Abstract
Recent studies suggest that NGAL (neutrophil gelatinase-associated lipocalin) is a novel iron transporter with functions distinct from that of transferrin and mediates a new iron-delivery pathway. To get a better understanding of NGAL function in oesophageal carcinoma, we analysed the expression of NGAL receptors in oesophageal carcinoma cells and identified a novel spliced variant designated NgalR-3. When expressed in a heterologous system, the protein produced from this novel spliced variant exhibits the biochemical characteristics of interaction and co-localization with NGAL protein in vivo. This new finding suggests that NgalR-3 may act as a potential NGAL receptor and play a role in NGAL-mediated iron transport in oesophageal carcinoma.
Keywords: iron transport, lipocalin, neutrophil gelatinase-associated lipocalin (NGAL), NgalR-3, oesophageal carcinoma
Abbreviations: 24p3R, 24p3 receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HA, haemagglutinin; NGAL, neutrophil gelatinase-associated lipocalin; NgalR, NGAL receptor; RT, reverse transcription; TBS, Tris-buffered saline
INTRODUCTION
Lipocalins are a diverse family of proteins with low levels of overall sequence conservation, but share a common tertiary structure with an eight-stranded hydrogen-bonded antiparallel β-barrel surrounding a cup-shaped internal ligand-binding site [1–2]. This confers on lipocalins the ability to bind a wide variety of small, principally hydrophobic, molecules and specific cell-surface receptors, and form covalent and non-covalent complexes with other soluble macromolecules [3]. NGAL (neutrophil gelatinase-associated lipocalin), a member of the lipocalin superfamily, was originally found to be a protein stored in specific granules of the human neutrophil [4]. Besides neutrophils, NGAL is expressed in most tissues normally exposed to micro-organisms, and induced in epithelial cells during inflammation [5,6]. Previous studies have demonstrated that NGAL is involved in diverse cellular processes. NGAL is capable of protecting MMP-9 (matrix metalloproteinase 9) from degradation by interacting with this protein [7,8]. It has been reported that NGAL can bind bacterial catecholate-type ferric siderophores and act as a potent bacteriostatic agent by sequestering iron. NGAL may be involved in the innate immune system and acute-phase response to infection [9,10]. It has also been reported that NGAL can protect against acute ischaemic renal injury and has been implicated in apoptosis as a survival factor [11,12]. Also, NGAL may be important in delivering iron to cells during the formation of the tubular epithelial cells of the primordial kidney [13,14].
Elevated NGAL expression has been observed in several types of cancer, such as carcinomas of the colon, ovaries, pancreas and breast [15–19]. Our previous studies have demonstrated that NGAL overexpression plays an important role in the progress of malignant transformation of human immortalized oesophageal epithelial cells and is involved in the invasion of oesophageal carcinoma cells [20–22]. Recently, our research has shown that NGAL is involved in the transformation and development of oesophageal carcinoma [23]. We also found that the expression of NGAL in the oesophageal carcinoma cells can be induced following stimulation by PMA [24].
Recently, Hvidberg et al. [25] found that megalin, a member of the low-density lipoprotein receptor family expressed in polarized epithelia, bound NGAL with high affinity. Devireddy et al. [26] also isolated a specific cell-surface receptor of 24p3 (a highly conserved murine homologue of NGAL), 24p3R, from murine FL5.12 cells and demonstrated that 24p3R expression conferred on cells the ability to undergo either iron uptake or apoptosis. Sequence analysis revealed the presence of a highly conserved 24p3R human homologue (GenBank® accession number NM_016609) designated NgalR-2 (NGAL receptor 2), which is a member of the organic cation transporter family. It also contains another spliced variant designated NgalR-1. According to previous studies, we proposed that NgalRs might be present in oesophageal carcinoma and play a role in NGAL-mediated iron transport in oesophageal carcinoma cells.
In the present study, we found a shorter NgalR cDNA, NgalR-3, generated by alternative splicing in oesophageal carcinoma cells. This spliced isoform contains an internal deletion of 125 nucleotides and could produce a 207-amino-acid protein containing four putative membrane-spanning domains. Interestingly, NgalR-3 exhibits the expected functionality of co-localization and interaction with NGAL protein in vivo.
MATERIALS AND METHODS
Tissue samples and cell lines
Human oesophageal carcinoma tissues and adjacent normal oesophageal mucosa tissues were collected from patients undergoing surgery in the First Affiliated Hospital of Shantou University from 2003 to 2005 and were frozen immediately at −70 °C. The study was approved by the Ethical Committee of the First Affiliated Hospital of Shantou University and written informed consents were obtained from all patients undergoing surgery to use resected samples for research. Human non-carcinoma tissues were acquired from the Pathology Department of Medical College of Shantou University. Human oesophageal carcinoma cell lines EC109, EC18, EC171 and EC8712 were cultured in 199 medium (Gibco) supplemented with 10% (v/v) heat-inactivated bovine serum. Cells were cultured in flasks in a humidified atmosphere of 5% CO2 at 37 °C.
