Abstract
Naturally occurring polymorphisms in the protease of human immunodeficiency virus type 1 (HIV-1) subtype C would be expected to lead to adaptive (compensatory) changes in protease cleavage sites. To test this hypothesis, we examined the prevalences and patterns of cleavage site polymorphisms in the Gag, Gag-Pol, and Nef cleavage sites of C compared to those in non-C subtypes. Codon-based maximum-likelihood methods were used to assess the natural selection and evolutionary history of individual cleavage sites. Seven cleavage sites (p17/p24, p24/p2, NC/p1, NC/TFP, PR/RT, RT/p66, and p66/IN) were well conserved over time and in all HIV-1 subtypes. One site (p1/p6gag) exhibited moderate variation, and four sites (p2/NC, TFP/p6pol, p6pol/PR, and Nef) were highly variable, both within and between subtypes. Three of the variable sites are known to be major determinants of polyprotein processing and virion production. P2/NC controls the rate and order of cleavage, p6gag is an important phosphoprotein required for virion release, and TFP/p6pol, a novel cleavage site in the transframe domain, influences the specificity of Gag-Pol processing and the activation of protease. Overall, 58.3% of the 12 HIV-1 cleavage sites were significantly more diverse in C than in B viruses. When analyzed as a single concatenated fragment of 360 bp, 96.0% of group M cleavage site sequences fell into subtype-specific phylogenetic clusters, suggesting that they coevolved with the virus. Natural variation at C cleavage sites may play an important role, not only in regulation of the viral cycle but also in disease progression and response to therapy.
One of the most dramatic changes in the human immunodeficiency virus type 1 (HIV-1)-AIDS epidemic has been the rapid emergence and devastating spread of subtype C viruses (16; http://www.unaids.org/epidemicupdatedec01/report /index html). HIV-1 C now accounts for >56% of all circulating viruses and is the most commonly transmitted subtype worldwide (9). Subtype C predominates in southern Africa (11, 20, 40) and India (29) and is increasing in frequency in China (10, 28) and Brazil (4, 10a, 32). The disproportionate increase in C viruses relative to other HIV-1 strains suggests that subtype C may be more easily transmitted or that it has a higher level of “fitness” at the population level. One possible explanation is that founder effects relating to the ongoing introduction of subtype C into new population groups with different host factors, or different social and sexual practices, may be responsible for the rapid spread. However, founder and host effects cannot account for the fact that C viruses are overtaking preexisting virus subtypes in several different geographical regions, including Yunnan Province in China and the southern region of Brazil (4, 10, 28, 32). It is increasingly evident that additional (nonhost) viral factors are also contributing to the rapid spread of HIV-1 C.
Viral studies indicate that subtype C has distinct genetic and phenotypic properties that differentiate it from other HIV-1 subtypes. Various studies have postulated that an extra NF-κB binding site in the long terminal repeat (28), a prematurely truncated Rev protein (10, 28), or a 5-amino-acid insertion in Vpu (18) may influence viral gene expression, altering the transmissibility and pathogenesis of C viruses (34). Factors related to viral entry and pathogenesis, such as the CCR5 and non-syncytium-inducing properties of C isolates (2, 21, 25), may also contribute to the increased spread of C viruses. One area of research that is receiving consideration is the possibility that C viruses have a more active, catalytically efficient protease (41).
The C protease is highly conserved at the amino acid level and has a distinct signature sequence that differentiates it from those of subtypes A, B, and D (11, 41). A subset of these signature residues, present in the hinge (M36I/R41K/H69K) and α-helix (I89M) of C (and A) proteases, has been linked to increased catalytic activity (41). Another signature pattern, identified in >80% of C viruses from South Africa, is T12S/T15V/L19I/I93L (11). The 12S, 15V, and 19I residues of this motif are located near the N terminus of the protease in an extended β-chain. The 93L polymorphism is located within a hydrogen-bonded turn immediately upstream from the protease-reverse transcriptase (RT) cleavage site, in close proximity to 12S/15V/19I and the dimerization domain (22, 30, 33, 35). All of these polymorphisms lie outside the catalytic site of the protease, in regions that would be expected to alter the enzyme's activity toward its natural cleavage sites, leading to adaptive (compensatory) changes in the cleavage site itself.
Since protease inhibitors (PIs) are currently the most active antiretroviral drugs used for the treatment of HIV-1 (44), it is important to collect information, not only on the C protease but also on its drug responsiveness, substrate-inhibitor specificities, and cleavage site characteristics. This information is needed to design PIs that are maximally effective against C viruses and to obtain new insights into the mechanism of drug resistance. Studies have shown that resistance mutations in the B protease are associated with impaired proteolytic processing, decreased enzymatic activity, and a failure to produce mature infectious virions (6, 7, 44). Compensatory cleavage site mutations can partially compensate for these defects (47). In this report, we describe the natural variability of subtype C cleavage sites in viruses from Africa, India, and Brazil and compare the results to cleavage site patterns in representative B and group M viruses.
MATERIALS AND METHODS
Cleavage site characteristics.
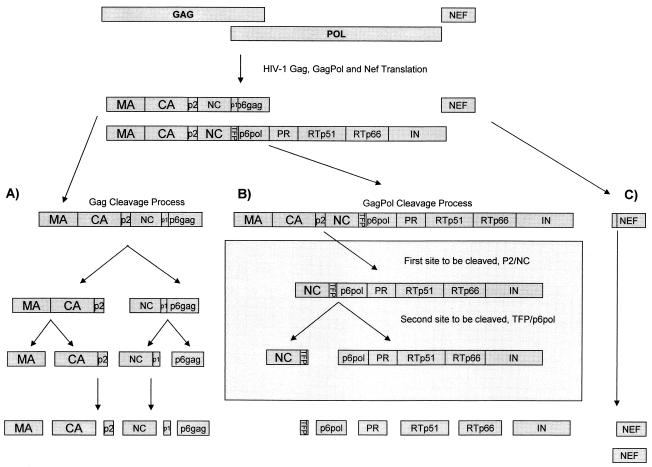
The HIV-1 protease is a small, 99-amino-acid aspartic enzyme that mediates the cleavage of Gag, Gag-Pol, and Nef precursor polyproteins. These reactions occur late in the viral life cycle, during virion assembly and maturation at the cell surface. The process is highly specific, temporally regulated, and essential for the production of infectious viral particles (13-15, 33). As shown in Fig. 1, the main structural proteins are formed by cleavage of the Pr55gag polyprotein into matrix (MA; p17), capsid (CA; p24), nucleocapsid (NC; p7), p6gag, and two spacer peptides, p2 and p1. The viral enzymes are formed by cleavage of Pr160gag-pol, a fusion protein derived by ribosomal frame shifting (13). Although Pr160gag-pol also contains p17, p24, and p2, its C-terminal cleavage products are NC, a transframe protein (TFP), p6pol, protease (PR), reverse transcriptase (RTp51), RNase H (RTp66), and integrase (IN) (12, 35). In total, 12 proteolytic reactions are required to generate a mature infectious virion. Each reaction occurs at a unique cleavage site that differs in amino acid composition (3). Some cleavage sites contain phosphorylated Ser/Thr or Tyr residues that alter the sites' susceptibilities to cleavage (39). P6gag, the major phosphoprotein of HIV-1, plays an essential role in the release of virus from the membranes of infected cells (19).
