Abstract
Amino acid differences at seven positions in the N termini of the glycoproteins D (gDs) specified by herpes simplex virus type 1 (HSV-1) and HSV-2 are largely responsible for the significantly higher cell fusion activity of HSV-2 gD with Chinese hamster ovary cells expressing human nectin-2 or only an endogenous hamster receptor.
Herpes simplex virus type 1 (HSV-1) and HSV-2 cause mucocutaneous lesions that are indistinguishable clinically, but the two serotypes exhibit differences in gene sequences, biology, pathogenesis, and epidemiology. In addition, the two serotypes exhibit some differences in the efficiency of cell entry, dependent on the entry receptors expressed, as outlined below.
Heparan sulfate chains on cell surface proteoglycans provide initial binding sites for both HSV-1 and HSV-2, but each serotype recognizes somewhat different structural features of heparan sulfate (8). After the binding of HSV-1 or HSV-2 to heparan sulfate, another viral glycoprotein, glycoprotein D (gD), engages any one of several cell surface receptors to initiate the process of viral entry (18). Viral entry occurs by fusion of the viral envelope with a cell membrane and requires the envelope glycoproteins gB, gH, and gL, as well as gD, and a gD receptor on the cell.
The human HSV gD receptors include herpesvirus entry mediator (HVEM) (14), a member of the tumor necrosis factor receptor family; nectin-1 (2, 7) and nectin-2 (11, 21), members of the immunoglobulin superfamily that are cell adhesion molecules present in cadherin-based adherens junctions (19); and sites in heparan sulfate generated by the action of specific 3-O-sulfotransferases (17). HSV-1 and HSV-2 differ in their ability to use the various gD receptors for viral entry. Human HVEM or nectin-1 can serve as entry receptors for both serotypes, whereas nectin-2 has a higher entry activity for HSV-2 strains than for HSV-1 strains, and 3-O-sulfated heparan sulfate has a higher entry activity for HSV-1 strains than for HSV-2 strains. Also, Chinese hamster ovary (CHO) cells express an endogenous receptor for HSV-2 entry that has only minimal activity for HSV-1 entry (14, 16). The activity of the HSV entry receptors can be assessed by quantitative viral entry assays (14), as well as by quantitative cell fusion assays in which cells expressing HSV gB, gD, gH, and gL are tested for their ability to fuse with cells bearing entry/fusion receptors (15). The activities of the viral glycoproteins can be assessed similarly by using mutant viruses in viral entry assays or mutant forms of the viral glycoproteins in cell fusion assays.
The purpose of the present study was to define the viral determinants responsible for the greater ability of HSV-2, compared to wild-type HSV-1, to interact functionally with human nectin-2 and the endogenous CHO receptor.
HSV-2 gD confers enhanced fusion activity with the CHO receptor and nectin-2.
Since gD is the ligand for all known HSV entry receptors, it seemed likely that HSV-2 gD determines the enhanced fusion activity of HSV-2 glycoproteins with the CHO receptor and nectin-2. To test this hypothesis, quantitative cell fusion assays were done with the homologous set of HSV-1 or HSV-2 glycoproteins or with mixtures in which each of the glycoproteins in the HSV-1 or HSV-2 set was substituted with its heterologous counterpart. CHO cells were transfected with plasmids expressing the appropriate mixtures of HSV glycoproteins and T7 polymerase (termed effector cells). The plasmids used for expression of the HSV-1(KOS) glycoproteins were previously described (15). A number of plasmids were generated for the present study, as described in Table 1, and the inserts were sequenced to confirm the desired coding of amino acid sequence. CHO cells stably expressing HVEM, nectin-1, or nectin-2 or control CHO cells were transfected with a plasmid carrying the firefly luciferase gene linked to a T7 promoter (termed target cells). The effector and target cells were then mixed in 1:1 ratios, plated, and incubated for 18 h prior to lysis for quantitation of luciferase as a measure of cell fusion (15).
TABLE 1.
