin fold. This side chain inserts into a pocket too small to accommodate an arginine residue (as in Q27R). The Q27P substitution would be expected to distort the β-strand interaction. New information presented here is that mutations in the same position in HSV-2 gD also disrupted physical and functional interactions with HVEM. It is not yet known how similar are the structures of HSV-1 and HSV-2 gD. An earlier study performed with soluble truncated forms of gD (306 amino acids instead of 285 in the form that was crystallized) used circular dichroism to estimate the secondary structure content of both HSV-1 and HSV-2 gDs (22). HSV-2 gD exhibited properties consistent with little or no α-helical content compared to 10% for HSV-1 gD. Also, the β-sheet content was estimated to be higher for HSV-2 gD than for HSV-1 gD. However, similar or different the structures prove to be, substitutions Q27P or Q27R in either HSV-1 or HSV-2 gD disrupt interactions with HVEM, both human and mouse.
The gD amino acid L25 also points toward the α-helix mentioned above and, although not part of the extended strand that forms hydrogen bonds with the β-strand in HVEM, the L25P substitution might be expected to affect this interaction. The carbonyl group of L25 forms a hydrogen bond via a water molecule with the hydroxyl group of Y23 in HVEM. Despite these structural findings, our results show that the L25P substitution in HSV-1 or HSV-2 gD had little or no effect on the binding of gD:Fc to HVEM or on fusion with cells expressing HVEM. This mutation in HSV-1(F)U10 did, however, reduce the efficiency of viral entry into cells expressing HVEM. Perhaps reduced efficiency of binding and cell fusion would also have been observed with the L25P mutation in HSV-1(F) gD, if tested as the HSV-1(KOS) gD mutants were tested. Alternatively, other factors may influence how the mutant form of gD functions in the context of the virion. The key point, however, is that not all of the identified contacts between HVEM and gD are equally important for a stable and functional interaction. This point was also recently documented in a study investigating the effects of amino acid substitutions in HVEM on interactions with HSV-1 gD (5). Interestingly, substitutions in two of the HVEM residues (T35 and V36) that make contacts with Q27 reduced binding to HSV-1 gD but had little effect on entry activity of the altered HVEM. Substitutions at Y23 completely abolished binding to gD and viral entry activity. Both findings seem contradictory to the ones presented here, but the latter finding, at least, can be explained by the fact that Y23 makes extensive contacts with other residues of gD and not just L25.
The most surprising findings were that the gD mutations had such different effects on functional interactions with human nectin-1 and nectin-2. These two proteins are related in sequence and structure, and homologous regions of the two receptors have been found to be critical for HSV entry (18, 19, 30). Amino acid substitutions in loops between the putative F and G β strands and the C′ and C" β strands of the N-terminal immunoglobulin folds of both nectin-1 and nectin-2 have been shown to reduce HSV entry activity. Also, other amino acid substitutions in the loop between the C′ and C" β strands of human and mouse nectin-2 can enhance or confer entry activity for both HSV-1 and HSV-2. It seems reasonable to predict that contact sites on HSV-2 gD for these two receptors are similar, and yet the N terminus of gD is required for fusion activity with nectin-2 but not with nectin-1. Moreover, the amino acid sequence in the N terminus influences the level of fusion activity with nectin-2. This is evident not only from the effects of mutations in the N terminus of HSV-1 gD but also from results obtained in a study designed to identify the regions of HSV-2 gD that permit fusion activity with nectin-2. The present study showed that as many as 7 amino acid differences between HSV-1 and HSV-2 in the first 53 amino acids, i.e., upstream of the immunoglobulin fold, are largely responsible for the differences in ability of HSV-1 and HSV-2 gDs to induce fusion with cells expressing human nectin-2 (36).
The results presented here show that contact regions for nectin-1 must be present downstream of amino acid 32 in gD. It seems likely, based on the considerations outlined above, that these regions would also make contact with nectin-2 but, in the case of nectin-2, would not be sufficient for fusion activity. Possibly there are additional contact regions for both nectin-1 and nectin-2 within the N-terminal 32 amino acids, and these are necessary for functional activity with nectin-2 but not with nectin-1. Alternatively, portions of gD within the first 32 amino acids may influence the conformation of the rest of gD in ways that are critical for nectin-2 binding and function for not for nectin-1. Dissociation constants for the interaction of HSV gD with nectin-1 are in the micromolar range (15), whereas it has not been possible to determine dissociation constants for gD-nectin-2 interactions. It seems likely that the affinity of HSV-2 or HSV-1 Rid gD for nectin-2 is lower than for nectin-1 or that the trans-homophilic interactions of nectin-2 dimers are of higher affinity than those of nectin-1 dimers and less easily disrupted to allow for gD binding or both. Physical interactions of HSV-1 Rid gD or HSV-2 gD with human nectin-2 have not been demonstrable by the CELISA used here, by ELISA, or by plasmon resonance, whereas all three methods have been used to demonstrate interactions of gD with nectin-1 (11, 15). Only by immunofluorescence have we been able to demonstrate a specific colocalization of HSV-1 Rid1 gD with human nectin-2, but not with a mutant of nectin-2 devoid of viral entry activity (30).
