Abstract
In order to cope with hostile host environments, many viruses have developed strategies to perturb the cellular machinery to suit their replication needs. Some herpesvirus genes protect cells from undergoing apoptosis to prolong the lives of infected cells, while others, such as Epstein-Barr virus Zta, slow down the G1/S transition phase to allow ample opportunity for transcription and translation of viral genes before the onset of cellular genomic replication. In this study, we investigated whether Kaposi's sarcoma-associated herpesvirus (KSHV) K-bZIP, a homologue of the Epstein-Barr virus transcription factor BZLF1 (Zta), plays a role in cell cycle regulation. Here we show that K-bZIP physically associates with cyclin-CDK2 and downmodulates its kinase activity. The association can be detected in the natural environment of KSHV-infected cells without artificial overexpression of either component. With purified protein, it can be shown that the interaction between K-bZIP and cyclin-CDK2 is direct and that K-bZIP alone is sufficient to inhibit CDK2 activity. The interacting domain of K-bZIP has been mapped to the basic region. The result of these associations is a prolonged G1 phase, accompanied by the induction of p21 and p27 in a naturally infected B-cell line. Thus, in addition to the previously described transcription and genome replication functions, a new role of K-bZIP in KSHV replication is identified in this report.
Herpesvirus replication is often broken down into early, delayed-early, late, and latent phases. The cellular environment required for each phase is unique, and a number of viral genes have evolved to interact with host factors to create a more favorable environment for viral replication. Regulation or deregulation of the cell cycle is one strategy often used by viruses to alter the cellular environment. In the literature, there is a preponderance of evidence showing that viral gene products interact with cell cycle proteins such as cyclins and CDKs or their regulators such as p53 and retinoblastoma protein (19, 20, 21, 22, 25, 41, 42, 49, 59). Some of these interactions lead to accelerated cell cycle progression and cellular proliferation, in some cases contributing to the transformation of target cells. Other herpesvirus gene products delay cell cycle progression and transiently arrest the infected cells at the G1 stage. This usually occurs among the first stages of the viral life cycle and involves immediate-early and/or early gene products (7, 10, 13, 15, 19, 25, 34), such as ICP0 (25, 34) and ICP27 (49) of herpes simplex virus, IE2 (54) and UL69 (35) of cytomegalovirus, and Zta (10) of Epstein-Barr virus (EBV).
It was postulated that lengthening the G1 phase would give additional time for the virus to complete transcription and translation of early genes and for viral DNA replication to occur before the onset of competing cellular genomic replication (see reference 19 for a review). The molecular mechanisms by which viral gene products interfere with G1 to S progression differ from virus to virus. Both EBV Zta and cytomegalovirus IE2 are known to bind p53 and stabilize it (19, 49, 59). On the other hand, cytomegalovirus IE2 also binds retinoblastoma protein and releases E2F, which transcriptionally activates cyclin E gene expression (22). By these means, cytomegalovirus blocks cellular DNA replication while at the same time it activates certain cell cycle pathways to provide a more suitable environment for viral DNA replication. The overall consequence of these interactions is to slow down the G1 to S transition (5, 10, 19).
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a member of the gammaherpesvirus family, which includes EBV and herpesvirus saimiri. KSHV infection is associated with all types of Kaposi's sarcoma, including AIDS-associated, endemic, and renal transplant-related Kaposi's sarcoma (3, 6, 26, 37, 39, 52). It has also been implicated in B-cell lymphoproliferative diseases such as primary effusion lymphoma and multicentric Castleman's disease (11, 50). Like other herpesviruses, KSHV encodes both latent and lytic genes (45). The latent genes are thought to be primarily responsible for the maintenance of latency and directly involved in cell transformation (30). The lytic genes are involved either directly in viral replication (genome replication, transcription, etc.) or in providing a cellular environment conducive for viral replication (e.g., B-cell activation, immune modulation of host response, target cell recruitment) or both (12, 45, 53). Products from both latent and lytic genes have been shown to interact with cell cycle proteins or their regulators. Perhaps the best known is v-cyclin, which directly interacts with and activates cellular CDK6, thereby accelerating cell cycle progression (14, 16, 29). Other viral gene products such as latency-associated nuclear antigen (LANA) interact with the cell cycle regulator p53 and thus can potentially perturb both the cell cycle and apoptosis programs (20).
Previously, we reported the identification of an early KSHV gene product, K-bZIP, which is the positional and structural homolog of EBV Zta (32). We show here that K-bZIP interacts directly with cyclin-CDK2 and downmodulates the kinase activity of CDK2, resulting in G1 growth arrest of the host cells. The association of K-bZIP and CDK2 can be detected in the naturally infected BCBL-1 line, and the expression of K-bZIP but not of a mutant lacking the interaction domain results in slower growth of cells. This growth arrest is accompanied by the increased expression of p21 and p27. Our results suggest that K-bZIP, in addition to being a transcriptional regulator, is also directly involved in cell cycle regulation.
MATERIALS AND METHODS
Plasmids.
Plasmids encoding the full-length K-bZIP (K-bZIP/wt, residues 1 to 273) were constructed in pcDNA3.1 (Invitrogen). Cloning introduced a CpoI site and either a hemagglutinin (HA) tag, a Flag tag, or a T7 tag to the N terminus as described previously (32). The resulting plasmids were denoted pHA-K-bZIP, pFlag-K-bZIP, and pT7-K-bZIP, respectively. A natural spliced variant of K-bZIP, K-bZIPΔLZ, which carries a deletion of the leucine zipper region, was inserted in-frame 3′ of the Flag tag sequence of modified pcDNA3.1 as described previously (32). The resulting plasmid was denoted pFlag-K-bZIPΔLZ. An expression plasmid with a deletion of the K-bZIP basic region (BR) (residues 123 to 189) (pFlag-K-bZIPΔBR) was constructed as described previously (28).
Deletion fragments of K-bZIP were amplified by PCR with Pfu Turbo (Stratagene) with primers as described previously (28) and then cloned into a modified pGEX4T-2 (Amersham-Pharmacia), which introduced a CpoI site to produce glutathione S-transferase (GST) fusion proteins. To prepare additional K-bZIP deletion mutants in this study, K-bZIP deletion fragments were amplified with the following primer pairs (italics designate CpoI sites and capital letters denote the viral sequence): K-bZIP 107F (5′-aac ggt ccg CTC TCT CAC ACA CCA CCA AGA-3′) and K-bZIP 157R (5′-aac gga ccg CTT AAC TAC AGA CGC AGG CAC-3′), and K-bZIP 158F (5′-aac ggt ccg GCC GAA GTA TGT GAT CAG TC-3′) and K-bZIP 208R (5′-aac gga ccg CTT ATG TGC CTC CAA TCT CGC-3′). Amplified fragments were then digested with CpoI (Fermentas) and inserted in frame downstream of the GST coding sequence. Expression plasmids for CDK2, cyclin A, and cyclin E were a generous gift from D. J. Templeton (Case Western Reserve University).