FIG. 1.
Schematic of the Gag and Gag-Pol processing sites showing the 12 individual protease cleavage sites: 5 cleavage sites in Gag (p17/p24, p24/p2, p2/NC, p7/p1, and p1/p6gag), 6 cleavage sites in Gag-Pol (NC/TFP, TFP/p6pol, p6pol/PR, PR/RT, RT/p66, and p66/IN), and a single site in Nef. The frequency of amino acid substitution at each of these cleavage sites is shown in Fig. 2 and Tables 3 and 4.
Sequence data and construction of cleavage site fragments.
A total of 84 full-length nucleotide sequences were selected for analysis. These sequences included two C isolates from South Africa, TV001 and TV002 (46), in addition to another 25 subtype C, 30 subtype B, and 27 representative group M reference sequences (including A [n = 3], B [n = 4], C [n = 5], D [n = 3], F1 and F2 [n = 4], G [n = 2], H [n = 2], J [n = 2], and K [n = 2] subtypes) extracted from the Los Alamos database (Table 1) (16). Sequences were selected based on the patient being treatment naive. Since the prevalence of drug resistance in untreated patients has been reported to range from 1 to 11% (17), sequences were also screened and excluded from the study if they were found to contain primary resistance mutations. The majority of the sequences were obtained by direct DNA PCR amplification and cloning of peripheral blood mononuclear cells. Nucleotide sequences were aligned by CLUSTAL W (37) and manually edited with the codon alignment of the Genetic Data Environment (GDE version 2.2) program (31). Calculation of the pairwise distance matrix, phylogenetic inference, and tree construction were performed on a dual-processor Linux computer by using the PAUP version 4.0b2a program (Sinauer Associates, Sunderland, Mass.) and a GDE for Linux HIV-1 interface (6a). Thirty-base-pair segments, consisting of 15 nucleotides (5 amino acids) on each side of the 12 cleavage sites, were extracted and concatenated into a 360-bp nucleotide sequence.
TABLE 1.
Sequences used for the analysis of protease cleavage sitesa
| Group or subtype | Data set | Accession no. |
|---|---|---|
| M | A1.KE.93.Q23-17 | AF004885 |
| A1.UG.85.U455 | M62320 | |
| A1.UG.92.92UG037 | U51190 | |
| B.FR.83.HXB2 | K03455 | |
| B.US.83.RF | M17451 | |
| B.US.86.JRFL | U63632 | |
| B.US.90.WEAU160 | U21135 | |
| C.BR.92.92BR025 | U52953 | |
| C.BW.96.96BW0502 | AF110967 | |
| C.ET.86.ETH2220 | U46016 | |
| D.CD.83.ELI | K03454 | |
| D.CD.83.NDK | M27323 | |
| D.CD.84.84ZR085 | U88822 | |
| F1.BE.93.VI850 | AF077336 | |
| F1.BR.93.93BR020.1 | AF005494 | |
| F2.CM.95.MP255 | AJ249236 | |
| F2.CM.95.MP257 | AJ249237 | |
| C.ZA.98.TV001c8.5 | AY16222 | |
| C.ZA.98.TV002c12 | AY16224 | |
| G.BE.96.DRCBL | AF084936 | |
| G.FI.93.HH8793-12.1 | AF061641 | |
| H.BE.93.VI991 | AF190127 | |
| H.BE.93.VI997 | AF190128 | |
| J.SE.93.SE7887 | AF082394 | |
| J.SE.94.SE7022 | AF082395 | |
| K.CD.97.EQTB11C | AJ249235 | |
| K.CM.96.MP535 | AJ249239 | |
| B | B.US.84.NY5CG | M38431 |
| B.US.-.AD8 | AF004394 | |
| B.CN.-.RL42 | U71182 | |
| B.US.90.WEAU160 | U21135 | |
| B.US.-.P896 | U39362 | |
| B.GA.-.OYI | M26727 | |
| B.US.-.DH123 | AF069140 | |
| B.GB.-.CAM1 | D10112 | |
| B.NL.86.3202A21 | U34604 | |
| B.AU.87.MBC925 | AF042101 | |
| B.ES.89.89SP061 | AJ006287 | |
| B.FR.83.HXB2 | K03455 | |
| B.US.83.SF2 | K02007 | |
| B.US.90.WCIPR9018 | U69591 | |
| B.US.84.MNCG | M17449 | |
| B.US.87.BC | L02317 | |
| B.TW.-.TWCYS | AF086817 | |
| B.US.86.JRFL | U63632 | |
| B.AU.86.MBC200 | AF042100 | |
| B.DE.86.HAN | U43141 | |
| B.US.83.RF | M17451 | |
| B.US.97.ARES2 | AB078005 | |
| B.AU.96.MBCC98 | AF042104 | |
| B.KO.97.WK | AF224507 | |
| B.AR.98.ARCH054 | AY037268 | |
| B.AR.00.ARMS008 | AY037269 | |
| B.AR.99.ARMA132 | AY037282 | |
| B.DE.86.D31 | U43096 | |
| B.US.88.WR27 | AF286365 | |
| B.US.86.YU2 | M93258 | |
| C | C.ET.86.ETH2220 | U46016 |
| C.BR.92.BR025 | U52953 | |
| C.IN.93.93IN999 | AF067154 | |
| C.IN.93.93IN904 | AF067157 | |
| C.IN.93.93IN905 | AF067158 | |
| C.IN.94.94IN11246 | AF067159 | |
| C.IN.94.94IN476 | AF286223 | |
| C.IN.95.95IN21068 | AF067155 | |
| C.BW.96.96BW01B03 | AF110959 | |
| C.BW.96.96BW0402 | AF110962 | |
| C.BW.96.96BW0502 | AF110967 | |
| C.BW.96.96BW1104 | AF110969 | |
| C.BW.96.96BW1210 | AF110972 | |
| C.BW.96.96BW17B03 | AF110980 | |
| C.BW.96.96BW1626 | AF110978 | |
| C.BW.96.MJ4 | AF321523 | |
| C.ZM.96.ZM651 | AF286224 | |
| C.ZM.96.ZM751 | AF286225 | |
| C.ZA.97.ZA012 | AF286227 | |
| C.BR.98.98BR004 | AF286228 | |
| C.IL.98.98IS002 | AF286233 | |
| C.IN.98.98IN012 | AF286231 | |
| C.IN.98.98IN022 | AF286232 | |
| C.TZ.98.98TZ013 | AF286234 | |
| C.TZ.98.98TZ017 | AF286235 | |
| C.ZA.98.TV001c8.5 | AY16222 | |
| C.ZA.98.TV002c12 | AY16224 |
Reconstruction of ancestral cleavage site sequences.