Negative effects of the N-terminal mutations in HSV-1 gD on fusion activity with cells expressing 3-OST-3B are consistent with the tentative identification of putative heparan sulfate-binding domains in gD (3). The electron density maps of the gD crystals included two large spherical features tentatively identified as sulfate ions. These features were found in basic domains that are candidates for heparan sulfate-binding sites. One such region was a positively charged pocket formed by β strands of the N-terminal hairpin and including amino acids K1, R35, and R36.
The new information presented here includes the effect of the L25P substitution in HSV-1 gD on functional interactions with HVEM, the effects of amino acid substitutions in the N terminus of HSV-2 gD on interactions with all of the HSV-2 fusion/entry receptors, the effects of deletions in the N termini of both HSV-1 and HSV-2 gDs on interactions with all of the HSV-1 and HSV-2 fusion/entry receptors, and the testing of wild-type and mutant forms of HSV-1 and HSV-2 gDs for functional interactions with all of the mouse fusion/entry receptors. The latter results extend our findings that mouse nectin-1 is a functional entry receptor for both HSV-1 and HSV-2, as well as for PRV and BHV-1 (25). Viral entry results for mouse HVEM have not previously been reported. We show here that mouse HVEM is indistinguishable from human HVEM in its ability to mediate entry of HSV-1(KOS), HSV-1(F), and HSV-1(F)U10 and in its inability to mediate entry of HSV-1(KOS)Rid1. In a previous discussion and review (25, 29), we had mentioned unpublished preliminary results suggesting that mouse HVEM could mediate the entry of HSV-1 Rid mutants. It is possible that mouse HVEM is not quite as inactive for HSV-1 Rid entry as human HVEM, a finding consistent with the cell fusion results shown in Fig. 5.
The next stage of this work is to introduce some of the mutant forms of gD into the viral genome to test whether the receptor specificities observed in the cell fusion assay also apply to viral entry. We predict that some of the HSV-1 and HSV-2 deletion mutants will be able to infect cells expressing only nectin-1, among the known HSV entry receptors. Such mutants will be invaluable for various studies such as identifying the key receptors for entry of virus into various cell types in culture and in vivo, and for investigating viral signaling via various receptors.
Acknowledgments
We thank N. Susmarski and M. L. Parish for excellent technical assistance; G. Campadelli-Fiume (University of Bologna, Bologna, Italy) for the HSV-1(F)U10 virus; G. Cohen and R. Eisenberg (University of Pennsylvania) for the R7 antibody; M. Lopez (INSERM [Paris, France]) for the R1.302 antibody; D. Myscofski for the pDM19 plasmid; R. Rosenberg (Massachusetts Institute of Technology) for the 3-OST-3B plasmid; Y. Kawaoka (University of Wisconsin) for the pCAGGS/MCS plasmid; and Y. Matsuura (National Institute of Infectious Disease, Tokyo, Japan) for the pCAGT7 and pT7EMLuc plasmids. We also thank C. Waltenbaugh (Northwestern University) and N. Clipstone (Northwestern University) for the use of the ELISA reader and luminometer.
This work was supported by grants from the National Institutes of Health (R37 AI36293, R01 CA21776, and U19 AI31494) and the Mizutani Foundation.