Cell culture, transient transfection, establishment of K-bZIP stable transfectants, and growth rate analysis.
Epithelial human embryonic kidney cell lines 293 and 293T, as well as a cervical carcinoma cell line, HeLa cells, were grown in monolayer cultures in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum in the presence of 5% CO2. To establish stable expression of Flag-control, Flag-K-bZIP, Flag-K-bZIPΔBR, and Flag-K-bZIPΔLZ, 293 cells were transfected with 10 μg of each expression plasmid, which contained the G418 resistance gene. After 2 to 3 weeks of selection with medium containing 400 μg of G418 per ml (Gibco-BRL), individual G418-resistant colonies were cloned and tested for the expression of K-bZIP and its mutants by Western blot analysis. These stable cell lines were grown at 37°C in DMEM supplemented with 10% fetal bovine serum and 200 μg of G418 per ml in the presence of 5% CO2.
To establish BCBL-1 cells stably expressing HA-K-bZIP, cells were transfected with 20 μg of pHA-K-bZIP, which contains a G418 resistance gene. Stable cells were collected as described previously (28). These stable cell pools were maintained in RPMI 1640 supplemented with 15% fetal bovine serum and 200 μg of G418 per ml in the presence of 5% CO2. Both viral lytic replication and HA-K-bZIP expression were induced by treatment of log-phase HA-K-bZIP stable BCBL-1 cells with tetradecanoyl phorbol acetate (TPA) (20 ng/ml; Sigma).
For determination of growth rate, 5 × 104 cells from each of the cell clones were seeded in 60-mm plates. Cell numbers were counted at the indicated time postseeding. Triplicate plates were seeded for each cell clone.
Cell cycle analysis.
For cell cycle analysis; BCBL-1, GA10, and BCP-1 cells were collected, washed once with phosphate-buffered saline (PBS), suspended in 0.5 ml of cold (4°C) PBS-0.1% glucose, fixed with 5 ml of ice-cold 70% ethanol for at least 45 min at 4°C, washed with PBS, and treated with RNase A (0.1 mg/ml) in a propidium iodide (69 mM; Sigma)-sodium citrate (38 mM) solution. Cell cycle analysis was performed by fluorescence-activated cell sorting (FACScan; Becton Dickinson) with the ModFit LT version 2.0 software (Verity Software House Inc.).
Immunoprecipitation and immunoblot analyses.
BCBL-1 cells were rinsed in ice-cold PBS, and 107 cells were lysed in EBC lysis buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% NP-40, 50 mM NaF, 200 μM Na2VO4, 1 mM phenylmethylsulfonyl fluoride) supplemented with a protease inhibitor cocktail (Roche). After centrifugation (15,000 × g for 10 min at 4°C), 20 μl of protein A- and protein G-Sepharose beads (Upstate) were added to the supernatants and incubated overnight at 4°C. Then 500 μg of each of the cleared supernatants was reacted with 3 μg of anti-CDK2 (SC-163; Santa Cruz), anti-cyclin A (SC-751; Santa Cruz), anti-cyclin E (SC-481; Santa Cruz), anti-cyclin D (Upstate), anti-CDK4 (SC-260-G; Santa Cruz), anti-p53 (SC-126; Santa Cruz), anti-p107 (SC-318; Santa Cruz), anti-cdc2 (SC-54; Santa Cruz), anti-p21 (SC-397; Santa Cruz), anti-p27 (SC-1641; Santa Cruz), antitubulin (SC-8035; Santa Cruz), or anti-HA (Babco) for 3 h to overnight at 4°C with gentle rotation.
The immune complex was captured by the addition 20 μl of a protein A- and protein G-Sepharose bead mixture and rocked for an additional 2 h at 4°C. Beads were washed four times with EBC buffer and then boiled for 5 min in 20 μl of 2× sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.6% bromophenol blue). 293T cells were cotransfected with 2 μg of pT7-K-bZIP and 3 μg of pFlag-CDK2 or pFlag-empty expression plasmids with FuGene 6 (Roche) according to the supplier's recommendations. The cells were harvested 48 h after transfection and lysed in EBC buffer. Then 500 μg of cell lysates was immunoprecipitated with the addition of 25 μl of anti-Flag antibody-conjugated agarose (Sigma). The beads were washed four times with EBC buffer and then boiled for 5 min in 20 μl of 2× SDS sample buffer.
Protein samples from total cell lysates (50 μg/lane) or immunoprecipitates were subjected to SDS-10% polyacrylamide gel electrophoresis (PAGE) and then transferred to a polyvinylidene difluoride membrane (Biotechnology Systems) with a semidry transfer apparatus (Amersham Pharmacia). After blocking for 1 h at room temperature with 5% skim milk in TBST (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, 0.05% Tween 20), the membranes were incubated with primary antibodies for 2 h at room temperature. The membranes were subsequently washed with TBST three times for 10 min each at room temperature and then incubated with horseradish peroxidase-conjugated antibodies for 1 h at room temperature. The membranes were then washed three times with TBST and visualized with enhanced chemiluminescence reagents (Amersham-Pharmacia). Final dilutions of the first antibodies for immunoblotting were 1:4,000 for the rabbit anti-K-bZIP antibody (28), 1:3,000 for the anti-T7 antibody (Novagen), 1 μg of antiactin (SC-1615; Santa Cruz), 1 μg of anti-CDK2, 1 μg of anti-p21, 1 μg of anti-p27 and 1 μg of anti-p53 per ml in TBST containing 5% skim milk.
Immunofluorescence assay.
Forty-eight hours post-TPA treatment, BCBL-1 cells were fixed with 3.7% paraformaldehyde in PBS for 5 min at room temperature and subsequently treated with 1.0% Triton X-100 followed by 1.0% NP-40 in PBS for 10 min each at room temperature. After being washed twice with 0.2% Tween 20 in PBS, cells were smeared on a coverslip (Fisher). After being blocked with PBS-2% bovine serum albumin (Fisher), cells were incubated with anti-K-bZIP rabbit serum (1:2,000) and anti-CDK2 mouse monoclonal antibody (5 μg/ml; B.D. Transduction Labs) in PBS-2% bovine serum albumin for 1 h at room temperature. After being washed four times with PBS, Alexa Fluor 555-conjugated goat anti-rabbit immunoglobulin G F(ab′)2 (1:3,000) (Molecular Probes) and Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G F(ab′)2 (1:2,000) (Molecular Probes) in PBS-2% bovine serum albumin were applied as secondary antibodies and allowed to react for 1 h at room temperature. DNA was stained with 1 μM To-Pro3 (Molecular Probes) in PBS for 1 min at room temperature. Imaging was performed with a confocal microscope equipped with an argon-krypton laser (LSM510-MicroSystem; Carl Zeiss Co., Ltd).
Preparation and purification of GST fusion proteins.
