Abstract
The RNA packaging process for retroviruses involves a recognition event of the genome-length viral RNA by the viral Gag polyprotein precursor (PrGag), an important step in particle morphogenesis. The mechanism underlying this genome recognition event for most retroviruses is thought to involve an interaction between the nucleocapsid (NC) domain of PrGag and stable RNA secondary structures that form the RNA packaging signal. Presently, there is limited information regarding PrGag-RNA interactions involved in RNA packaging for the deltaretroviruses, which include bovine leukemia virus (BLV) and human T-cell leukemia virus types 1 and 2 (HTLV-1 and -2, respectively). To address this, alanine-scanning mutagenesis of BLV PrGag was done with a virus-like particle (VLP) system. As predicted, mutagenesis of conserved basic residues as well as residues of the zinc finger domains in the BLV NC domain of PrGag revealed residues that led to a reduction in viral RNA packaging. Interestingly, when conserved basic residues in the BLV MA domain of PrGag were mutated to alanine or glycine, but not when mutated to another basic residue, reductions in viral RNA packaging were also observed. The ability of PrGag to be targeted to the cell membrane was not affected by these mutations in MA, indicating that PrGag membrane targeting was not associated with the reduction in RNA packaging. These observations indicate that these basic residues in the MA domain of PrGag influence RNA packaging, without influencing Gag membrane localization. It was further observed that (i) a MA/NC double mutant had a more severe RNA packaging defect than either mutant alone, and (ii) RNA packaging was not found to be associated with transient localization of Gag in the nucleus. In summary, this report provides the first direct evidence for the involvement of both the BLV MA and NC domains of PrGag in viral RNA packaging.
The RNA packaging process for retroviruses involves a recognition event of the genome-length viral RNA by the viral Gag polyprotein precursor (PrGag), which acts to initiate the morphogenesis of virus particles (see reference 17 for review). The mechanism underlying this genome recognition event is poorly understood, but many biochemical and genetic analyses have revealed that this event involves the interaction between stable RNA secondary structures at the 5′ end of the viral genome and, in many cases, amino acids in the nucleocapsid (NC) domain of PrGag (1, 2, 4, 6, 7, 9, 10, 13, 15, 29, 30, 32, 36, 37).
The genome recognition event is an important step in the morphogenesis of infectious retrovirus particles. This recognition event leads to the predominant packaging (encapsidation) of the genome-length viral RNA into assembling particles (38). This discrimination process, which is primarily a viral RNA-protein interaction, is known to strongly favor the full-length viral RNA over spliced viral RNAs and cellular mRNAs (38). In general, the RNA sequences necessary and sufficient for the RNA packaging process are located in a region that includes the 5′-noncoding region along with the 5′ half of the gag gene (38). These sequences are referred to as the “packaging” or “encapsidation” signal. The packaging signal can enhance the packaging of RNAs containing the signal over those not containing the packaging signal. However, in the absence of RNAs containing a packaging signal, cellular RNAs can be packaged into particles, which supports the conclusion that RNA is a structural element in retrovirus particles (31).
The Gag polyprotein precursor alone is sufficient for the formation of particles, and the presence of RNA aids in efficient particle production (5). Since activation of the viral protease does not typically occur until after virus particle release, the genome recognition event involves full-length viral RNA and unprocessed Gag polyprotein (38). Gag includes three main domains (matrix [MA], capsid [CA], and NC). Attempts to address the RNA packaging process have focused on identifying RNA-binding domains within unprocessed Gag (and the processed Gag products) and demonstrating the specificity of binding of viral RNA to Gag either in cell-free reactions or in the uptake of viral RNA into virus particles.
The NC proteins of all retroviruses share the characteristics of a high percentage of basic residues and zinc binding domains involved in RNA packaging and in many instances alter the specificity of RNA binding (8, 12, 14, 17, 21, 23, 33, 36, 37). It is not clear whether NC actually confers all of the selective recognition of the viral genomic RNA (4, 32). The p2 spacer peptide between CA and NC of human immunodeficiency virus type 1 (HIV-1) PrGag has been suggested to play a role in selective RNA packaging (20). However, previous studies with HIV-1 have indicated that MA does not play a role in RNA packaging (32).