REFERENCES
- 1.Brandimarti, R., T. Huang, B. Roizman, and G. Campadelli-Fiume. 1994. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc. Natl. Acad. Sci. USA 91:5406-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH, and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 3.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 4.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis-Poynter, N., S. Bell, T. Minson, and H. Browne. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 68:7586-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean, H. J., S. Terhune, M.-T. Shieh, N. Susmarski, and P. G. Spear. 1994. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell type-dependent alterations in infectivity. Virology 199:67-80. [DOI] [PubMed] [Google Scholar]
- 8.Dean, H. J., M. S. Warner, S. S. Terhune, R. M. Johnson, and P. G. Spear. 1995. Viral determinants of the variable sensitivity of herpes simplex virus strains to gD-mediated interference. J. Virol. 69:5171-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feenstra, V., M. Hodaie, and D. C. Johnson. 1990. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J. Virol. 64:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geraghty, R. J., A. Fridberg, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 2001. Use of chimeric nectin-1(HveC)-related receptors to demonstrate that ability to bind alphaherpesvirus gD is not necessarily sufficient for viral entry. Virology 285:366-375. [DOI] [PubMed] [Google Scholar]
- 11.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147-158. [DOI] [PubMed] [Google Scholar]
- 12.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 13.Kobasa, D., M. E. Rodgers, K. Wells, and Y. Kawaoka. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J. Virol. 71:6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krummenacher, C., A. V. Nicola, J. C. Whitbeck, H. Lou, W. Hou, J. D. Lambris, R. J. Geraghty, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpes virus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 72:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krummenacher, C., A. H. Rux, J. C. Whitbeck, M. Ponce de Leon, H. Lou, I. Baribaud, W. Hou, C. Zou, R. J. Geraghty, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1999. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 73:8127-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez, M., F. Jordier, F. Bardin, L. Coulombel, D. Chabannon, and P. Dubreuil. 1997. Identification of a new class of IgG superfamily antigens expressed in hemopoiesis, p. 1081-1083. In T. Kishomoto, H. Kikutani, A. von dem Borne, D. Y. Mason, M. Miyasaka, A. Moretta, K. Okumura, S. Shaw, T. A. Springer, K. Sugumura, and H. Zola (ed.), Leukocyte typing IV: white cell differentiation antigens. Garland Publishing, Inc., New York, N.Y.
- 18.Martinez, W. M., and P. G. Spear. 2002. Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex viruses 1 and 2 but not for pseudorabies virus or bovine herpesvirus 1. J. Virol. 76:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez, W. M., and P. G. Spear. 2001. Structural features of nectin-2 (HveB) required for herpes simplex virus entry. J. Virol. 75:11185-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 21.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH, and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 22.Nicola, A. V., S. H. Willis, N. N. Naidoo, R. J. Eisenberg, and G. H. Cohen. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J. Virol. 70:3815-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pertel, P., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 24.Shieh, M.-T., D. WuDunn, R. I. Montgomery, J. D. Esko, and P. G. Spear. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 116:1273-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla, D., M. Dal Canto, C. L. Rowe, and P. G. Spear. 2000. Striking similarity of murine nectin-1α to human nectin-1α (HveC) in sequence and activity as a gD receptor for alphaherpesvirus entry. J. Virol. 74:11773-11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 27.Shukla, D., C. L. Rowe, Y. Dong, V. R. Racaniello, and P. G. Spear. 1999. The murine homolog (Mph) of human herpesvirus entry protein B (HveB) mediates entry of pseudorabies virus but not herpes simplex virus types 1 and 2. J. Virol. 73:4493-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press, Inc., Boca Raton, Fla.
- 29.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Struyf, F., W. M. Martinez, and P. G. Spear. 2002. Mutations in the N-terminal domains of nectin-1 and nectin-2 reveal differences in requirements for entry of various alphaherpesviruses and for nectin-nectin interactions. J. Virol. 76:12940-12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi, K., H. Nakanishi, M. Miyahara, K. Mandai, K. Satoh, A. Satoh, H. Nishioka, J. Aoki, A. Nomoto, A. Mizoguchi, and Y. Takai. 1999. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J. Cell Biol. 145:539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terry-Allison, T., R. I. Montgomery, M. S. Warner, R. J. Geraghty, and P. G. Spear. 2001. Contributions of gD receptors and glycosaminoglycan sulfation to cell fusion mediated by herpes simplex virus 1. Virus Res. 39-45. 74: [DOI] [PubMed]
- 33.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2 and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 34.Whitbeck, J. C., C. Peng, H. Lou, R. Xu, S. H. Willis, M. Ponce de Leon, T. Peng, A. V. Nicola, R. I. Montgomery, M. S. Warner, A. M. Soulika, L. A. Spruce, W. T. Moore, J. D. Lambris, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 71:6083-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon, M., and P. G. Spear. 2002. Disruption of adherens junctions liberates nectin-1 to serve as receptor for herpes simplex virus entry. J. Virol. 76:7203-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zago, A., and P. G. Spear. 2003. Differences in N termini of herpes simplex virus type 1 and 2 gDs that influence functional interactions with the human entry receptor nectin-2 and an entry receptor expressed in Chinese hamster ovary cells. J. Virol. 77:9695-9699. [DOI] [PMC free article] [PubMed]