GST fusion proteins were expressed in Escherichia coli strain BL21 transformed with the following plasmids encoding distinct domains of K-bZIP: pGEX-K-bZIP F.L. (residues 1 to 237), pGEX-K-bZIP I (residues 1 to 121), pGEX-K-bZIP II (residues 107 to 157), pGEX-K-bZIP III (residues 122 to 189), pGEX K-bZIP IV (residues 158 to 208), pGEX K-bZIP V (residues 190 to 237), or pGEX4T-2. The GST fusion proteins were then purified with glutathione-Sepharose beads (Amersham-Pharmacia) by a standard procedure. Bacterial cells (500 ml) were cultured in Luria broth for each construct. Protein expression was induced with 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG). Bacterial cells were washed once in PBS and then lysed by sonication in PBS containing 1% Triton X-100 and 1% Sarkosyl. After clearing by centrifugation at 7,000 × g for 10 min at 4°C, glutathione-Sepharose beads (500 μl of a 1:1 slurry in PBS) were added to the lysates for affinity purification. After incubation overnight at 4°C with rotation, the beads were washed four times in PBS containing 1% Triton X-100 and 1% Sarkosyl. The proteins immobilized on the glutathione-agarose beads were quantified by Coomassie blue staining, with bovine serum albumin as a protein standard.
In vitro interaction assay.
GST-protein beads containing approximately 2.0 μg of proteins were resuspended in binding buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol, 0.1% NP-40, and 10% glycerol, supplemented before use with 1 mg of bovine serum albumin per ml, 0.5 mM phenylmethylsulfonyl fluoride, and 1 × protease inhibitor cocktail), and then incubated for 30 min at 4°C with 10 μl of in vitro translated proteins, which were labeled with [35S]methionine with the TNT coupled transcription and translation system (Promega). The beads were washed four times with binding buffer and then resuspended and boiled in 2× SDS sample buffer. After proteins were separated by SDS-PAGE, radiolabeled polypeptides retained on the beads were visualized by autoradiography.
In vitro kinase assay.
cyclin-CDK2 complexes were immunoprecipitated from 250 μg of total cell lysate from 293 stable cells, BCBL-1 cells, or HA-BCBL-1 cells in EBC buffer with anti-CDK2 antibody (SC-163; Santa Cruz). Immunocomplexes were washed three times with kinase buffer (20 mM HEPES [pH 7.5], 10 mM MgCl2, 10 mM MnCl2, 1 mM dithiothreitol, 1 mM NaF, 0.1 mM Na2VO4). Kinase reactions were performed for 15 min at 30°C with histone 1 (Upstate) as a substrate.
In vitro CDK2 kinase inhibition assays were performed with the full-length GST fusion with K-bZIP. The full-length GST-K-bZIP protein was recovered from bacterial lysates with glutathione-Sepharose beads, separated by SDS-PAGE, excised, equilibrated in SDS electrophoresis buffer, and electroeluted into a cellulose tube. The purified protein in SDS electrophoresis buffer was dialyzed against PBS overnight. Then 2 μl of CDK2-cyclin A (Upstate) was incubated with purified GST-K-bZIP for 1 h at 4°C in kinase buffer (10 μl of kinase buffer and 10 μl of PBS containing 0.5 μg of purified GST-K-bZIP as indicated in the figure legends) without ATP. Total protein amounts were adjusted with bovine serum albumin. Kinase reactions were performed for 5 min at 25°C in a volume of 30 μl (20 μl of kinase buffer and 10 μl of PBS containing GST-K-bZIP, 10 μCi of [γ-32P]ATP [Amersham-Pharmacia], 50 μM cold ATP, and 0.5 μg of histone 1 as a substrate). Reactions were stopped by addition of 30 μl of SDS sample buffer, and the mixtures were boiled, fractionated by SDS-PAGE, and visualized with a phosphoimager. Kinase activity was measured with Quantity One software (Bio-Rad).
RESULTS
KSHV lytic replication prolongs the G1 phase of naturally infected B cells.
In our investigation of TPA-induced reactivation of KSHV, we observed a general increase in G1 phase of treated cells. These results are illustrated in Table 1. KSHV latent cell lines BCP-1 and BCBL-1 were used in this study, with GA10, a KSHV-free B-cell line, as a control. We measured the fraction of cells in G1 phase by flow cytometry at different times after TPA treatment. TPA treatment of BCP-1 resulted in a marked increase in the G1 fraction, from 47% before treatment to 71% at 48 h after treatment. TPA treatment of BCBL-1 cells had a less robust effect on the G1 fraction, from 37% before treatment to 46% at 72 h, perhaps reflecting the fact that only 10 to 20% of BCBL-1 cells respond to TPA treatment.
TABLE 1.
G0/G1 phase after TPA treatmenta
| Cells | % of cells in G0/G1 at time (h) posttreatment:
|
||||
|---|---|---|---|---|---|
| 0 | 12 | 24 | 48 | 72 | |
| BCP-1 | 46.60 | 54.48 | 64.34 | 70.83 | 55.65 |
| BCPL-1 | 37.27 | 33.98 | 32.15 | 38.18 | 45.55 |
| GA10 | 36.83 | 41.77 | 35.33 | 36.52 | 39.31 |
Cell cycle distribution of KSHV-positive (BCP-1 and BCBL-1) and negative (GA10) cell lines was analyzed after TPA treatment.
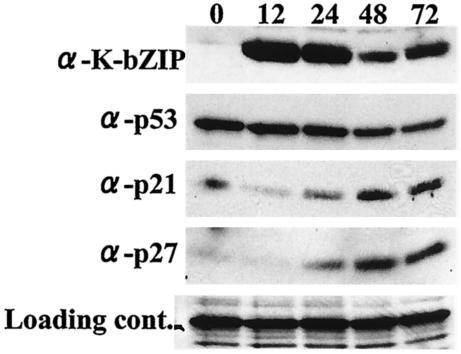
We also monitored the molecular events of BCP-1 cells after TPA treatment by Western blot analysis. As shown in Fig. 1, there was a rapid rise of the level of K-bZIP within 12 h, consistent with its classification as an early-lytic gene product. This was followed by a gradual rise of p21 and p27 protein levels. The amount of p53 did not seem to change significantly in TPA-treated BCP-1, likely due to the high basal level of p53 in these cells. Thus, at least for this cell line, the increase in p21 may not all be attributed to p53 activation. We wished to elucidate the mechanism underlying the cell cycle delay and hypothesized that, in a manner similar to EBV Zta, KSHV K-bZIP may play a role in this process.
FIG. 1.
Effect of KSHV reactivation on p53, p21, and p27 levels. After TPA treatment, BCP1 cells were lysed with EBC buffer. Protein extract (100 μg/lane) from each time point (in hours) was loaded. K-bZIP was detected with anti-K-bZIP rabbit serum. p53, p21, and p27 antibodies were purchased commercially. Nonspecific reaction of antibody was used as a loading control. Protein expression levels of both p21 and p27 were increased after viral reactivation.