Bovine leukemia virus (BLV), as well as other deltaretroviruses, replicates to low titers in animals and is poorly infectious in cell culture. Cocultivation is typically used to infect permissive host cells. Because of these difficulties, information regarding the molecular details of the virus life cycles, including virus assembly, is limited. To more easily study the RNA packaging step in the virus assembly pathway of BLV, we have used a virus-like particle (VLP) model system to identify the protein determinants of BLV RNA packaging (39). As expected, many of the basic amino acid residues and all zinc finger residues mutated in the NC domain of PrGag led to significant reductions in RNA packaging. Interestingly, many of the basic residues in MA were also found to significantly influence RNA packaging. Together, these observations provide the first evidence that both the MA and NC domains of BLV PrGag are involved in viral RNA packaging.
MATERIALS AND METHODS
VLP vector and mutagenesis.
The vector used to produce VLPs, pGag-HA, has a hemagglutinin (HA) epitope tag fused to the C terminus of the BLV PrGag. The construction of this vector and the ability of the vector to produce VLPs have been described previously (39). This vector transcribes a single RNA transcript and no spliced RNA. The RNA is translated to express PrGag in the absence of the viral protease. Therefore, the VLPs do not have mature cores. The primary BLV RNA packaging signal, consisting of two stable RNA stem-loop structures (i.e., SL1 and SL2), are located just downstream of the gag start codon (24-26). Due to the presence of the packaging signal, this RNA transcript can be packaged into VLPs through the interaction with PrGag. In this system, the VLP expression vector transcribes only one RNA species due to the absence of the BLV splice donor and splice acceptor sites.
Site-directed mutations were introduced into pGag-HA by using the QuickChange XL kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. All derivatives made of pGag-HA were sequenced to verify the correct introduction of the desired mutation and the absence of undesired mutations.
Cells and transfections.
To express BLV PrGag in mammalian cells, COS-1 cells were transfected with pGag-HA or a derivative and placed under G418 selection to obtain stable cells expressing PrGag. Cells were grown in 100-, 60-, or 35-mm-diameter dishes in Dulbecco's modified Eagle's medium (GIBCO BRL, Gaithersburg, Md.) supplemented with 10% Fetal Clone III serum (HyClone, Logan, Utah). Superfect (Qiagen, Valencia, Calif.) was used for transfection of cells. Two days posttransfection, cells were placed under G418 selection until resistant colonies formed (∼3 weeks). Approximately 100 G418-resistant colonies were pooled and used for VLP analysis.
Quantitation of VLP production.
Supernatant from pooled stable cell clones producing VLPs was harvested and clarified by low-speed centrifugation (5 min at 700 × g) and then subjected to ultracentrifugation for 1 h at 40,000 × g at 4°C. Pelleted VLPs were then resuspended in radioimmunoprecipitation assay (RIPA) buffer (1% IGEPAL CA-630, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% deoxycholate [DOC], 5 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]).
Lysates prepared from VLP-producing cells and VLP samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose, and BLV Gag was detected with a primary antibody directed against the HA epitope tag (Covance, Princeton, N.J.) and an antimouse immunoglobulin (Ig) horseradish peroxidase-linked whole antibody (from sheep) as a secondary antibody (Amersham, Arlington Heights, Ill.). Quantitation of band intensities was done with the Quantity One software package with the Chemi Doc 2000 Documentation System (Bio-Rad, Richmond, Calif.).
Quantitation of viral RNA in cells and VLPs and determination of RNA packaging efficiencies.
Total RNA was extracted from transfected cells by using a cellular RNA extraction kit (Qiagen) according to the manufacturer's instructions. Viral RNA was extracted with a QIAamp viral RNA kit (Qiagen) according to the manufacturers' instructions. To control for DNA contamination, the RNAs isolated were treated with DNase I (Invitrogen) and used in reverse transcription-PCR (RT-PCR) without the addition of reverse transcriptase to confirm the absence of DNA.
Detection of the RNA transcript that expresses PrGag in cells and its incorporation into VLPs due to the RNA packaging signal was done by quantitative real-time RT-PCR analysis. Quantitative real-time RT-PCR was carried out with the Qiagen RT-PCR Master SYBR Green kit in a 20-μl reaction volume. RNA templates were reverse transcribed at 50°C for 25 min followed by a denaturing step at 95°C for 15 min. PCR was then performed with the primers +BLV979 (5′-AACCGCCATCGTGCTTGGGCA-3′) and −BLV1183 (5′-CGCTTCAGCGGCGGCTATTGC-3′). These primers amplify an approximately 200-bp-long fragment within the gag gene.