To examine the evolutionary histories of individual cleavage sites, Phylogenetic Analysis under Maximum Likelihood (PAML) software (26) was used to identify amino acid and nucleotide substitutions along each branch of the tree. Branch lengths were estimated using a nucleotide substitution model; amino acid sequences were deduced from the reconstructed nucleotide triplets. The analyses involved the use of maximum-likelihood methods and a time-reversible model which assume different substitution rates, base frequencies, and transition/transversion rate ratios (kappa) (42, 43). Using this approach, we were able to reconstruct the ancestral sequences and internal nodes for each of the 12 protease cleavage sites in the B, C, and M group data sets. The number of proximal ancestors for each data set was n − 1, which translated into 29 ancestral sequences for subtype B, 26 sequences for subtype C, and 26 sequences for the group M viruses. The most recent common ancestor (MRCA) nucleotide sequence for each virus in the three data sets was saved and translated into its corresponding amino acids.
Diversity and cleavage site polymorphisms.
Nucleotide diversity at cleavage sites was measured using a Kimura 2-α parameter model with a distance matrix implemented in the MEGA program version 2.0 (Arizona State University, Tempe). Amino acid diversity was measured using a Poisson distribution method implemented in the same MEGA package. P values for diversity measurements were calculated by applying the t test to the distance matrix of each data set. To determine whether the sequences had evolved over time, amino acid profiles for individual Gag, Gag-Pol, and Nef cleavage sites were compared to the inferred MRCA for that site.
Assessment of positive selection pressure.
Nucleotide sequences were also analyzed with Codeml, a program from the PAML software package (26). The likelihood ratio test (1) and recently developed codon-based models (42, 43) were used to assess natural selection and adaptive evolution at the amino acid level. These selection models use maximum-likelihood scores to account for variation in the dn/ds (nonsynonymous/synonymous) ratio (ω) at individual codons along the length of the sequence. High rates of synonymous mutation are indicative of conservation and a strict requirement for biological function, while high rates of nonsynonymous substitution are indicative of adaptive change in response to host selection pressure. An individual amino acid was considered to be positively selected if the dn/ds ratio was significantly greater than 1.0.
RESULTS
Viral characteristics.
No primary RT- or PI-resistant mutations were detected among the 84 full-length sequences selected for study. Although attempts were made to include only sequences amplified directly from HIV-1 proviral DNA and to match these sequences based on duration of infection, plasma viral load, and CD4 count, this proved difficult. A surprisingly small amount of full-length sequence data was available from treatment-naive patients infected with subtype B, and when available, it was often poorly annotated. Despite these limitations, the frequency and pattern of naturally occurring polymorphisms observed in this study were remarkably similar to those reported for a control group of subtype B infections treated with nucleoside reverse transcriptase inhibitors but not with PIs or nonnucleoside reverse transcriptase inhibitors (5). Most of the non-C sequences came from regions of the world where treatment is not yet readily available.
Genetic diversity and patterns of amino acid variability at individual cleavage sites.
Variation at the 12 cleavage sites of subtypes B and C and group M is shown in Table 2. Seven (58.3%) sites (p17/p24, p24/p2, NC/p1, NC/TFP, PR/RT, RT/p66, and p66/IN) were found to be relatively well conserved, both over time and between subtypes, with a mean intrasubtype distance ranging from 0.40 ± 0.20% to 7.49 ± 5.62%. The remaining five (41.7%) sites exhibited moderate (p1/p6gag) to extensive (p2/NC, TFP/p6pol, p6pol/PR, and Nef) variation, with mean intrasubtype diversities reaching levels as high as 42.42 ± 15.16%. For the purposes of this study, we have referred to these three patterns as conserved, moderately variable, and variable. Polymorphisms were more common among C than B viruses (P < 0.0001). Overall, six cleavage sites (p17/p24, p2/NC, NC/p1, PR/RT, RT/p66, and Nef) had signficantly higher levels of diversity among C viruses (P < 0.0001); five sites (p24/p2, p1/p6gag, NC/TFP, p6pol/PR, and p66/IN) had similar levels of diversity in both subtypes, and one site, TFP/p6pol, was highly variable among B and group M viruses (mean distances, 16.7 and 24.7%, respectively) but relatively conserved in C viruses (mean distance, 7.6%) (P < 0.0001).
TABLE 2.