Expression plasmids generated for this study
| Plasmid | Protein expressed | Insert in the expression plasmid pCAGGS |
|---|---|---|
| pAZB2 | gB2 | Nucleotides 56219 to 54368 and 54617 to 53374 from HSV-2(333)a were amplified by PCR, joined by the unique SphI site in the overlapping regions, and cloned into the EcoRI and KpnI restriction sites |
| pAZD2 | gD2 | PCR amplification of nucleotides 140992 to 142323 from HSV-2(333)a cloned into the EcoRI and KpnI restriction sites |
| pAZH2 | gH2 | Nucleotides 46608 to 45131 and 45157 to 43933 from HSV-2(333)a were amplified by PCR, joined by the unique XhoI site in the overlapping regions, and cloned into the EcoRI and KpnI restriction sites |
| pAZL2 | gL2 | PCR amplification of nucleotides 9382 to 10225 from HSV-2(333)a cloned into EcoRI and KpnI restriction sites |
| pAZCH5 | gD2(1-66)/gD1(67-369)b | Nucleotides 140992 to 141288 from HSV-2(333)a and nucleotides 138692 to 139626 from HSV-1(KOS)c were amplified by PCR and combined in a final PCR by using only the outside primers |
| pAZCH1 | gD2(1-98)/gD1(99-369) | gD2(1-98) was joined to gD1(99-369) by a natural restriction site (Van91I) |
| pAZCH16 | gD1(1-66)/gD2(67-368) | Nucleotides 138398 to 138691 from HSV-1(KOS)c and nucleotides 141289 to 142323 from HSV-2(333)a were amplified by PCR and combined in a final PCR using only the outside primers |
| pAZCH35 | gD1(1-98)/gD2(99-368) | gD1(1-98) was joined to gD2(99-368) by a natural restriction site (Van91I) |
| pAZMUT1.1 | gD1(A7P) | Codon GCC was changed to CCC, causing an Ala→Pro substitution in position 7 |
| pAZMUT2.1 | gD1(A42P,G43S) | Codons GCG and GGC were changed to CCG and AGC, causing Ala→Pro and Gly→Ser substitutions in positions 42 and 43 |
| pAZMUT3.1 | gD1(P45E) | Codon CCG was changed to GAG, causing a Pro→Glu substitution in position 45 |
| pAZMUT4.1 | gD1(A7P,D21N) | Codons GCC and GAC were changed to CCC and AAC, causing Ala→Pro and Asp→Asn substitutions in positions 7 and 21 |
| pAZMUT5.1 | gD1(A7P,P45E) | Codons GCC and CCG were changed to CCC and GAG, causing Ala→Pro and Pro→Glu substitutions in positions 7 and 45 |
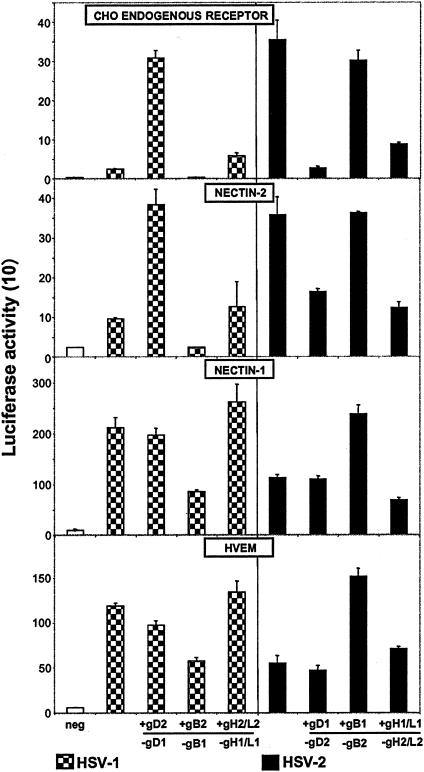
Figure 1 shows that the homologous mixtures of HSV-1 and HSV-2 glycoproteins induced levels of cell fusion with each receptor consistent with viral entry assays done with the same receptors (7, 12, 14, 21). The cell fusion assay may be more sensitive in that HSV-1 glycoproteins exhibited low but detectable activities with the endogenous CHO receptor and nectin-2, whereas HSV-1(KOS) entry with these receptors is more difficult to detect. Figure 1 also shows that substitution of HSV-1 gD (gD1) with gD2 in the otherwise HSV-1 set significantly enhanced cell fusion with target cells expressing the CHO receptor only or also nectin-2 but not with target cells expressing nectin-1 or HVEM. In the converse combination, substitution of gD2 with gD1 in the otherwise HSV-2 set significantly reduced cell fusion with the CHO receptor and nectin-2 but not with nectin-1 or HVEM. Thus, gD is the major determinant of the differential ability of HSV-1 and HSV-2 glycoproteins to induce cell fusion with the CHO receptor and nectin-2.