The PCR protocol consisted of 50 cycles of denaturation (95°C for 15 s), annealing (60°C for 20 s), and extension (72°C for10 s). For each step, the temperature transition rate was 20°C/s. The PCR was monitored with a Light Cycler (Roche Diagnostics) after each elongation step by SYBR Green I dye binding to amplified products. Quantitation was carried out with an external standard curve. Standard RNA was synthesized by in vitro transcription of a linearized plasmid containing a BLV proviral DNA representing the 5′ end of the gag gene. All in vitro transcripts were DNase I treated and tested for DNA contamination.
Standard curves were constructed from 10-fold serial dilutions of synthetic RNA transcripts. After real-time RT-PCR was completed, logarithmic values of fluorescence for each dilution were plotted against cycle number. A baseline was set just above the fluorescence background, and a crossing point was determined with amplification curves obtained during the initial exponential phase of amplification. The specificity of the amplified product was then determined by melting curve analysis. Melting curve acquisitions were done immediately after PCR was completed, by heating at 95°C for 0 s, cooling to 75°C for 15 s, and heating slowly at 20°C/s until 95°C with continuous fluorescence recording.
RNA packaging efficiencies were determined by the ratio of viral RNA detected from VLPs to viral RNA detected from total RNA recovered from VLP-producing cells divided by the ratio of Gag protein detected from VLPs to Gag protein detected from VLP-producing cells. RNA packaging efficiencies of mutants were then compared relative to that of the parental pGag-HA vector.
Confocal microscopy.
Transfected cells were grown on coverslips, fixed with 4% paraformaldehyde, and permeabilized with Triton X-100, both diluted in phosphate-buffered saline. The cells were then incubated with anti-HA antibody followed by incubation with Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes). Images were collected with a Bio-Rad MRC 600 confocal microscope.
RESULTS
Effects of mutations in the NC domain of PrGag on RNA packaging.
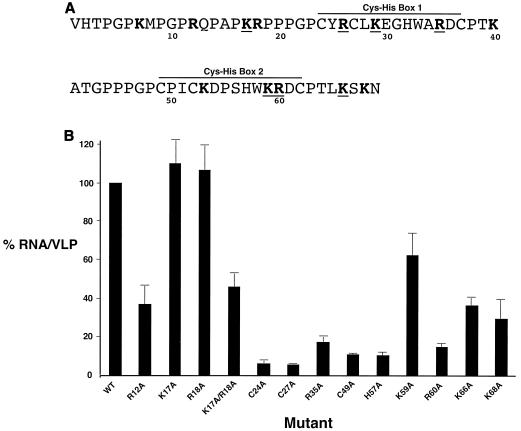
In order to determine the influence of NC on RNA packaging, a series of NC mutants were made in pGag-HA and tested for their ability to influence RNA packaging. Alanine residues were introduced in place of basic amino acid residues and in place of the conserved residues in each of the two zinc finger binding domains (Fig. 1A). The panel of NC mutants was analyzed in parallel with the parental vector. Figure 1B shows the RNA packaging efficiencies of the mutants. There were three groups of mutant phenotypes. The first group of mutants had no RNA packaging defects compared to the parental vector. These mutants included K17A and R18A. A second group of mutants had mild RNA packaging defects (40 to 70% reduction in VLP production) (i.e., R12A, K17A/R18A, K59A, K66A, and K68A). The third group of mutants had severe RNA packaging defects compared to the parental vector (85 to 95% reduction) (i.e., C24A, C27A, R35A, C49A, H57A, and R60A). For many of the mutants, VLP production was reduced two- to threefold that of the parental vector (i.e., R12A, K17A, R18A, K17A/R18A, R35A, and C49A). This reduction is within the linear detection range for the immunoblot assay used (described below). As predicted, many of the basic amino acid residues that were mutated led to significant reductions in RNA packaging efficiencies. The mutated basic residues that resulted in the greatest reductions were R35A and R60A. Interestingly, each of these residues lies between the conserved histidine and cysteine residues in the first (i.e., R35A) and second (i.e., R60A) zinc finger domains.
FIG. 1.
Mutagenesis of basic residues and the zinc fingers in the BLV NC domain of PrGag results in RNA packaging defects. (A) The BLV NC domain of PrGag. Amino acid residues in boldface indicate basic residues, and boldface residues that are underlined indicate basic and/or charged residues conserved among BLV, HTLV-1, and HTLV-2. The locations of the two zinc finger (Cys-His box) domains within NC are indicated. (B) Relative RNA packaging efficiencies of BLV NC mutants. RNA packaging efficiencies were determined by the ratio of viral RNA to the amount of Gag from VLPs (see Materials and Methods for details). The RNA packaging efficiency with wild-type BLV NC was set at 100, and the values for the NC mutants (± standard deviation) are relative to that of the wild type (WT). Mutants were tested in parallel with the parental construct, and each experiment was done in triplicate.