Inter- and intrasubtype diversity at 12 cleavage sites, expressed as amino acid distances between sequences
| Protease sitea | Mean % distance
|
P value
|
||||
|---|---|---|---|---|---|---|
| Subtype B | Subtype C | M group | B vs C | B vs M | C vs M | |
| p17/p24 | 2.31 | 5.15 | 2.23 | <0.0001 | 0.806 | <0.0001 |
| p24/p2 | 2.41 | 3.07 | 2.67 | 0.077 | 0.454 | 0.267 |
| p2/NC | 18.66 | 42.42 | 39.22 | <0.0001 | <0.0001 | 0.062 |
| NC/p1 | 0.40 | 5.42 | 5.22 | <0.0001 | <0.0001 | 0.7244 |
| p1/p6gag | 8.93 | 9.81 | 14.47 | 0.318 | <0.0001 | <0.0001 |
| NC/TFP | 5.30 | 3.90 | 6.30 | 0.0012 | 0.0215 | <0.0001 |
| TFP/P6pol | 16.69 | 7.60 | 24.70 | <0.0001 | <0.0001 | <0.0001 |
| p6pol/PR | 17.66 | 16.28 | 16.55 | 0.15 | 0.019 | 0.365 |
| PR/RT | 0.70 | 1.56 | 2.93 | <0.0001 | <0.0001 | <0.0001 |
| RT/p66 | 0.81 | 7.49 | 6.03 | <0.0001 | <0.0001 | 0.004 |
| p66/IN | 2.44 | 2.22 | 2.24 | 0.524 | 0.61 | 0.961 |
| Nef | 11.89 | 24.52 | 25.68 | <0.0001 | <0.0001 | 0.33 |
| Gag (501 aa) | 6.99 | 9.75 | 15.72 | <0.001 | <0.001 | <0.001 |
| Pol (1,004 aa) | 6.02 | 5.83 | 5.89 | 0.376 | 0.419 | 0.875 |
| Nef (207 aa) | 14.66 | 16.50 | 18.50 | 0.07 | <0.001 | <0.001 |
| 12 protease cleavage sites (360 bp; 120 aa) | 4.80 | 10.10 | 12.10 | <0.001 | <0.001 | <0.001 |
aa, amino acids.
Compared to the M data set, the level of polymorphism at C cleavage sites was as wide ranging as that observed for the entire M group, a data set containing nine different HIV-1 subtypes. One cleavage site, p17/p24, was significantly more diverse in subtype C (mean divergence, 5.15%) than in group M (mean divergence, 2.23%) and subtype B (mean divergence, 2.31%) viruses (<0.0001 for both comparisons). Seven sites (p24/p2, p2/NC, NC/p1, p6pol/PR, RT/p66, p66/IN, and Nef) exhibited similar levels of diversity in both data sets (P = 0.004 to 0.961). Only four group M cleavage sites (p1/p6gag, NC/TFP, TFP/p6pol, and PR/RT) had mean diversities that were significantly greater than that observed for subtype C (P < 0.0001). For >50% of the sites, the variability of B viruses was significantly lower than that observed for subtype C or group M (P < 0.0001), despite the fact that the B viruses covered a broader time frame.
The observed polymorhpisms were not randomly distributed across the variable cleavage sites but were confined to specific amino acids, most of which were positively selected in C but not in B viruses (Fig. 2). The least variable residues were the P1 positions of p1/p6gag, TFP/p6pol, and Nef and the P1′ positions of p6pol/PR and Nef. As shown, these positions flank the scissile bond, with P1 located immediately upstream and P1′ located immediately downstream of the cleavage junction. The most variable positions were the P1, P3, P4, P5, P3′, and P5′ residues of p2/NC; the p3′ residue of p1/p6gag; the P1′ and P4′ residues of TFP/p6pol; the P1 to P4 residues of p6pol/PR; and the P3 to P5 and P2′ residues of Nef.
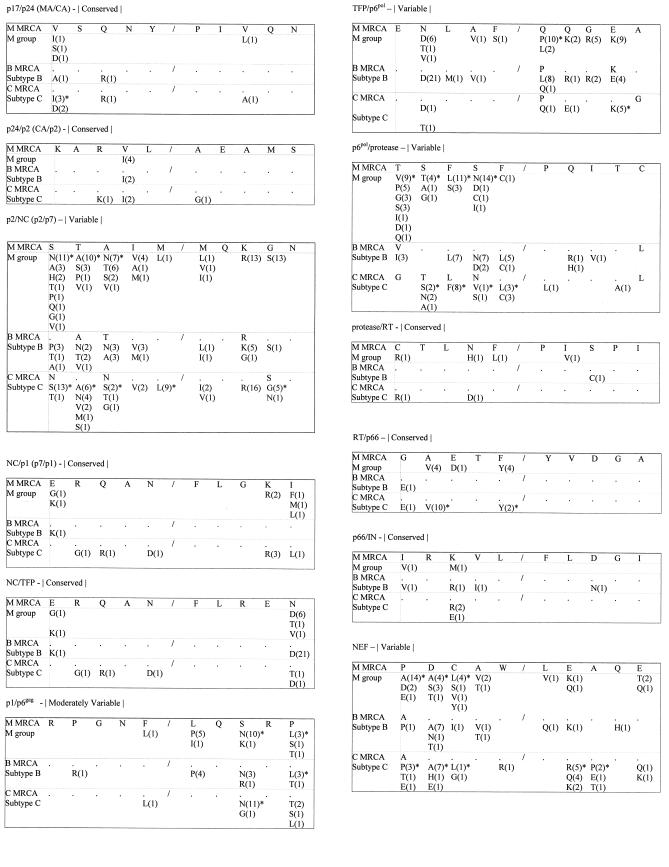
FIG.2.
Amino acid polymorphisms at Gag, Gag-Pol, and Nef cleavage sites. The letters refer to the amino acid substitutions; the numbers in parentheses refer to the number of times the substitution was observed. Each cleavage site sequence consists of the 5 amino acids upstream and the 5 amino acids downstream of the scissile bond, indicated by a shill. The labeling of amino acids is according to the convention of P1 to P5 going from the scissile bond toward the amino terminus and P1′ to P5′ going toward the carboxy terminus. Positively selected amino acids are marked with asterisks. Dots represent amino acids that are identical to those in the M MRCA.
Subtyping and phylogenetic-tree analysis.