FIG. 1.
Cell fusion induced by plasmids expressing homologous or heterologous mixtures of the HSV-1 or HSV-2 glycoproteins. The cell fusion assay was performed by transfecting CHO effector cells with plasmids expressing the indicated glycoproteins or vector alone (neg). The effector cells were mixed with CHO target cells expressing only the CHO endogenous receptor or also nectin-2, nectin-1, or HVEM. Effector cells transfected with an homologous set of HSV-1 or HSV-2 glycoproteins are the controls. The glycoprotein substituted is indicated with a minus (−), and the substituting protein is indicated with a plus (+). Cell fusion was quantified by measurements of luciferase activity (arbitrary units divided by 10 to give the numbers shown on the y axis). The values shown are means and standard deviations from one experiment performed in triplicate. The results presented are representative of at least two other additional experiments.
Other findings emerged from the results presented in Fig. 1. Substitution of gB2 with gB1 in the HSV-2 set significantly enhanced fusion, but only with target cells expressing nectin-1 and HVEM, whereas substitution of gB1 with gB2 in the HSV-1 set reduced cell fusion with target cells expressing any of the receptors. It has been reported that deletions from the cytoplasmic C terminus of gB2 can enhance cell fusion induced by HSV-2 glycoproteins with cells in which the fusion receptors were not identified and that the effect was not necessarily correlated with altered cell surface expression of this glycoprotein (5). We have confirmed these findings and showed further that substitution of wild-type gB2 with truncated gB2 had the same effect on cell fusion with nectin-1 and HVEM (A. Zago and P. G. Spear, unpublished results), as did substitution with gB1 (Fig. 1). It seems likely that the cytoplasmic tail of wild-type gB2 has a domain that is inhibitory for cell fusion, at least with certain fusion receptors, whereas such an inhibitory activity in wild-type gB1 is not so evident. The greater ability of HSV-1 glycoproteins, compared to HSV-2 glycoproteins, to induce cell fusion with nectin-1 and HVEM is probably accounted for by this inhibitory domain in gB2. We chose to use wild-type gB2 in the present study.
Substitutions of gH and gL were not done separately because these glycoproteins are known to function as a heterodimer (9). With nectin-1 and HVEM, substitutions of gH/gL in either combination with the other glycoproteins had little effect on cell fusion (Fig. 1). The same was true for the CHO receptor and nectin-2 when gH1/gL1 was substituted with gH2/gL2. However, substitution of gH2/gL2 with gH1/gL1 in the otherwise HSV-2 mixture reduced cell fusion activity with the CHO receptor and nectin-2. This phenomenon requires further exploration beyond the scope of the present study.
Mapping of domains in gD2 that confer cell fusion activity with the CHO receptor and nectin-2.