Effects of mutations in the MA domain of PrGag on RNA packaging.
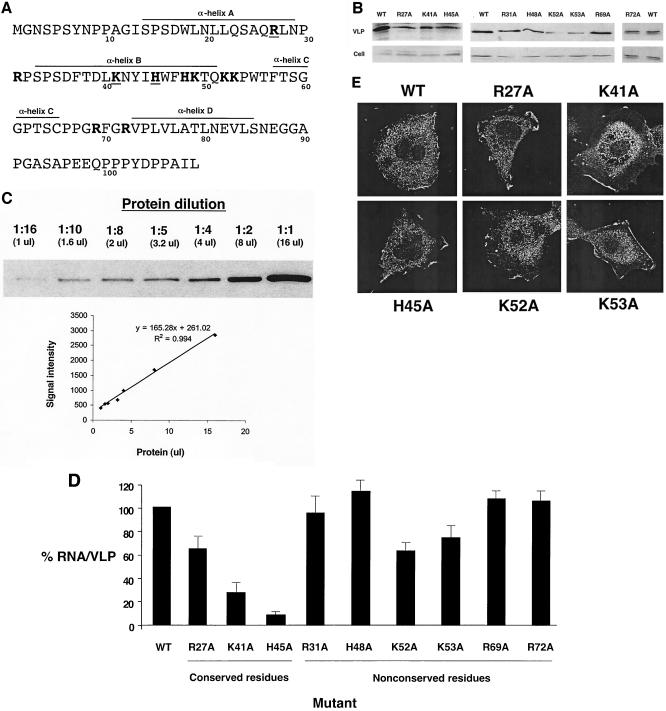
Previous studies with other retroviruses have provided little support for a role of the MA domain in retroviral RNA packaging (32). To test whether basic residues in the BLV MA domain influence RNA packaging, a panel of mutants that changed basic residues in MA to alanine was created (Fig. 2A). The RNA packaging efficiencies of the mutants were then analyzed in parallel with the parental vector. Figure 2B shows a representative protein analysis of Gag from cells and VLPs. The K52A and K53A MA mutants led to the lowest levels of VLPs (8 and 17%, respectively). These reductions are within 13-fold of the level of the parental vector and are within the linear range of detection for the assay, which was at least 16-fold (Fig. 2C). Figure 2D shows the resulting RNA packaging efficiencies for the panel of MA mutants tested. As with the NC mutants, there were three general groups of mutant phenotypes. Several basic residues that are not conserved among BLV, human T-cell leukemia virus type 1 (HTLV-1), and HTLV-2 led to no or small changes in RNA packaging efficiencies (i.e., R31A, H48A, K53A, R69A, and R72A). A second group led to modest reductions in RNA packaging efficiencies (i.e., R27A and K52A). Finally, a third group had significant reductions in RNA packaging (66 to 92%) compared to that of the parental vector (K41A and H45A). The H45A MA mutant had a reduction in RNA packaging efficiency (92%) that was as great as any of those in the individual NC mutants, indicating that BLV MA plays an important role in RNA packaging. The K49 residue was not analyzed in this study.
FIG. 2.
Mutagenesis of basic residues in the BLV MA domain of PrGag results in RNA packaging defects. (A) The BLV MA domain of PrGag. Amino acid residues in boldface indicate basic residues, and boldface residues that are underlined indicate basic and/or charged residues conserved among BLV, HTLV-1, and HTLV-2. The α-helices in MA are indicated, based upon nuclear magnetic resonance solution structure analysis of BLV MA (28). (B) Quantitation of VLP production. Preparation of lysates from VLP-producing cells and VLPs and immunoblot analysis are described in Materials and Methods. Representative results from the immunoblot analysis are shown. WT, wild type. (C) Linear range of detection for immunoblot analysis. Protein was diluted (1:1, 1:2, 1:4, 1:5, 1:8, 1:10, and 1:16) and was subjected to immunoblot analysis. The band intensity (arbitrary units) for each dilution was determined with the Quantity One software package of the Chemi Doc 2000 Documentation System (Bio-Rad) and plotted against the amount of protein used for immunoblot analysis (16, 8, 4, 3.2, 2, 1.6, or 1 μl) to determine if protein detection within the dilution range was linear. (D) Relative RNA packaging efficiencies of BLV MA mutants. RNA packaging efficiencies were determined by the ratio of viral RNA to the amount of Gag from VLPs (see Materials and Methods for details). The RNA packaging efficiency with wild-type BLV MA was set at 100, and the values for the MA mutants (± standard deviation) are relative to that for the wild type. Mutants were tested in parallel with the parental construct, and each experiment was done in triplicate. (E) Cellular distribution of PrGag for selected MA mutants. Cells stably transfected with VLP constructs containing mutations in MA were grown on coverslips, fixed, and incubated with an anti-HA Ig followed by incubation with Alexa Fluor 488-conjugated antimouse Ig. Images were obtained with a confocal microscope.