K-bZIP physically interacts with cyclin-CDK2 in naturally infected B cells.
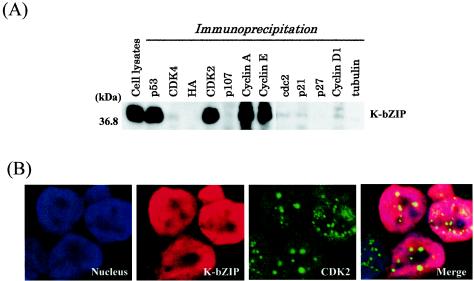
To explore the possible role of K-bZIP in cell cycle regulation, we asked whether K-bZIP interacts directly with cell cycle regulators in BCBL-1 cells without artificial overexpression of the K-bZIP gene product. To this end, BCBL-1 cells were treated with TPA to induce the expression of K-bZIP, and cell extracts isolated at 48 h after treatment were subjected to immunoprecipitation-Western blot analyses with antibodies against various CDKs, cyclins, and additional unrelated molecules as controls. Among the CDKs and cyclins tested, K-bZIP only coprecipitated with CDK2 and its associated cyclins, cyclins E and A, as shown in Fig. 2A. This is interesting, as Polson et al. (42) demonstrated that K-bZIP is phosphorylated by cyclin-CDK2. p53, which is also known to interact with K-bZIP (41), was used as a positive control, and antibody against the irrelevant epitope HA was used as a negative control.
FIG. 2.
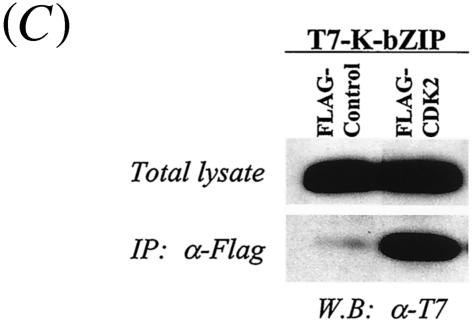
Coimmunoprecipitation assay. (A) Coimmunoprecipitation assay with the KSHV-positive BCBL-1 cell line. BCBL-1 cells induced to viral lytic replication with TPA (48 h) were harvested with EBC buffer, and the same amounts of lysates (500 μg) were precipitated with antibodies against different molecules and then immunoblotted with anti-K-bZIP rabbit serum. (B) Colocalization of K-bZIP with CDK2. Confocal analysis was performed with anti-K-bZIP rabbit serum and anti-CDK2 mouse monoclonal antibody 48 h after TPA induction. K-bZIP (red) and CDK2 (green) were detected with Alexa Fluor 555-conjugated goat F(ab′)2 anti-rabbit immunoglobulin G and Alexa Fluor 488-conjugated goat F(ab′)2 anti-mouse immunoglobulin G. The nucleus was counterstained with To-Pro-3 (blue). This panel is representative of 10 different fields. (C) Association between K-bZIP and CDK2-cyclins in 293T cells. 293T cells were cotransfected with the indicated plasmids. Cell lysates were precipitated with Flag antibody-conjugated agarose, and coimmunoprecipitation of K-bZIP was detected with anti-T7 antibody. The expression of T7-tagged K-bZIP in total lysates is shown in the same blots as a control. IP, immunoprecipitation; W.B, Western blotting; α, anti.
To further establish the interaction between K-bZIP and cyclin-CDK2 in vivo, we studied their subcellular colocalization in BCBL-1 cells. For this experiment, we used highly specific mouse monoclonal antibodies against CDK2, allowing them to be distinguished from the rabbit immunoglobulin G antibody against K-bZIP. Alexa Fluor 555-conjugated anti-rabbit immunoglobulin G and Alexa Fluor 488-conjugated anti-mouse immunoglobulin G were used to detect K-bZIP (red) and CDK2 (green), respectively. As shown in Fig. 2B, K-bZIP was localized in the nucleus in a diffuse pattern, and CDK2 was primarily present in Cajal bodies, as we showed previously (33). The merged image indicates that a fraction of K-bZIP and CDK2 were colocalized in the Cajal bodies in this environment.
K-bZIP interacts with CDK2 in the absence of other viral factors.
The studies described above were conducted in physiologically relevant KSHV-infected BCBL-1 cells. However, the situation was complicated by the presence of other KSHV gene products in these cells. Additionally, to rule out the possibility that the observed association in BCBL-1 cells was due to nonspecific effects of the polyclonal antibodies used, we constructed CDK2 and K-bZIP expression vectors tagged with Flag and T7 sequences, respectively. These constructs were linked to a cytomegalovirus promoter and cotransfected into 293T cells. Forty-eight hours after transfection, the cells were harvested and lysed. CDK2 was immunoprecipitated with Flag antibody-coated beads, and the coprecipitates were probed with anti-T7 antibody to detect K-bZIP. As shown in Fig. 2C, T7-K-bZIP coprecipitated with Flag-CDK2 but not with the Flag-tagged vector control. Reciprocal experiments with the T7-CDK2 and Flag-K-bZIP expression vectors confirmed this result (data not shown). These data showed that K-bZIP without other viral products is able to interact with CDK2.
K-bZIP interacts directly with cyclin-CDK2 via the basic region and downmodulates CKD2 kinase activity.
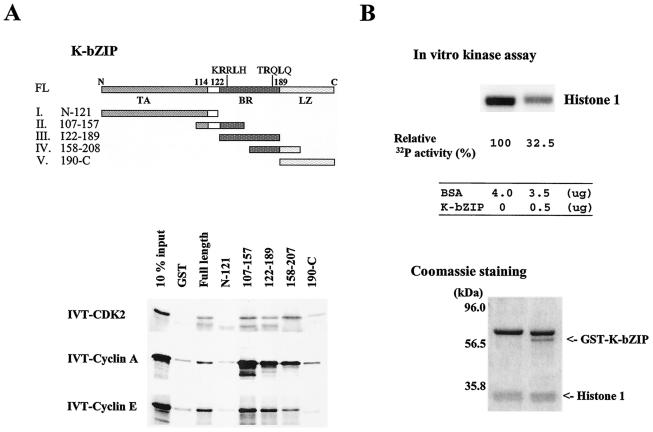
To determine whether the interaction between K-bZIP and cyclin-CDK2 is direct, we incubated in vitro-translated [35S]CDK2, cyclin A, or cyclin E with purified GST-K-bZIP. As shown in Fig. 3A, full-length GST-K-bZIP pulled down CDK2, cyclin A, and cyclin E. There was some background association of GST with in vitro-translated cyclins A and E, which however could be distinguished from the increased intensities of the GST-K-bZIP pulldowns.
FIG. 3.