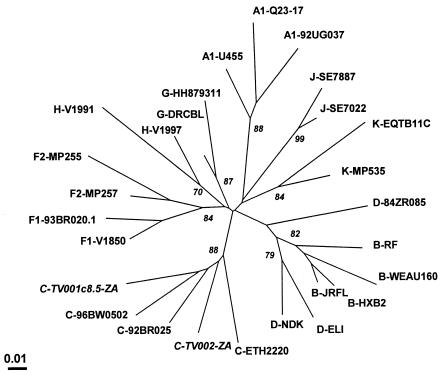
When subjected to phylogenetic analysis (Fig. 3), the concatenated 360-bp fragments of the group M data set fell into eight subtype-specific clusters representing subtypes A, B and D, C, F, G, H, J, and K, with the cleavage sequences for subtypes B and D segregating together in the same subcluster. This pattern was supported by high bootstrap values, by high-score maximum-likelihood trees, and by phylogenetic analysis of the deduced amino acids. These findings reveal the subtype-specific nature of protease cleavage sites and suggest that the evolution of cleavage sites parallels that of the full-length genome. With the exception of C.98IN022, B.AR00.ARMS008, and B.US.P896, all of the cleavage site fragments in the B and C data sets segregated into two distinct monophyletic groups representing either subtype B or subtype C viruses (data not shown). The longer branch lengths in the C subcluster were reflective of the increased diversity of C viruses relative to subtype B.
FIG. 3.
Phylogenetic relationships of the South African Tygerberg virology (TV) cleavage site sequences relative to other subtypes in the group M data set. This representative maximum-likelihood tree is based on concatenation and analysis of the 12 protease site nucleotide sequences as a single segment of 360 bp. An indication of the degree of sequence dissimilarity is given by the distance from the central node. The percentage of bootstrap trees out of 1,000 replications supporting a particular phylogenetic group is shown alongside the node considered.
Identification and dating of common ancestors.
Maximum-likelihood methods were next used to reconstruct the internal nodes of the phylogenetic tree and to estimate the times of divergence of individual sequences from their MRCA. These estimates were determined by measuring the number of substitutions along each branch of the tree. MRCAs for the B, C, and group M data sets are shown in Fig. 2. Two different patterns were observed based on the relationship between a given sequence and its MRCA. Conserved (n = 7) and moderately variable (n = 1) cleavage sites shared the same (identical) MRCA among all three data sets. The proximal location of the MRCA relative to the root of the tree suggests that, for these sequences, cleavage site diversification occurred after subtype divergence. In contrast, variable cleavage sites (n = 4) showed a high degree of divergence both from their subtype-specific MRCA and from the group M MRCA. Ancestral nodes for the variable sites were located closer to the tips of the tree (data available upon request).
Variability of cleavage sites relative to other regions of the HIV-1 genome.
For C viruses, the average intersequence divergence among the concatenated cleavage site fragments was higher (10.1%) than those observed for the Gag (9.8%) and Pol (5.8%) proteins but lower (16.5%) than that observed for Nef. In contrast, B cleavage sites were significantly less diverse (4.8%; P < 0.0001) than those of the Gag (7.0%), Pol (6.0%), and Nef (14.7%) proteins of subtype B. Among group M viruses, cleavage site diversity was significantly higher (12.1%) than that calculated for Pol (5.9%) but lower than that determined for Gag (15.7%) and Nef (18.5%). These results are presented in more detail in Table 2.
Physical-chemical properties of amino acids at P1-P1′ cleavage junctions.
Overall, excluding the highly conserved asparagine (N) residue at the P1 position of NC/p1 and NC/TFP, >97.0% of P1-P1′ amino acids in group M were nonpolar. Of these, 77.8% were hydrophobic, 21.5% were small amino acids (78.9% proline, 20.8% alanine, and 0.3% glycine), 0.3% were polar uncharged (one serine and three glutamine), 0.2% were polar charged (one arginine and one aspartic acid), and 0.3% were ungrouped (cysteine) residues. These amino acids were localized at specific positions within the cleavage sequence. The small amino acids were localized primarily to the P1′ position of p17/p24, p24/p2, TFP/p6pol, p6pol/PR, and PR/RT. Hydrophobic amino acids were concentrated at the P1-P1′ junction. As previously reported for HIV-1 B (23), the P1-P1′ amino acids of subtype C fell into two different patterns defined by the size of the P1′ amino acid: type I, represented by p2/NC and p1/p6gag, and type II, represented by p17/p24 and p24/p2. Both types carried large nonpolar, hydrophobic amino acids (leucine, tyrosine, phenylalanine, and methionine) at position P1 and either a large (type I) or small (type II) hydrophobic amino acid (proline, alanine, or glycine) at P1′.
MRCAs and subtype-specific signature patterns.
A summary of amino acid signature patterns relative to the subtype B and C and group M MRCAs is shown in Tables 3 and 4. Mutations at cleavage sites defining the enzymatic (PR/RT, RT/p66, and p66/IN) and structural (p17/24, p24/p2, and NC/p1) components of HIV-1 were relatively uncommon and, when detected, were found at greater frequencies among C versus B viruses. In total, only 1.2, 0.8, and 1.1% of the 840 amino acids at each of the PR/RT, p66/IN, and p24/p2 cleavage sites carried substitutions. As a result, the majority of sequences at conserved sites (81.5 to 96.7%) were identical to both the subtype-specific MRCA and the common ancestor of the group M viruses. Several cleavage sites involved in the regulation of polyprotein processing and protease activation, p2/NC, TFP/p6pol, and p6pol/PR, were highly variable and differed significantly from both the subtype-specific and group M MRCAs. With the exception of the TFP/p6pol site, which was more variable in subtype B, divergence from the MRCA was greatest for C and M viruses. None of the p2/NC and p6pol/PR sequences in the group M data set was identical to the M MRCA signatures for p2/NC and p6pol/PR, STAIM/MQKGN and TSFSF/PQITC, respectively.
TABLE 3.
Relationship between cleavage site signature patterns and common ancestorsa
| Group or subtype | No. (%) of sequence with same MRCA
|
|||||||
|---|---|---|---|---|---|---|---|---|
| p17/p24 (MA/CA) VSQNY/PIVQN | p24/p2 (CA/p2) KARVL/AEAMS | NC/p1 (p7/p1) ERQAN/FLGKI | NC/TFP ERQAN/FLREN | p1/p6gag RPGNF/LQSRP | PR/RT CTLNF/PISPI | RT/p66 GAETF/YVDGA | p66/IN IRKVL/FLDGI | |
| M | 23 (85.2) | 23 (85.2) | 21 (77.7) | 17 (63.0) | 13 (48.1) | 22 (81.5) | 19 (70.4) | 25 (92.6) |
| B | 28 (93.3) | 28 (93.3) | 29 (96.7) | 8 (26.7) | 23 (76.7) | 29 (96.7) | 29 (96.7) | 26 (86.7) |
| C | 21 (77.7) | 23 (85.2) | 21 (77.7) | 22 (81.5) | 15 (55.6) | 25 (92.6) | 14 (51.9) | 24 (88.9) |
Conserved and moderately variable cleavage sites (group M MRCA is identical to subtype B and C MRCAs.