In other retroviruses, such as HIV-1, basic amino acid residues in MA can influence PrGag membrane targeting and are proposed to form a positively charged patch that promotes membrane binding (16, 27, 40, 41). To determine whether MA mutants that significantly reduced RNA packaging also influenced PrGag membrane localization, the cellular distribution of Gag was analyzed by confocal microscopy. Figure 2E shows the effects of several basic amino acid mutants on Gag distribution in cells. These mutants (i.e., R27A, K41A, H45A, K52A, and K53A) had a cellular distribution of Gag that was comparable to that observed with the wild type. Specifically, Gag expression yielded a stippled appearance along the plasma membrane for the parental Gag and for these mutants (Fig. 2E).
Importance of charged amino acid residues in MA on RNA packaging.
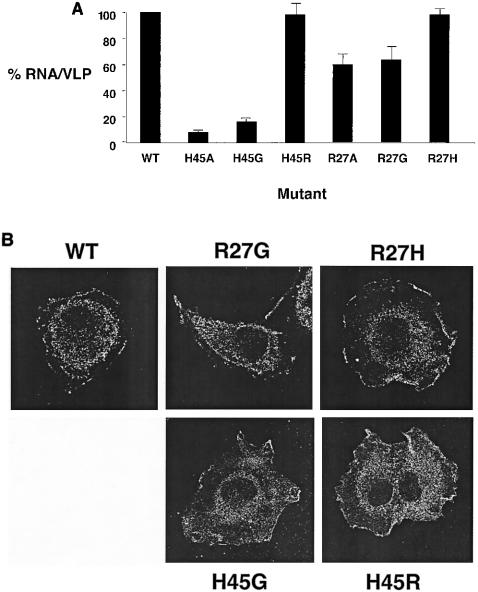
The MA residues R27 and H45, when mutated, revealed significant reductions in RNA packaging efficiencies. To further analyze the importance of these positively charged amino acid residues on RNA packaging, mutations were made at either R27 or H45 to create either a conservative (i.e., R27H or H45R) or nonconservative (i.e., R27G or H45G) change at these residues. Figure 3A shows the effects of these amino acid substitutions on RNA packaging. R27G had a phenotype comparable to that of R27A, while R27H had a phenotype that was comparable to that of the parental vector. Furthermore, H45G had an RNA packaging defect similar to that of H45A, while H45R had a phenotype equivalent to that of the parental vector. These data indicate that there is a correlation between the absence of a positively charged amino acid at positions 27 or 45 and a defect in RNA packaging. The cellular distribution of Gag was not influenced by these mutations, indicating there was no correlation between the RNA packaging defects and Gag membrane localization (Fig. 3B).
FIG. 3.
Requirement of charged amino acid residues in BLV MA for RNA packaging. RNA packaging efficiencies were determined by the ratio of viral RNA to the amount of Gag from VLPs (see Materials and Methods for details). The RNA packaging efficiency with wild-type (WT) BLV MA was set at 100, and the values for the MA mutants (± standard deviation) are relative to that for the wild type. (A) Mutants were tested in parallel with the parental construct, and each experiment was done in triplicate. (B) Cellular distribution of PrGag. Cells stably transfected with VLP constructs were grown on coverslips, fixed, and incubated with antibodies prior to confocal microscopy.
Combined effect of MA and NC mutations on RNA packaging.
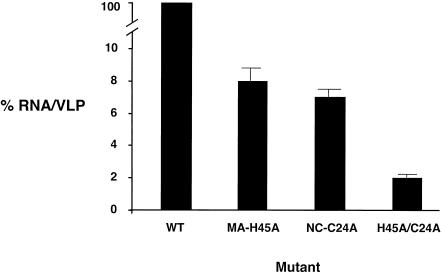
To test for the potential interplay between MA and NC on RNA packaging, a mutant construct was created in which both MA and NC mutations were introduced into the same vector and then assayed for the ability to package RNA. In particular, the MA/NC double mutant MA H45A/NC C24A was tested. The combined mutant had a 10-fold reduction in VLP production and a severe defect in RNA packaging (98% reduction compared to that of the parental strain) that was significantly lower than the RNA packaging efficiency of either mutant alone (Fig. 4). This indicates that the combination of mutations together further reduced the efficiency of RNA packaging.