In vitro interaction and interacting domain of K-bZIP. (A) Domains of K-bZIP and GST-K-bZIP mutants I to V are indicated in the panel. The results of GST pulldown assays for in vitro-translated (IVT), 35S-labeled full-length CDK2, cyclin A, and cyclin E are shown. FL, full length; TA, transactivation domain; BR, basic region; LZ, leucine zipper domain. (B) Inhibition of CDK2-cyclin kinase activity by K-bZIP. CDK2-cyclin kinase activity was measured by in vitro kinase assay with histone 1 as a substrate. The kinase activity of half of the reaction was measured by Quantity One. Relative kinase activity is shown. Protein amounts used in this assay are indicated at the bottom of the panel. The Coomassie-stained gel of the other half of the reaction is shown as a loading control. BSA, bovine serum albumin.
To map the interaction domain of K-bZIP, we constructed a series of K-bZIP deletion mutants (Fig. 3A, upper panel) and subjected them to the GST pulldown assay. The results in Fig. 3A, lower panel, showed that the N-terminal half of K-bZIP was ineffective in pulling down CDK2, cyclin A, or cyclin E. By contrast, the basic region appeared to bind CDK2, cyclin A, and cyclin E with an intensity even higher than that of the full-length K-bZIP. The leucine zipper region showed limited affinity toward cyclin A and CDK2. These data suggest that the interaction between K-bZIP and the cyclin-CDK2 complex is direct and that the basic region of K-bZIP is the primary domain interacting with this complex, consistent with the presence of a cyclin-binding motif in this region (see Discussion).
This direct interaction has a functional impact on CDK2. In an in vitro kinase assay of the CDK2 complex with histone 1 as a substrate, the addition of GST-K-bZIP significantly reduced the kinase activity (Fig. 3B, upper panel). The lower panel provides the proper reaction control, which indicates that the amounts of histone 1 were similar and the only difference in these two reactions was the addition of purified GST-K-bZIP. These results suggest that direct association of K-bZIP with the cyclin-CDK2 complex leads to inhibition of the kinase activity.
K-bZIP expression affects cell growth kinetics.
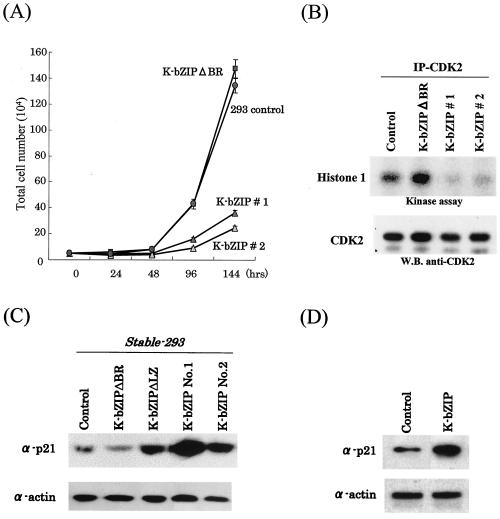
To study the biological consequence of K-bZIP interaction with cyclin-CDK2, we established stable clones of 293 cells bearing Flag-K-bZIP. Cells transfected with Flag-K-bZIPΔBR, a mutant devoid of the basic region, were used as controls. As shown in Fig. 4A, the growth rate of cells bearing wild-type K-bZIP (K-bZIP #1) was significantly lower than the parental line. An independently isolated K-bZIP-293 line (K-bZIP #2) behaved similarly, suggesting that the results are not due to clonal variations of selectants. 293 cells bearing a mutation that deletes the basic region, expected to be incapable of interacting with CDK2, grew with kinetics nearly identical to that of the parental 293 cells. In cells expressing the full-length construct, CDK2 activity was barely detectable, as shown in Fig. 4B. In agreement with the kinetics data, the CDK2 activity in K-bZIPΔBR-transfected cells remained similar to that of vector-transfected cells.
FIG. 4.
(A) K-bZIP influence on cell cycle division. For determination of cell growth rate, 5 × 104 cells of Flag-293-K-bZIPwt 1, Flag-293-K-bZIPwt 2, Flag-K-bZIPΔBR, and Flag-293-vector were seeded in 60-mm plates and cultured in 10% fetal bovine serum-DMEM. Cell numbers were counted at the indicated time points postseeding. Triplicate plates were prepared for each cell clone. (B) Inhibition of CDK2 kinase activity in Flag-293-K-bZIPwt cell lines. CDK2 kinase activity was determined by in vitro kinase assay after immunoprecipitation (IP) of CDK2 from stable cell lines. Histone 1 was used as a substrate for the assay. The amounts of the CDK2 were measured by immunoblotting. W.B., Western blotting. (C) Upregulation of p21 in Flag-293-K-bZIPwt and Flag-K-bZIPΔLZ cell lines. Protein expression levels of p21 in stably K-bZIP-expressing cell lines were measured. Total cell lysates were prepared from the stable cell lines. Actin served as an internal control for the amounts of protein on the membrane. (D) Upregulation of p21 in transiently transfected HeLa cells. HeLa cells were transfected pFlag-empty vector or pFlag-K-bZIPwt. At 72 h posttransfection, total cell lysates were obtained, and 50 μg of protein was loaded in each lane and probed with anti-p21 antibody. Actin served as an internal control for the amount of protein on the membrane.
The downmodulation of CDK2 was also accompanied by an increase in the levels of p21 (Fig. 4C). It has been reported that phosphorylation by CDK2 targets p21 for degradation (48). Inhibition of CDK2 activity by K-bZIP would thus be expected to increase the p21 protein level, as we observed here. To offer an independent confirmation of this notion, HeLa cells were transiently transfected with K-bZIP, and the levels of p21 protein were measured. As shown in Fig. 4D, K-bZIP expression led to an increased level of p21. These data suggest that the significant growth-inhibitory effect of K-bZIP is caused by a combined effect of binding of K-bZIP to cyclin-CDK2 and the increased expression of CDK2 inhibitor p21. Recently, Wu et al. (58) showed that K-bZIP enhanced p21 expression through interaction with C/EBPα, a transcriptional factor for the p21 locus. These authors mapped the interacting domain to be primarily in the leucine zipper domain of K-bZIP.
To distinguish the relative contributions of p21 increase by CDK2 and C/EBPα, we constructed a K-bZIP mutant, K-bZIPΔLZ, devoid of the leucine zipper region (amino acids 189 to 237), and developed a corresponding, stable 293 clone. K-bZIPΔLZ bound cyclin-CDK2 but failed to interact with C/EBPα (58) (data not shown). As shown in Fig. 4C, increased p21 expression was observed in a K-bZIPΔLZ clone, albeit at a lower level, compared to the wild-type K-bZIP clones. These data suggest that K-bZIP mediated the increase in p21 expression through both CDK2 and other pathways, such as C/EBPα.
K-bZIP overexpression inhibits growth of BCBL-1 and extends G1 phase of reactivated cells.