TABLE 4.
Relationship between cleavage site signature patterns and common ancestorsa
| Group or subtype | p2/NC (p2/p7)
|
TFP/p6pol
|
p6pol/PR
|
NEF
|
||||
|---|---|---|---|---|---|---|---|---|
| Sequence | No. (%) same | Sequence | No. (%) same | Sequence | No. (%) same | Sequence | No. (%) same | |
| Group M | STAIM/MQKGN | 0 (0) | ENLAF/QQGEA | 6 (22.2) | TSFSF/PQITC | 0 (0) | PDCAW/LEAQE | 5 (18.5) |
| Subtype B | SATIM/MQRGN | 13 (43.3) | ENLAF/PQGKA | 5 (16.7) | VSFSF/PQITL | 13 (43.3) | ADCAW/LEAQE | 18 (60.0) |
| Subtype C | NTNIM/MQKSN | 3 (11.1) | ENLAF/PQGEG | 21 (77.8) | GTLNF/PQITL | 10 (37.0) | ADCAW/LEAQE | 6 (22.2) |
Variable cleavage sites (subtype B and C MRCAs differ from the group M MRCA.
Positive selection of amino acids at protease cleavage sites.
The concatenated 360-bp cleavage site fragments were next compared internally to assess the mutational behavior of P1-P5 and P1′-P5′ sites in the absence of drug therapy. As described in Materials and Methods, the analyses were performed using codon-based maximum-likelihood methods that test for the variation in selection pressure (dn/ds) at individual amino acids along the length of the sequence. Application of the likelihood ratio test indicated that the best-fit model for subtype C and group M sequences was the positive-selection (discrete) model (χ2 = 54.47; P < 0.0001 and 62.34 and P < 0.0001, respectively), while for B viruses the neutral model performed as well as the positive model (χ2 = 1.2; P > 0.05). Overall, when analyzed as a single concatenated fragment, all three datasets were found to be under negative (purifying) selection, with dn/ds (ω) values ranging from 0.26 to 0.31 for all sites. Only 20 (16.6%) of the 120 amino acids within the 12 cleavage sites of subtype C were found to be under positive selection, with a ω3 value of 1.7. Group M and subtype B cleavage sites had fewer positively selected amino acids: 11.1 and 6.7%, respectively (Fig. 2).
DISCUSSION
The presence of polymorphisms in the protease of subtype C would be expected to result in adaptive (compensatory) changes in the natural cleavage sites that are recognized and cleaved by the C enzyme. To test this hypothesis, we examined the prevalences and patterns of cleavage site mutations in the Gag, Gag-Pol, and Nef proteins of subtype C compared to those of non-C viruses. Using phylogenetic and ancestral reconstruction methods, we identified two groups of natural cleavage site sequences. The first group consisted of eight sequences, seven of which were highly conserved in all HIV-1 subtypes, and an eighth sequence which was moderately variable. Mutations at these sites were uncommon and, when present, were observed at relatively equivalent frequencies among different subtypes. These cleavage sites, which defined the main structural (MA, CA, and NC) and enzymatic (RT, RNase H, and integrase) proteins of HIV-1, were under strong negative (purifying) selection pressure, had a common ancestor, and showed little genetic evolution over time. The second group consisted of five cleavage sites that were under positive (diversifying) selection pressure, exhibited extensive inter- and intrasubtype variability, and showed little (or no) resemblance to the common ancestor of group M or to the subtype-specific MRCA.
Our data showing that the majority (58.3%) of cleavage sites are highly conserved in all subtypes were not unexpected, given the narrow specificity of the HIV-1 protease relative to cellular proteases, such as pepsin (27). The strong purifying selection pressure exerted on these sites is presumably a reflection of the need to maintain the spatial configuration of the enzyme-substrate complex, conserve the hydrophobic nature of the scissile bond, and retain the biological activity of functionally important sites, such as the P1′ proline of p17/p24 and the P1 and P1′-P5′ residues of NC/p1. Cleavage of the P17/p24 site is known to play an important role in virion maturation, while processing of NC/p1 is required for ribosomal frame shifting and Gag-Pol expression (8, 13-15, 24). In B viruses, cleavage of p17/24, p24/p2, and NC/p1 has been shown to be suboptimal, with the NC/p1 site being rate limiting (23). It has been suggested that the slow, regulated cleavage of these structural proteins may represent a common strategy to ensure that the assembled virions have the full complement of proteins needed to bud from the cell surface, bind to a new cell, and initiate a new round of viral replication (35).
The carboxyl terminus of NC is particularly interesting. Unlike other cleavage sites, which carry an aromatic amino acid at P1 and either a leucine or proline residue at P1′, the C termini of NC/p1 and NC/TFP carry an asparagine (N) residue at P1 opposite a phenylalanine (F) residue at P1′ (5, 22-24). In this study of 84 untreated patients, no mutations were detected at the P2 or P1′-P3′ positions of NC/p1 or NC/TFP and only a single N→C mutation was detected at P1. Taken together, these findings underscore the unique nature and limited mutability of the NC/p1 and NC/TFP cleavage junctions. Although these sites were strongly conserved in natural infection, recent studies have shown that an A→V substitution at the P2 positions of NC/p1 and NC/TFP is a common adaptive change, occurring in 29% of PI-resistant patients taking indinavir, saquinavir, and/or ritonavir for the treatment of subtype B (5, 7, 44, 45). This valine substitution is frequently associated with an M46 I or L mutation (and possibly a V82 mutation) in the protease and leads to altered polyprocessing and improved growth of protease-mutated viruses. Whether similar “second-locus” mutations will be observed during the treatment of non-B subtypes remains to be established. The identification of common patterns may facilitate the development of broad-based inhibitors with increased specificity and improved binding to the mutated protease. These secondary inhibitors might preempt (or delay) the emergence of resistance.