FIG. 4.
Combined influence of NC and MA mutations on RNA packaging. RNA packaging efficiencies were determined by the ratio of viral RNA to the amount of Gag from VLPs (see Materials and Methods for details). The RNA packaging efficiency with wild-type BLV was set at 100, and the values for the mutants (± standard deviation) are relative to that for the wild type (WT). Mutants were tested in parallel with the parental construct, and these experiments were done in triplicate.
Lack of transient nuclear localization of Gag.
A recent study with Rous sarcoma virus (RSV) revealed that RSV Gag protein enters the nucleus by a nuclear-targeting sequence in the MA domain and is subsequently transported to the cytoplasm by using a CRM1-mediated nuclear export pathway (35). Transient expression of a dominant-negative CRM1 or by treating cells with leptomycin B (LMB; a drug that attaches to the central domain of CRM1 to disrupt its interaction with nuclear export signals) resulted in the redistribution of Gag from the cytoplasm to the nucleus. The MA mutant, Myr1E, was insensitive to the effects of LMB treatment, apparently because it bypassed the nuclear compartment during virus assembly (35). Myr1E has a defect in RNA packaging, which implies that RSV nuclear localization of Gag might be involved in viral RNA-Gag interactions.
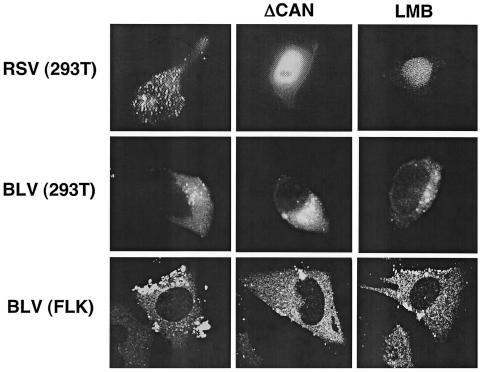
Since we discovered BLV MA mutants with RNA packaging defects like Myr1E, we hypothesized that the viral RNA-Gag interactions involved in BLV RNA packaging might occur in the nucleus after transient nuclear localization of Gag. To test whether BLV Gag is localized in the nucleus, 293T cells transiently transfected with the BLV VLP construct PR+ (39) were treated with either LMB or transiently cotransfected with a dominant-negative CRM1 (ΔCAN). In addition, fetal lamb kidney cells chronically infected with BLV (FLK-BLV) were either treated with LMB or transiently transfected with ΔCAN. When 293T cells were either cotransfected with an RSV Gag-green fluorescent protein (GFP) construct and ΔCAN or transfected with Gag-GFP and then treated with LMB, there was a distinct nuclear localization of Gag-GFP, comparable to that originally reported by Scheifele et al. (35). When 293T cells were either cotransfected with the BLV vector and ΔCAN or treated with LMB after transfection with the BLV vector, no BLV Gag was observed in the nucleus of cells (Fig. 5). Comparable results were obtained with LMB treatment or transfection of ΔCAN into FLK-BLV cells. One interpretation of these data is that BLV Gag does not enter the nucleus, implying that the genome recognition event occurs in the cytoplasm. However, it is possible that BLV Gag does enter the nucleus and is exported by a CRM1-independent pathway.
FIG. 5.
Lack of transient nuclear localization of BLV Gag. 293T cells were transiently transfected alone with an RSV Gag-GFP construct or a BLV vector (PR+) and treated with LMB (10 ng/μl) or were transiently cotransfected with the RSV or BLV construct and a dominant-negative CRM1 (ΔCAN) (10 μg per approximately 5 × 105 cells). In parallel, FLK cells chronically infected with BLV were treated with LMB (10 ng/μl) or were transiently transfected with ΔCAN (10 μg per approximately 5 × 105 cells). Forty-eight hours posttransfection, cells were fixed and then analyzed by confocal microscopy.
DISCUSSION
This is the first report to provide direct genetic evidence that both the MA and NC domains of BLV PrGag are protein determinants of RNA packaging. The unique observation in this study is the identification of several basic amino acid residues in MA that when mutated, led to RNA packaging defects as great as, if not greater than, those observed for single-amino-acid changes in NC. These observations imply that BLV, and perhaps other deltaretroviruses, use distinct viral RNA-protein interactions in genome recognition and RNA packaging.