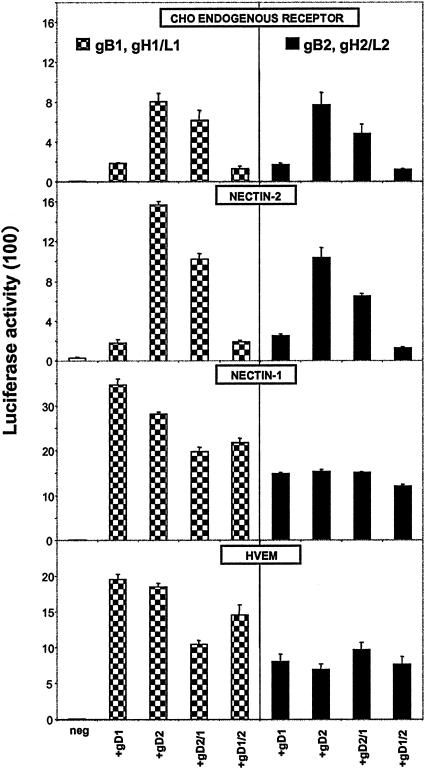
Plasmids expressing hybrid forms of gD were constructed (Table 1). These hybrids had the first 66 or 98 amino acids from gD1 or gD2 and the remainder from gD2 or gD1, respectively. Plasmids expressing the hybrid proteins were used in cell fusion assays, along with plasmids expressing the wild-type forms of gD1 or gD2, in combination with the other HSV-1 or HSV-2 glycoproteins. The results presented in Fig. 2 show that the gD2/1 hybrid having the first 66 amino acids from HSV-2 resembled gD2 in its enhancement of fusion with target cells expressing the CHO receptor only or also nectin-2 when combined with the HSV-1 glycoproteins. The activity of the gD2/1 hybrid was ca. 60 to 80% that observed with gD2 regardless of whether the other glycoproteins were from HSV-1 or HSV-2 and regardless of receptor (except that the gD2/1 hybrid had activity comparable to that of gD2 when the receptors were nectin-1 or HVEM and the other glycoproteins were from HSV-2). Also, the converse gD1/2 hybrid resembled gD1 in its poor cell fusion activity with target cells expressing the CHO receptor only or also nectin-2 when combined with either the HSV-1 or HSV-2 glycoproteins. The gD1/2 hybrid was slightly more active than the gD2/1 hybrid with the other HSV-1 glycoproteins and slightly less active with the other HSV-2 glycoproteins when the receptors were nectin-1 and HVEM. Results similar to those shown in Fig. 2 for CHO cells were also obtained with hybrids in which the first 98 amino acids were switched. We conclude from these results (i) that the hybrids had no gross conformational abnormalities and had fusion activities comparable to those of gD1 and gD2, at least when tested with gB2 and gH2/gL2 and the fusion receptors, nectin-1 and HVEM, and (ii) that the superior activity of gD2 in fusion activity with the CHO receptor and nectin-2 is largely due to amino acid differences between gD2 and gD1 within the first 66 amino acids.
FIG. 2.
Cell fusion induced by gD2/1 or gD1/2 hybrid molecules in combination with the other HSV-1 or HSV-2 glycoproteins. These hybrids have the first 66 amino acids from gD1 or gD2 and the remainder from gD2 or gD1, respectively. The CHO effector cells were transfected with plasmids expressing the indicated glycoproteins or vector alone (neg). Effector cells were mixed with CHO target cells expressing only the CHO endogenous receptor or also nectin-2, nectin-1, or HVEM. Cell fusion was quantified by measurements of luciferase activity (arbitrary units divided by 100 to give the numbers shown on the y axis). The values shown are means and standard deviations from one experiment performed in triplicate. The results presented are representative of at least two other additional experiments.