Our analyses also revealed important differences among HIV-1 subtypes. Particularly intriguing was the identification of five cleavage sites that exhibited extensive variability across all subtypes, with C viruses being significantly more variable than subtype B. Variation was restricted to a few specific amino acids, most of which were positively selected in C but not in B viruses. In contrast to conserved sites, variable cleavage sites tended to be those with regulatory rather than structural or enzymatic functions. At least four of the variable sites (p2/NC, p1/p6gag, TFP/p6pol, and p6pol/PR) are known to play major roles in the regulation of polyprotein processing and, in the case of TFP/p6pol, in the activation of the protease enzyme (22-24, 27, 30). Studies of subtype B have shown that p2/NC is the initial and most rapidly processed cleavage site, controlling both the rate and the order of Gag and Gag-Pol polyprocessing (30). Our results indicate that p2/NC is by far the most variable cleavage site, with intrasubtype diversity ranging from 18.7% in subtype B to levels of 42.4% in subtype C.
The p1/p6gag cleavage product, p6gag, is a major phosphoprotein that is critical to the release of mature, infectious virions (19). Although not well studied, phosphorylation of Gag and Gag-Pol sequences has been shown to alter susceptibility to cleavage, attenuating or even preventing the proteolytic process (38). The TFP/p6pol cleavage site, defining the N terminus of p6pol, was the only site to have a significantly higher level of diversity among B than among C viruses. TFP/p6pol is a novel cleavage site located 8 amino acids downstream from NC in the TFP domain of Gag-Pol (22, 35). Although TFP/p6pol lies outside (and upstream) of the protease, the EDL tripeptide of this cleavage site (ENL in the case of C viruses) has been postulated to have a major influence on protease activation and on the timing and specificity of Gag-Pol cleavage, delaying the release of the protease until after the viral particle has budded from the cell membrane. Such a mechanism may protect the cell from the cytotoxic effects of proteolysis (36, 38). The observed subtype variation in the cleavage sites controlling the initiation and rate of Gag and Gag-Pol processing (p2/NC) and the activation of protease (TFP/p6gag) suggests that there may be important differences in the way that B and C viruses regulate polyprocessing and virion assembly. Subtle cleavage site differences could, over time, have a major differential impact on the pathogenesis of HIV-1 subtypes and on response to therapy. Early treatment studies suggest that C viruses give an excellent initial response to highly active antiretroviral therapy but that the duration of the response may be less than that reported for B viruses.
In summary, our results point to important inter- and intrasubtype differences in protease cleavage sites, especially in the p2/NC, TFP/p6pol, and p6pol/PR sites. The main limitations of our study relate to the cross-sectional nature of the data sets and the limited availability of well-matched pretreatment controls for use in the B data set. Despite these limitations, the potential impact of our findings on HIV-1 disease progression and response to therapy warrants further investigation, both at the patient level and in vitro using site-directed mutagenesis. The separate monophyletic clustering of B and C cleavage sites suggests that cleavage sites have evolved in a subtype-specific manner. The divergence between ancestral and contemporary sequences in the C data set and the location of an ancestral node distal to the group M MRCA suggest that variation in C cleavage sites began early, prior to the diversification of HIV-1 subtypes. A more detailed investigation of C cleavage sites, both over time and in response to therapy, is in progress. The present study forms the baseline for these ongoing studies.
Acknowledgments
This work was supported by grants from the Wellcome Trust (United Kingdom) (grant number 061238/2/00/2 to S.C.), the NIH HIV Vaccine Design and Development Team (contract number NO1-AI-05396 to S.W.B.), and the Poliomyelitis Research Foundation (PRF) and the Harry Crossley Foundation (to S.E.).
REFERENCES
- 1.Anisimova, M., J. P. Bielawski, and Z. Yang. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18:1585-1592. [DOI] [PubMed] [Google Scholar]
- 2.Ball, S. C., A. Abraha, K. R. Collins, A. J. Marozsan, H. Baird, M. E. Quinones-Mateu, A. Penn-Nicholson, M. Murray, N. Richard, M. Lobritz, P. A. Zimmerman, T. Kawamura, A. Blauvelt, and E. J. Arts. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:1021-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billich, S., M. T. Knoop, J. Hansen, P. Strop, J. Sedlacek, R. Mertz, and K. Moelling. 1988. Synthetic peptides as substrates and inhibitors of human immunodeficiency virus-1 protease. J. Biol. Chem. 263:17905-17908. [PubMed] [Google Scholar]
- 4.Brindeiro, R., B. Vanderborght, F. Caride, L. Correa, R. M. Oravec, O. Berro, L. Stuyver, and A. Tanuri. 1999. Sequence diversity of the reverse transcriptase of human immunodeficiency virus type 1 from Brazilian untreated individuals. Antimicrob. Agents Chemother. 43:1674-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cote, H. C. F., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.De Oliveira, T., R. Miller, M. Tarin, and S. Cassol. 2002. An integrated Genetic Data Environment (GDE)-based LINUX interface for the analysis of HIV-1 and other microbial sequences. Bioinformatics 19:1-2. [DOI] [PubMed]
- 7.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3736-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ericson-Viitanen, S., J. Manfredi, P. Viitanen, D. E. Tribe, R. Tritch, C. A. Hutchison III, D. D. Loeb, and R. Swanstrom. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retrovir. 5:577-591. [DOI] [PubMed] [Google Scholar]
- 9.Esparza, J., and N. Bhamarapravati. 2000. Accelerating the development and future availability of HIV-1 vaccines: why, when, where and how? Lancet 355:2061-2066. [DOI] [PubMed] [Google Scholar]
- 10.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Gonzales, L. J. M., R. M. Brindeiro, M. Gordon, A. Calazans, M. A. Soares, S. Cassol, and A. Tanuri. The subtype C of human immunodeficiency virus type 1 circulating in Brazil and South Africa presents a hypersusceptibility to the protease inhibitor Lopinavir. J. Antimicrob. Chemother. in press. [DOI] [PMC free article] [PubMed]