An interesting observation made with the MA mutants analyzed in this study is that the basic residues in MA that influenced RNA packaging had no effect on Gag membrane localization. This is in contrast to studies with HIV-1 that indicate a role for the basic amino residues of MA in membrane localization (by the formation of a charged patch that is thought to promote membrane binding) (11, 16, 27, 40, 41). However, our data support those from a previous study of the basic residues in the HTLV-1 MA, which found that HTLV-1 MA basic residue mutants led to normal transport of Gag to the membrane as well as cleavage of PrGag (22). Many of these HTLV-1 mutants had reduced infectivity. Our study found that several of the BLV MA basic residue mutants affected RNA packaging and did not influence Gag membrane targeting (although they did lead to reductions in virus particle production), suggesting that the reduced infectivity observed with the HTLV-1 mutants may be (in some cases) related to an RNA packaging defect.
The locations of the basic residues that led to defects in RNA packaging help to provide some mechanistic insights into potential viral RNA-protein interactions involved in genome recognition. Among the NC mutants, the residues that lie within the first and second zinc finger domains, including R35 and R60, had a significant impact on RNA packaging. Similar residues in the HIV-1 NC zinc finger have been implicated in interactions with SL3 RNA and have been shown to be important for RNA packaging by mutagenesis studies (9). The basic residues in the BLV MA that caused the greatest defects in RNA packaging (i.e., K41 and H45) are located in α-helix B (28). Presently, no structural data are available regarding the potential viral RNA-MA protein interactions. However, it has been previously reported that BLV MA specifically interacts with the viral RNA in a region (18, 19) that spans the locations of SL1 and SL2 (26). This suggests that the contacts made by BLV MA with the viral RNA may also involve an interaction with SL1 and/or SL2. Interestingly, the HIV-1 MA protein has been found to bind to the viral RNA in the pol gene, via the basic amino acids in MA, and virus mutants that disrupted this RNA-protein interaction had delayed replication kinetics, although no role in RNA packaging was reported (34). The BLV MA mutants in this study that influenced RNA packaging do not overlap with the known RNA sequence of gag known to be involved in RNA packaging (25, 26). This indicates that the mutations introduced into the VLP vector influence RNA packaging by the amino acid change in MA and not the nucleotide changes in the RNA. This conclusion is supported by the data in Fig. 3, which show the importance of charged amino acid residues (and not just nucleotide changes) in RNA packaging.
The magnitude of the defects in RNA packaging described here in this study for the most extreme MA and NC mutants (i.e., a MA-NC double mutant) are in the range of those observed previously when SL1 or SL2 was mutated (24, 26). Taken together, these observations suggest that residues in both MA and NC make direct contacts with the packaging signal or perhaps adjacent sequences that would influence genome recognition. The interaction of NC with RNA sequences outside of those RNA secondary structures of the RNA packaging signal has been shown directly or implied with murine leukemia virus and spleen necrosis virus and is believed to play an important role in RNA packaging (3, 10).
The data in this report provide evidence for a role of both the MA and NC domains of Gag in BLV RNA packaging. This observation implies that distinct viral RNA-protein interactions occur in BLV genome recognition and RNA packaging to that of other retroviruses. Further studies will determine the precise RNA-protein interactions involved in BLV RNA packaging, the location in the cell at which genome recognition occurs, and whether both MA and NC play a role in RNA packaging of other deltaretroviruses.
Acknowledgments
We thank M. Knowlton for technical assistance; K. Wolken for assistance with confocal microscopy; J. Wills, L. Parent, and T. Hope for sharing reagents; and M. Summers for helpful discussions.
This work was supported by the American Cancer Society (RPG 0027801).