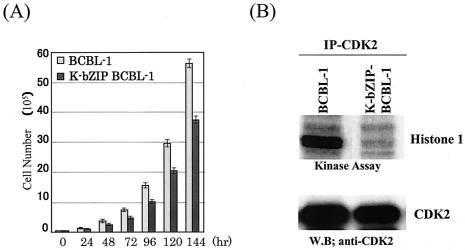
Having demonstrated that K-bZIP expression leads to growth inhibition of 293 cells, we wished to extend this analysis to the more appropriate context of BCBL-1 cells. BCBL-1 cells were transfected with HA-K-bZIP, which carries the G418 resistance marker gene. Stable clones selected in medium supplemented with G418 were pooled, and their growth rate was compared to that of vector-transfected BCBL-1 cells which had been similarly selected. As shown in Fig. 5A, the K-bZIP-BCBL-1 pool displayed a significantly slower growth rate than the vector-transfected BCBL-1 cells. This slower growth was accompanied by much reduced CDK2 kinase activity, as shown by the in vitro kinase assay (Fig. 5B).
FIG. 5.
(A) K-bZIP slows down cell cycle division. A total of 5 × 104 K-bZIP-BCBL-1 cells or parental BCBL-1 cells were seeded in 60-mm plates and cultured in 15% fetal bovine serum-RPMI 1640. Cell numbers were counted at the indicated time points. Triplicate plates were seeded for each cell clone. (B) Inhibition of CDK2 kinase activity in K-bZIP-BCBL-1 cells. CDK2 kinase activity was determined by the in vitro kinase assay after induction of K-bZIP expression (72 h). CDK2 was immunoprecipitated from K-bZIP-BCBL-1 or parental BCBL-1 cells. Histone 1 was used as a substrate for the assay. The amounts of CDK2 were measured by immunoblotting. W.B, Western blotting.
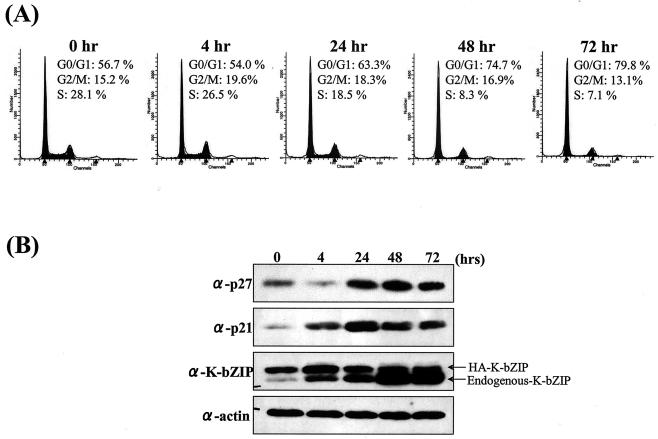
We also studied the G1 fraction of K-bZIP-BCBL-1 cells after TPA treatment and compared it with that of the parental BCBL-1 cells. As shown in Fig. 6A, the constitutive expression of K-bZIP increased the fraction of cells in G1 before TPA treatment (0 h) to 57% (from the original 37% in parental BCBL-1 cells, Table 1). The HA epitope tag allowed us to distinguish between the ectopic and endogenous forms of K-bZIP by size, and their levels through the course of TPA treatment are shown in Fig. 6B. Because TPA treatment induces the expression of the endogenous viral K-bZIP, we would expect it to work in concert with the ectopically expressed HA-K-bZIP to further augment the G1 fraction increase. Indeed, the G0/G1 fraction increased from 54% at 4 h to 63% at 24 h and to 80% at 72 h after TPA treatment. At the same time, the S fraction was reduced from 27% at 4 h posttreatment to only 7% at 72 h. In parallel, the levels of both p21 and p27 increased significantly. This showed that a combined action of exogenous and endogenous K-bZIP expression has a devastating effect on the cell cycle progression of the infected cell.
FIG. 6.
KSHV reactivation and K-bZIP expression prolong G0/G1 phase. (A) Cell cycle analysis of TPA-treated K-bZIP-BCBL-1 cells. Cell cycle analysis after induction by TPA was carried out with flow cytometry. Cell cycle analysis was performed with ModFit LT. The percentage of cells in G0/G1 phase is indicated. (B) Upregulation of p21 and p27 in K-bZIP-BCBL-1 cells after induction. Protein extract (100 μg/lane) from each time point (in hours) was loaded. Protein expression levels of p21, p27, and total K-bZIP in HA-K-bZIP-BCBL-1 cells were measured by immunoblotting. Actin served as an internal control for the amount of protein on the membrane. α, anti.
DISCUSSION
Previously, we and others (32, 61) independently reported the identification of a basic leucine zipper protein of KSHV, and we designated this gene K-bZIP. K-bZIP is the structural and positional analogue of the EBV Zta gene. EBV Zta functions as a potent transcriptional factor which triggers lytic replication when overexpressed in latent EBV cell lines, and it, together with Rta, represents the main latent to lytic replication switch for the virus. There is considerable evidence indicating that KSHV Rta, the ortholog of EBV Rta, is a potent activator of KSHV replication. Overexpression of KSHV Rta in the BCBL-1 cell line leads to reactivation of the latent KSHV genome (36, 40, 53). In contrast, overexpression of K-bZIP has not yielded lytic replication (42) and, as shown in this report, it results in a reduction of the growth rate in the latent cells.
We recently reported that K-bZIP is a transcriptional coregulator of KSHV Rta (28). Through association, K-bZIP suppresses the transactivation ability of KSHV Rta in a promoter-dependent manner. Others have shown that K-bZIP is a coregulator of CBP, p53, and C/EBPα (27, 41, 58). Thus, K-bZIP's transcriptional role seems at present to be limited to its activity as a coactivator or a corepressor. By analogy to EBV Zta, K-bZIP may have other nontranscriptional functions in the KSHV life cycle. EBV Zta is known to bind the viral DNA replication origin and is involved in EBV genomic replication (18, 46, 57). K-bZIP was similarly shown to assist in the formation of a potential KSHV DNA prereplication complex (57), though its specific role in genome replication remains elusive. EBV Zta has also been shown to mediate cellular growth arrest by stabilizing p53, inducing the expression of p21 and p27 (10, 19, 44), thereby contributing to the inhibition of CDK2 activity. In this study, we explored the role of K-bZIP in modulating cell cycle progression.
CDK2 plays a pivotal role in cell cycle progression. Phosphorylation by CDK2 of retinoblastoma protein or its related molecules, the p130 and p107 pocket proteins, sets the stage for entry into the S phase by releasing and activating E2F, a transactivator of cyclin E and DNA polymerase (9, 24, 47, 55). This is viewed as a major pathway by which CDK2 advances G1 to S transition (4). Recently, it was shown that CDK2 has additional substrates which facilitate this transition in a retinoblastoma protein-independent manner. These substrates include E2F1, E2F5, histone H1, HIRA, and NPAT (23, 60). HIRA and NPAT are both involved in the histone biosynthesis which accompanies DNA replication (17, 23). CDK2 also phosphorylates its own inhibitors, p21 and p27, targeting them for degradation (38, 48). Thus, CDK2 activity controls multiple pathways leading to S-phase progression. It is therefore no surprise that multiple signal transduction pathways such as growth factors, oncogenes, and DNA repair machinery all target CDK2 activity to control cell cycle.