- 11.Gordon, M., T. De Oliveira, K. Bishop, H. M. Coovadia, L. Madurai, S. Engelbrecht, E. Janse van Rensburg, A. Mosam, and S. Cassol. 2003. Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implications for vaccine and antiretroviral control strategies. J. Virol. 77:2587-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikuta, K., S. Suzuki, H. Horikoshi, T. Mukai, and R. B. Luftig. 2000. Positive and negative aspects of the human immunodeficiency virus protease: development of inhibitors versus its role in AIDS pathogenesis. Microbiol. Mol. Biol. Rev. 64:725-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, H. E. Varmus. 1998. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of HIV-1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 68:6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krausslich, H., F. H. Ingraham, M. Skoog, E. Wimmer, P. V. Pallai, and C. A. Carter. 1989. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc. Natl. Acad. Sci. USA 86:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuiken, C., B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. Mullins, J. Sodroski, S. Wolinsky, and B. Korber. 2002. HIV sequence compendium. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 17.Little, S. J. 2000. Transmission and prevalence of HIV resistance among treatment-naïve subjects. Antivir. Ther. 5:33-40. [DOI] [PubMed] [Google Scholar]
- 18.McCormick-Davis, C., S. B. Dalton, D. K. Singh, and E. B. Stephens. 2000. Comparisons of Vpu sequences from diverse geographical isolates of HIV type 1 identifies the presence of highly variable domains, additional invariant amino acids and a signature sequence motif common to subtype C isolates. AIDS Res. Hum. Retrovir. 16:1089-1095. [DOI] [PubMed] [Google Scholar]
- 19.Muller, B., T. Patschinsky, and H. G. Krausslich. 2002. The late-domain-containing protein p6 is the predominant phosphoprotein of human immunodeficiency virus type 1 particles. J. Virol. 76:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung'u, M. Rahman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peeters, M., R. Vincent, J. L. Perret, M. Lasky, D. Patrel, F. Liegeois, V. Courgnaud, R. Seng, T. Matton, S. Molinier, and E. Delaporte. 1999. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:115-121. [DOI] [PubMed] [Google Scholar]
- 22.Pettit, S. C., S. Gulnik, L. Everitt, and A. H. Kaplan. 2003. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 77:366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettit, S. C., G. J. Henderson, C. A. Schiffer, and R. Swanstrom. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by viral protease. J. Virol. 76:10226-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenetics. Bioinformatics 16:395-399. [DOI] [PubMed] [Google Scholar]
- 27.Ridky, T. W., C. E. Cameron, J. Cameron, J. Leis, T. Copeland, A. Wlodawer, I. T. Weber, and R. W. Harrison. 1996. Human immunodeficiency virus, type 1 protease substrate specificity is limited by interactions between substrate amino acids in adjacent enzyme subsites. J. Biol. Chem. 271:4709-4717. [DOI] [PubMed] [Google Scholar]
- 28.Rodenburg, C. M., Y. Li, and S. A. Trask. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retrovir. 17:161-168. [DOI] [PubMed] [Google Scholar]
- 29.Shankarappa, R., R. Chatterjee, and C. Learn. 2001. Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. J. Virol. 75:10479-10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shehu-Xhilaga, M., H. G. Kraesslich, S. Pettit, R. Swanstrom, J. Y. Lee, J. A. Marshall, S. M. Crowe, and J. Mak. 2001. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 75:9156-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, S. W., R. Overbeek, C. R., Woese, W. Gilbert, and P. M. Gillevet. 1994. The Genetic Data Environment: an expandable GUI for multiple sequence analysis. Comput. Appl. Sci. 10:671-675. [DOI] [PubMed] [Google Scholar]
- 32.Soares, M. A., T. De Oliveira, R. M. Brindeiro, R. S. Diaz, E. C. Sabino, L. Brigido, I. L. Pires, M. G. Morgado, M. C. Dantas, D. Barreira, P. R. Teixeira, S. Cassol, A. Tanuri, and the Brazilian Network for Drug Resistance Surveillance. 2003. A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS 17:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Swanstrom, R., and J. W. Wills. 1997. Retroviral gene expression. II. Synthesis, processing, and assembly of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [PubMed]
- 34.Tatt, I. D., K. L. Barlow, A. Nicoll, and J. P. Clewley. 2001. The public health significance of HIV-1 subtypes. AIDS 15:S59-S71. [DOI] [PubMed] [Google Scholar]
- 35.Tessmer, U., and H.-G. Karausslich. 1998. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J. Virol. 72:3459-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, E. K., R. J. Connelly, S. Pennathur, L. Dubrovsky, O. K. Haffar, and M. I. Bukrinsky. 1996. Anti-idiotypic antibody to the V3 domain of gp120 binds to vimentin: a possible role of intermediate filaments in the early steps of HIV-1 infection cycle. Viral Immunol. 9:73-87. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomasselli, A. G., and R. L. Heinrickson. 1994. Specificity of retroviral proteases: an analysis of viral and nonviral protein substrates. Methods Enzymol. 241:279-301. [DOI] [PubMed] [Google Scholar]
- 39.Tozser, J., P. Bagossi, P. Boross, J. M. Louis, E. Majerova, S. Oroszlan, and T. D. Copeland. 1999. Effect of serine and tyrosine phosphorylation on retroviral proteinase substrates. Eur. J. Biochem. 265:423-429. [DOI] [PubMed] [Google Scholar]
- 40.Van Harmelen, J. H., E. Van der Ryst, and S. A. Loubser. 1999. A predominantly HIV type 1 subtype C-restricted epidemic in South African urban populations. AIDS Res. Hum. Retrovir. 15:395-398. [DOI] [PubMed] [Google Scholar]
- 41.Velazquez-Campoy, A., M. J. Todd, S. Vega, and E. Freire. 2001. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc. Natl. Acad. Sci. USA 98:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, Z. 2000. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J. Mol. Evol. 51:423-432. [DOI] [PubMed] [Google Scholar]
- 43.Yang, Z., R. Nielsen, N. Goldman, and A.-M. K. Pedersen. 2000. Codon-substitution models for variable selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zur Megede, J., S. Engelbrecht, T. De Oliveira, S. Cassol, T. J. Scriba, E. Janse van Rensburg, and S. W. Barnett. 2002. Novel evolutionary analyses of full-length HIV-1 subtype C molecular clones from Cape Town, South Africa. AIDS Res. Hum. Retrovir. 18:1327-1332. [DOI] [PubMed] [Google Scholar]
- 47.Zybarth, G., and C. Carter. 1995. Domains upstream of the protease (PR) in human immunodeficiency virus type I Gag-Pol influence PR autoprocessing. J. Virol. 69:3878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]