REFERENCES
- 1.Amarasinghe, G. K., J. Zhou, M. Miskimon, K. J. Chancellor, J. A. McDonald, A. G. Matthews, R. R. Miller, M. D. Rouse, and M. F. Summers. 2001. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein: structural studies and implications for genome recognition. J. Mol. Biol. 314:961-970. [DOI] [PubMed] [Google Scholar]
- 2.Banks, J. D., and M. L. Linial. 2000. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 74:456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beasley, B. E., and W.-S. Hu. 2002. cis-Acting elements important for retroviral RNA packaging specificity. J. Virol. 76:4950-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz, R. D., Å. Ohagen, S. Höglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Certo, J. L., T. O. Kabdulov, M. L. Paulson, J. A. Anderson, and W. S. Hu. 1999. The nucleocapsid domain is responsible for the ability of spleen necrosis virus (SNV) Gag polyprotein to package both SNV and murine leukemia virus RNA. J. Virol. 73:9170-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Certo, J. L., B. F. Shook, P. D. Yin, J. T. Snider, and W.-S. Hu. 1998. Nonreciprocal pseudotyping: murine leukemia virus proteins cannot efficiently package spleen necrosis virus-based vector RNA. J. Virol. 72:5408-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 10.D'Souza, V., J. Melamed, D. Habib, K. Pullen, K. Wallace, and M. F. Summers. 2001. Identification of a high affinity nucleocapsid protein binding element within the Moloney murine leukemia virus Psi-RNA packaging signal: implications for genome recognition. J. Mol. Biol. 314:217-232. [DOI] [PubMed] [Google Scholar]
- 11.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 12.Fu, W., and W.-S. Hu. 2003. Functional replacement of nucleocapsid flanking regions by heterologous counterparts with divergent primary sequences: effects of chimeric nucleocapsid on the retroviral replication cycle. J. Virol. 77:754-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison, G. P., G. Miele, E. Hunter, and A. M. L. Lever. 1998. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 72:5886-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jewell, N. A., and L. M. Mansky. 2000. In the beginning: genome recognition, RNA encapsidation, and the initiation of complex retrovirus assembly. J. Gen. Virol. 81:1889-1899. [DOI] [PubMed] [Google Scholar]
- 18.Katoh, I., H. Kyushiki, Y. Sakamoto, Y. Ikawa, and Y. Yoshinaka. 1991. Bovine leukemia virus matrix-associated protein MA(p15): further processing and formation of a specific complex with the dimer of the 5′-terminal genomic RNA fragment. J. Virol. 65:6845-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh, I., T. Yasunaga, and Y. Yoshinaka. 1993. Bovine leukemia virus RNA sequences involved in dimerization and specific gag protein binding: close relation to the packaging sites of avian, murine, and human retroviruses. J. Virol. 67:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaye, J. F., and A. M. L. Lever. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 72:5877-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klug, A. 1999. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 293:215-218. [DOI] [PubMed] [Google Scholar]
- 22.Le Blanc, I., A. R. Rosenberg, and M.-C. Dokhélar. 1999. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73:1860-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, E.-G., A. Alidina, C. May, and M. L. Linial. 2003. Importance of basic residues in binding of Rous sarcoma virus nucleocapsid to the RNA packaging signal. J. Virol. 77:2010-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansky, L. M., and L. C. Gajary. 2002. The primary nucleotide sequence of the bovine leukemia virus RNA packaging signal can influence efficient RNA packaging and virus replication. Virology 301:272-280. [DOI] [PubMed] [Google Scholar]
- 25.Mansky, L. M., A. E. Krueger, and H. M. Temin. 1995. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J. Virol. 69:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansky, L. M., and R. M. Wisniewski. 1998. The bovine leukemia virus encapsidation signal is composed of RNA secondary structures. J. Virol. 72:3196-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massiah, M. A., M. R. Starich, C. Paschall, M. F. Summers, A. M. Christensen, and W. I. Sundquist. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198-223. [DOI] [PubMed] [Google Scholar]
- 28.Matthews, S., M. Mikhailov, A. Burny, and P. Roy. 1996. The solution structure of the bovine leukemia virus matrix protein and similarity with lentiviral matrix proteins. EMBO J. 15:3267-3274. [PMC free article] [PubMed] [Google Scholar]
- 29.McBride, M. S., and A. T. Panganiban. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70:2963-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride, M. S., and A. T. Panganiban. 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 71:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poon, D. T. K., G. Li, and A. Aldovini. 1998. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J. Virol. 72:1983-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poon, D. T. K., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purohit, P., S. Dupont, M. Stevenson, and M. R. Green. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmalzbauer, E., B. Strack, J. Dannull, S. Guehmann, and K. Moelling. 1996. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J. Virol. 70:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz, M. D., D. Fiore, and A. T. Panganiban. 1997. Distinct functions and requirements for the Cys-His boxes of the human immunodeficiency virus type 1 nucleocapsid protein during RNA encapsidation and replication. J. Virol. 71:9295-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 39.Wang, H., K. M. Norris, and L. M. Mansky. 2002. Analysis of bovine leukemia virus Gag membrane targeting and late domain function. J. Virol. 76:8485-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, X., X. Yu, T.-H. Lee, and M. Essex. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 67:6387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]