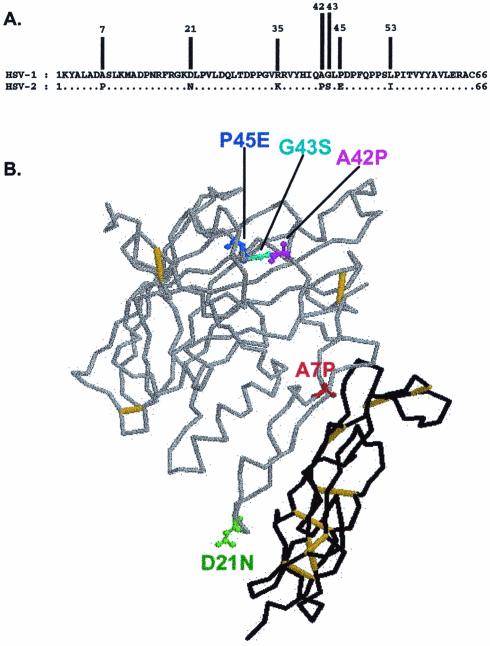
Figure 3 shows an alignment of the amino acid sequences of gD1 and gD2 in the first 66 amino acids. There are seven differences in sequence, five of which are also noted on a backbone trace of the structure of gD1 (1). For the five positions most likely to affect secondary and tertiary structure (i.e., positions 7, 21, 42, 43 and 45), gD1 was mutated by substituting the amino acid present in gD1 for the one present in gD2. These substitutions were made individually and in pairs (A7P, A7P/P45E, A7P/D21N, A42P/G43S, and P45E). When these mutant forms of gD1 were tested for ability to induce the fusion of target cells expressing the CHO receptor only or also nectin-2, none exhibited as high as 50% of the activity observed with gD2, using either the HSV-1 or HSV-2 forms of gB and gH/gL, except in the case of the HSV-1 set of glycoproteins tested with target cells expressing only the CHO receptor. Only the gD1 mutants carrying the A7P substitution, either alone or in combination with P45E or D21N, exhibited enhanced cell fusion activity (50 to 60% that of gD2) under these conditions (data not shown). None of the gD1 mutants tested with either HSV-1 or HSV-2 gB and gH/gL differed significantly from wild-type gD1 in cell fusion activity when the receptors were nectin-1 or HVEM (data not shown). We conclude that the A7P substitution in gD2 contributes to the higher level of fusion activity of gD2 but that multiple substitutions within the first 53 amino acids of gD2 (the domain containing all of the differences between gD1 and gD2 in the first 66 amino acids), including A7P, are required for the gD2-like levels of cell fusion activity with the CHO receptor and nectin-2.
FIG. 3.
(A) Alignment of the 66 N-terminal amino acids of HSV-1(KOS) gD and HSV-2(333) gD. Dots indicate identity. The positions where the amino acids differ are indicated. (B) Backbone trace of the crystal structure of gD1 bound to HVEM (1). HVEM is shown in black, and gD1 is shown in gray. Residues that have been substituted in the gD1 mutants are displayed in different colors.
The levels of gD expressed on the surfaces of glycoprotein-expressing cells used in the cell fusion assays were assessed by an immunoassay, in which the live cells in the monolayer were incubated with a type-common anti-gD rabbit serum, R#7, and then fixed and incubated with a detection system as previously described (6). Comparable levels of cell surface expression were consistently noted for gD1, gD2, all of the gD1/gD2 hybrids, and all HSV-1 mutants (data not shown), indicating that the differences in cell fusion observed could not be explained by expression levels of gD.
The results presented here and elsewhere show that the amino acid sequence in the N-terminal region of gD, outside of the Ig-fold, influences whether human nectin-2 and an endogenous CHO receptor can serve as entry/fusion receptors. We showed here (i) that gD2 is more active than gD1 in inducing the fusion of cells expressing human nectin-2 or only an endogenous CHO receptor, regardless of whether the viral glycoproteins gB, gH, and gL were from HSV-1 or HSV-2 and consistent with the greater activity of these receptors for HSV-2 entry (16, 21), and (ii) that as many as 7 amino acid differences between gD1 and gD2 in the first 53 amino acids of the N terminus are largely responsible for the greater activity of gD2. It was previously shown that specific amino acid substitutions in the N terminus of gD1 enable functional interactions with human nectin-2. Substitutions Q27P or Q27R in gD1 confer the ability of HSV-1 to use nectin-2 as an entry or fusion receptor (15, 20, 21). Also, substitution L25P in gD1 enables HSV-1 to use nectin-2 as an entry receptor (11). Interestingly, the mutations at position 27 were also shown to enhance, by about 10-fold, the limited ability of HSV-1 strain KOS to infect CHO cells (3). Other effects of the mutations at position 27 in gD1 include enhanced affinity of isolated soluble mutant gD1 for nectin-1 (10) and significantly reduced ability of HSV-1 to use HVEM as an entry or fusion receptor (14, 15, 20), a finding consistent with the location of amino acid 27 in a contact region identified in the X-ray structures (1). It should be noted that L25 and Q27, and in fact all of the positions in the contact region from positions 24 to 32, are identical in gD1 and gD2. Thus, substitutions at positions 25 or 27 enable gD1 to interact functionally with nectin-2 but are not necessary for gD2 to interact with nectin-2.