Viruses have also evolved ways to target cyclin-CDK2 to modulate the cell cycle to suit their particular needs during replication. Some of these viral gene products, primarily those involved in latency or transformation, accelerate G1 to S transition, while others do the opposite. A number of factors interact directly with cell cycle regulators such as CDKs and cyclins or interact with checkpoint proteins such as the CDK inhibitors p53 and retinoblastoma protein; others transcriptionally modulate CDK inhibitors (5, 10, 19, 22, 51, 56, 58, 59). There are yet other viral gene products, such as v-cyclin of KSHV, which represent hyperactive homologs of the cell cycle proteins themselves (14, 16, 21, 29). The consequences of all these interactions are either activation or inactivation of the cyclin-CDK complexes, which leads to acceleration or deceleration of progression through the cell cycle.
In this report, we have shown that KSHV K-bZIP targets CDK2 directly and induces growth delay at the G0/G1 phase. K-bZIP achieves this end by physically associating via its basic region with cyclin-CDK2. This interaction is direct, as demonstrated by in vitro studies with purified protein preparations, and can be detected in naturally infected B cells without overexpression of either component. In addition, some of the K-bZIP and CDK2 are found to be colocalized in the nucleus. The consequence of this interaction is the inhibition of CDK2 kinase activity. The inhibitory mechanism is likely to be similar to those of p21, p27, and p57 (1). This class of factors, which also include retinoblastoma protein, HIRA, and E2F1, bind cyclin-CDK2 tightly and are characterized by cyclin-binding RXL motifs, surrounded by S/TPXK/R motifs, which serve as sites of phosphorylation by CDK2 (1, 23).
We observed that the K-bZIP basic region contains two RXL motifs (KRRLH and TRQLQ), similar to p21, which are both deleted in our K-bZIPΔBR construct. These may account for the observed association with cyclins E and A. KRRLH is present in the N-terminal half of the basic region, where as TRQLQ is in the C-terminal half. As shown in Fig. 3A, both halves interact with the cyclin-CDK2 complex, in agreement with the above assertion. Interestingly, precisely 14 amino acids preceding each of the RXL motif is an S/TPXK/R motif (TPPR for RRL, and SPTR for RQL). Both sites were shown by Polson et al. (42) to be phosphorylated by CDK2. Our results also showed that K-bZIP binds in vitro-translated CDK2. We are not sure that this interaction is direct, due to the likely presence of cyclins in the reticulocyte lysates. In either case, we have presented strong evidence that K-bZIP is able to bind the cyclin-CDK2 complex. This tight binding mediates both a potent inhibition of cyclin-CDK2 kinase activity, presumably by blocking the interaction of substrates with cyclin-CDK2 complexes, and a profound reduction in cell growth, as seen in 293 cells overexpressing K-bZIP. Because this cell line is devoid of any other KSHV proteins, the data suggest that K-bZIP alone is sufficient to inhibit cell cycle progression. In BCBL-1 cells, the effect of overexpression of K-bZIP is less dramatic, likely due to the presence of KSHV latent proteins such as v-cyclin and LANA, both of which have been shown to have the potential to accelerate cell cycle progression (16, 20, 21, 29). The deceleratory effect of K-bZIP may be blunted by these latent viral proteins, which normally occupy a separate and distinct phase of the viral life cycle. After TPA treatment, which represses the expression of latent genes and activates viral lytic genes, including K-bZIP, a significant fraction (nearly 80%) of the cells were in the G0/G1 phase, while only 7% were in S phase. This rather dramatic effect was caused by a combination of the ectopically expressed HA-K-bZIP and the endogenous K-bZIP (Fig. 6A). The above experiments demonstrate the ability of K-bZIP to delay the G1 to S transition, presumably allowing ample time for virus transcription, translation, and initiation of viral DNA replication to take place.
Based on the present work and published reports, there are at least three nonmutually exclusive pathways by which K-bZIP can inhibit CDK2 and induce growth arrest. The first, as shown in this study, is direct binding to cyclin-CDK2. There are other indirect effects of K-bZIP on CDK2 activity. For instance, we observed increased protein levels of both p21 and p27 in cells that overexpressed K-bZIP. As described above, CDK2 phosphorylation of p21 and p27 targets these molecules for degradation. K-bZIP binding would thus have the potential to increase the protein levels of p21 and p27 by shielding them from this degradation signal. A second mechanism, recently reported by Wu et al. (58), is increased expression of p21 mediated by K-bZIP through binding C/EBPα, a factor known to drive the transcription of p21. These authors demonstrated that K-bZIP overexpression in mouse fibroblasts lead to a prolonged G0/G1 phase, accompanied by an increase in p21 transcripts. C/EBPα was a necessary component in this process. While their data were primarily based on studies in mouse fibroblasts, the cell cycle inhibition and p21 increase are consistent with what we observed in K-bZIP-overexpressing BCBL-1 cells. It is thus possible that this mechanism also operates in BCBL-1 cells. Finally, it was reported that K-bZIP interacts with and presumably stabilizes p53 (41). We were able to reproduce these data in KSHV-infected cells (Fig. 2A). As p53 is a known transcriptional activator of p21, it is conceivable that this interaction may stabilize p53 and thereby lead to increased transcription of p21 in cells overexpressing K-bZIP. The stabilization of p53 by cytomegalovirus IE2 and EBV Zta is a suggested mechanism by which these two viral gene products inhibit cell growth (19, 51, 54). All three mechanisms described above converge on the inactivation of CDK2, which results in the delayed cell cycle progression from G1 to S.
There are other implications of the interaction between CDK2 and K-bZIP. Polson et al. (42) showed that CDK2 phosphorylates K-bZIP at Thr111 and Ser167 in vitro. It is conceivable that K-bZIP inhibits CDK2 activity by serving as a high-affinity substrate. In return, CDK2 phosphorylates K-bZIP, potentially modifying its function. Phosphorylation of bZIP transcription factors may alter their transactivation activity, DNA binding ability, stability, and/or subcellular localization. We have previously shown that Meq, a bZIP protein encoded by Marek's disease herpesvirus, is colocalized with CDK2 in Cajal bodies and phosphorylated by cyclin-CDK2 (33). CDK2 phosphorylation at Ser42 of Meq weakens its DNA binding affinity and translocates Meq into the cytoplasm. Likewise, herpes simplex virus UL42 DNA synthesis processivity factor is known to bind and be phosphorylated by CDK1 (2). It is worth noting that CDK2 plays an important role in the replication of other DNA viruses such as human papillomavirus (31), cytomegalovirus (8), and polyomavirus (43). Whether CDK2 plays a direct role in KSHV replication and how CDK2 phosphorylation of K-bZIP may alter its function will require further investigation. The availability of CDK2 phosphoacceptor site mutants of K-bZIP (42) should facilitate such work.
In summary, we have shown that K-bZIP interacts directly with and inhibits cyclin-CDK2, thereby prolonging the G1 phase of KSHV-infected cells. Our study and that of Wu et al. (58) clearly establish a nontranscriptional role of K-bZIP in KSHV replication.
Acknowledgments
This work was supported by NIH grants CA91574 and CA46613 (to H.-J.K.) and Universitywide AIDS Research Program grant R00-D-034 (to H.-J.K). Y.I. was supported by a UEHARA memorial postdoctoral fellowship from Japan.
REFERENCES
- 1.Adams, P. D., W. R. Sellers, S. K. Sharma, A. D. Wu, C. M. Nalin, and W. G. Kaelin, Jr. 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2001. cdc2 cyclin-dependent kinase binds and phosphorylates herpes simplex virus 1 UL42 DNA synthesis processivity factor. J. Virol. 75:10326-10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkan, S., D. S. Karcher, A. Ortiz, S. Khalil, M. Akhtar, and M. A. Ali. 1997. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus in organ transplant patients with immunosuppression. Br. J. Haematol. 96:412-414. [DOI] [PubMed] [Google Scholar]
- 4.Bartek, J., and J. Lukas. 2001. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 490:117-122. [DOI] [PubMed] [Google Scholar]
- 5.Bonin, L. R., and J. K. McDougall. 1997. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J. Virol. 71:5861-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 7.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 8.Bresnahan, W. A., I. Boldogh, P. Chi, E. A. Thompson, and T. Albrecht. 1997. Inhibition of cellular Cdk2 activity blocks human cytomegalovirus replication. Virology 231:239-247. [DOI] [PubMed] [Google Scholar]
- 9.Bruce, J. L., R. K. Jr. Hurford, M. Classon, J. Koh, and N. Dyson. 2000. Requirements for cell cycle arrest by p16INK4a. Mol. Cell 6:737-742. [DOI] [PubMed] [Google Scholar]
- 10.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 11.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee, M., J. Osborne, G. Bestetti, Y. Chang, and P. S. Moore. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 13.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehmann, G. L., T. I. McLean, and S. L. Bachenheimer. 2000. Herpes simplex virus type 1 infection imposes a G1/S block in asynchronously growing cells and prevents G1 entry in quiescent cells. Virology 267:335-349. [DOI] [PubMed] [Google Scholar]
- 16.Ellis, M., Y. P. Chew, L. Fallis, S. Freddersdorf, C. Boshoff, R. A. Weiss, X. Lu, and S. Mittnacht. 1999. Degradation of p27(Kip) cdk inhibitor triggered by Kaposi's sarcoma virus cyclin-cdk6 complex. EMBO J. 18:644-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewen, M. E. 2000. Where the cell cycle and histones meet. Genes Dev. 14:2265-2270. [DOI] [PubMed] [Google Scholar]
- 18.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friborg, J. Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 21.Godden-Kent, D., S. J. Talbot, C. Boshoff, Y. Chang, P. Moore, R. A. Weiss, and S. Mittnacht. 1997. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J. Virol. 71:4193-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagemeier, C., R. Caswell, G. Hayhurst, J. Sinclair, and T. Kouzarides. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 13:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, C., D. M. Nelson, X. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 25.Hobbs, W. E. 2nd., and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y. Q., J. J. Li, M. H. Kaplan, B. Poiesz, E. Katabira, W. C. Zhang, D. Feiner, and A. E. Friedman-Kien. 1995. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet 345:759-761. [DOI] [PubMed] [Google Scholar]
- 27.Hwang, S., Y. Gwack, H. Byun, C. Lim, and J. Choe. 2001. The Kaposi's sarcoma-associated herpesvirus K8 protein interacts with CREB-binding protein (CBP) and represses CBP-mediated transcription. J. Virol. 75:9509-9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumiya, Y., S.-F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H.-J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of KSHV Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaldis, P., P. M. Ojala, L. Tong, T. P. Makela, and M. J. Solomon. 2001. CAK-independent activation of CDK6 by a viral cyclin. Mol. Biol. Cell 12:3987-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, B. Y., T. Ma, J. S. Liu, S. R. Kuo, G. Jin, T. R. Broker, J. W. Harper, and L. T. Chow. 2000. HeLa cells are phenotypically limiting in cyclin E/CDK2 for efficient human papillomavirus DNA replication. J. Biol. Chem. 275:6167-6174. [DOI] [PubMed] [Google Scholar]
- 32.Lin, S.-F., D. R. Robinson, G. Miller, and H.-J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J.-L., Y. Ye, Z. Qian, Y. Qian, D. J. Templeton, L. F. Lee, and H.-J. Kung. 1999. Functional interactions between herpesvirus oncoprotein MEQ and cell cycle regulator CDK2. J. Virol. 73:4208-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 37.Memar, O. M., P. L. Rady, and S. K. Tyring. 1995. Human herpesvirus-8: detection of novel herpesvirus-like DNA sequences in Kaposi's sarcoma and other lesions. J. Mol. Med. 73:603-609. [DOI] [PubMed] [Google Scholar]
- 38.Montagnoli, A., F. Fiore, E. Eytan, A. C. Carrano, G. F. Draetta, A. Hershko, and M. Pagano. 1999. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 13:1181-1189. [DOI] [PMC free article] [PubMed]
- 39.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in KSHV gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynisdottir, I., S. Bhattacharyya, D. Zhang, and C. Prives. 1999. The retinoblastoma protein alters the phosphorylation state of polyomavirus large T antigen in murine cell extracts and inhibits polyomavirus origin DNA replication. J. Virol. 73:3004-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, A., M. Armstrong, D. Dwyer, and E. Flemington. 1999. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J. Virol. 73:9029-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherr, C. J. 2000. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 60:3689-3695. [PubMed] [Google Scholar]
- 48.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 49.Song, B., K. C. Yeh, J. Liu, and D. M. Knipe. 2001. Herpes simplex virus gene products required for viral inhibition of expression of G1-phase functions. Virology 290:320-328. [DOI] [PubMed] [Google Scholar]
- 50.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 51.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 52.Sun, R., S.-F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, R., S.-F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 56.Wu, F. Y., H. Chen, S. E. Wang, C. M. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein alpha interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, F. Y., J.-H. Ahn, D. J. Alecendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication comparetments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, F. Y., Q. Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. D. Lane, and G. S. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Q., D. Gutsch, and S. Kenney. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, J., B. Dynlacht, T. Imai, T. Hori, and E. Harlow. 1998. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 12:456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]