Attempts were made to isolate the endogenous CHO receptor, without success. Oligonucleotides matching conserved sequences in nectin-1 and nectin-2 were tested for the ability to prime amplification of homologous sequences from reverse transcripts of RNA extracted from CHO cells or from plasmids extracted from a library of CHO expression clones. Antibodies raised against human or mouse nectin-2 were tested for their ability to bind to CHO cells by immunofluorescence. Identification of the endogenous CHO receptor would probably require expression cloning, which would in turn require a cell line with a high level of resistance to HSV-2 entry. Some pilot experiments done with the B78H1 cell line were not satisfactory. We abandoned the effort to identify the receptor because the results obtained in the present study were so similar for human nectin-2 and the endogenous CHO receptor, suggesting that the endogenous CHO receptor could be the Chinese hamster homolog of nectin-2 despite our inability to amplify nectin-2 gene sequences by PCR. Human nectin-2 and the endogenous CHO receptor are distinct because certain mutations in gD2 can inhibit cell fusion to a much greater extent with control CHO cells than with CHO cells expressing nectin-2 (22).
Although the results summarized here indicate that the N-terminal regions of gD1 and gD2 influence entry and cell fusion with nectin-2 or the endogenous CHO receptor, further studies are required to determine whether the N terminus makes direct contact with these receptors, influences the conformation of other regions in gD that are involved in direct contacts, or influences interactions with the other viral glycoproteins that are required for cell fusion.
Acknowledgments
We thank Nanette Susmarski for excellent technical assistance and Gary Cohen and Roselyn Eisenberg for antiserum.
This work was supported by National Institutes of Health grants R37 AI36293, R01 CA21776, and U19 AI31494.
REFERENCES
- 1.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 2.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean, H. J., S. Terhune, M.-T. Shieh, N. Susmarski, and P. G. Spear. 1994. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell type-dependent alterations in infectivity. Virology 199:67-80. [DOI] [PubMed] [Google Scholar]
- 4.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan, Z., M. L. Grantham, M. S. Smith, E. S. Anderson, J. A. Cardelli, and M. I. Muggeridge. 2002. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J. Virol. 76:9271-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147-158. [DOI] [PubMed] [Google Scholar]
- 7.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 8.Herold, B. C., S. I. Gerber, B. J. Belval, A. M. Siston, and N. Shulman. 1996. Differences in the susceptibility of herpes simplex virus types 1 and 2 to modified heparin compounds suggest serotype differences in viral entry. J. Virol. 70:3461-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krummenacher, C., A. H. Rux, J. C. Whitbeck, M. Ponce de Leon, H. Lou, I. Baribaud, W. Hou, C. Zou, R. J. Geraghty, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1999. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 73:8127-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez, W. M., and P. G. Spear. 2001. Structural features of nectin-2 (HveB) required for herpes simplex virus entry. J. Virol. 75:11185-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 15.Pertel, P., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 16.Shieh, M.-T., D. WuDunn, R. I. Montgomery, J. D. Esko, and P. G. Spear. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 116:1273-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 18.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi, K., H. Nakanishi, M. Miyahara, K. Mandai, K. Satoh, A. Satoh, H. Nishioka, J. Aoki, A. Nomoto, A. Mizoguchi, and Y. Takai. 1999. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J. Cell Biol. 145:539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terry-Allison, T., R. I. Montgomery, M. S. Warner, R. J. Geraghty, and P. G. Spear. 2001. Contributions of gD receptors and glycosaminoglycan sulfation to cell fusion mediated by herpes simplex virus 1. Virus Res. 74:39-45. [DOI] [PubMed] [Google Scholar]
- 21.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2 and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 22.Yoon, M., A. Zago, D. Shukla, and P. G. Spear. 2003. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors, HVEM, nectin-2, and 3-O-sulfated heparan sulfate, but not with nectin-1. J. Virol. 77:9221-9231. [DOI] [PMC free article] [PubMed]