Human NgalR cDNA cloning and plasmid constructions
NgalR cDNAs were cloned by RT (reverse transcription)–PCR. Briefly, RNA was extracted from oesophageal carcinoma cell line EC18 cells with the TRIzol® Reagent (Invitrogen) and reverse-transcribed using the Reverse Transcription System (Promega). NgalRs were amplified using the following primers [designed according to the published sequence (GenBank® accession number NM_016609)]: primer 1, 5′-CGGAATTCGGGCCTCGGACCCCATCTT-3′, as forward (initiation codons ATG were deleted), and primer 2, 5′-CGCTCGAGCTCAGAGGGCAGGGTTGG-3′, as reverse (containing an EcoRI and an XhoI restriction site respectively). Conditions for PCR amplification were: 30 cycles of 30 s at 95 °C, 30 s at 55 °C and 2 min at 72 °C, and a last cycle of elongation at 72 °C for 5 min. PCR products were purified and ligated into the pGEM-T vector (Promega). The insert fragment was verified by complete sequencing.
To construct the Myc-tagged form of NgalRs, two alternatively spliced variants of NgalR products were obtained, NgalR-2 and NgalR-3, digested with EcoRI and XhoI and subcloned into the pCMV-Myc mammalian expression vector (BD Biosciences) that expresses proteins containing the N-terminal c-Myc epitope tag to finally obtain Myc–NgalR-2 and Myc–NgalR-3.
RT–PCR analyses
Total RNA was extracted from frozen stored tissues and cells with TRIzol® reagent. RT was performed in a total volume of 20 μl using 1 μg of total RNA, oligo-dT primer, random primers and AMV (avian myeloblastosis virus) reverse transcriptase (Promega). PCR was then performed for 35 cycles with 1 μl of cDNA as a template. To avoid amplification of contaminating genomic DNA, amplified fragments of each gene were intron-spanning. The primers used for novel NgalR-3 variant identification were primer 3, 5′-TGTGCTCACCACCAACGC-3′, as forward, and primer 4, 5′-CCTCAATCTGCCGCTTCACT-3′, as reverse. Additional primers were as follows: forward primer 5′-GTGGATCCTTCCTCGGCCCTGAAATCATG-3′ and reverse primer 5′-GGGAATTCTCAGCCGTCGATACACTGGTC-3′ for NGAL cDNA; and forward primer 5′-CCCCTACTGCCTATATCGAC-3′ and reverse primer 5′-AATGATACGGGTGCTCTGAG-3′ for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Cell culture and transfection
COS-7 cells were cultured in 199 medium (Gibco) supplemented with 10% (v/v) heat-inactivated bovine serum. pCMV-Myc plasmid vector DNA containing NgalR-2 or NgalR-3 cDNA was purified using the Qiagen Plasmid Midi kits. At 1 day before transfection, 1.5×106 COS-7 cells were seeded on to 10-cm-diameter tissue-culture dishes. The cells were transfected with 10 μg of total plasmid DNA per dish using SuperFect transfection reagent (Qiagen). Cells were cultured for 24 h after transfection and were used for subsequent studies.
Western blot analysis
Total cell lysates were prepared in a lysis buffer (150 mM NaCl, 0.1% Nonidet P40 and 50 mM Tris/HCl, pH 7.5) supplemented with 1 mM PMSF and Complete™ protease inhibitor cocktail tablets (Santa Cruz Biotechnology). Lysates were cleared by centrifugation at 10000 g for 10 min at 4 °C, separated by SDS/PAGE (12% gels) and transferred on to a PVDF membrane (Millipore). The membrane was incubated in 10 ml of blocking buffer [TBS (Tris-buffered saline), containing 0.1% (v/v) Tween 20 and 5% (w/v) non-fat dried milk powder] for 1 h at room temperature (25 °C) followed by the addition of the anti-Myc antibody (BD Biosciences) for 2 h at room temperature. Then, the membrane was rinsed three times with TBS/Tween 20 and incubated with horseradish-peroxidase-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology) for 2 h at room temperature. Immunoreactive bands were revealed by Western blotting with luminol reagent (Santa Cruz Biotechnology) and photographed by FluorChem 8900 (Alpha Innotech).
Immunofluorescence cell staining analysis
Cells seeded on coverslips were co-transfected with either Myc-tagged NgalR-2 or Myc-tagged NgalR-3 with HA (haemagglutinin)-tagged NGAL respectively, and HA-tagged and Myc-tagged empty vectors as control. Cells were incubated for 24 h. After being washed with PBS, cells were fixed with 4% (w/v) formaldehyde for 15 min and then treated with 0.1% (v/v) Triton X-100 in PBS for 15 min. Cells were subsequently blocked with Image-iT FX signal enhancer (Invitrogen) for 30 min at room temperature and incubated with an anti-HA antibody or an anti-Myc antibody overnight at 4 °C. The cells were washed and incubated with an Alexa Fluor® 594-conjugated donkey anti-rabbit IgG (Molecular Probes) or fluorescein-conjugated goat anti-mouse IgG (Zymed) as a secondary antibody for 90 min at 37 °C. The cells were washed and mounted in ProLong Gold antifade reagent (Invitrogen) and examined with a Nikon Eclipse TE200-E fluorescence microscope.
Co-immunoprecipitation
For co-immunoprecipitation studies, COS-7 cells were co-transfected with either HA-tagged NGAL or HA-tagged empty control vector with Myc-tagged NgalR-3/2 respectively, and grown for 24 h in 10-cm-diameter tissue culture dishes. Cells were harvested by lysis as described above. The resulting supernatants were incubated on a rocker with 50 μl of anti-HA affinity matrix (Roche) overnight at 4 °C. Immunoprecipitates were collected by centrifugation at 1000 g for 1 min at 4 °C, washed three times in lysis buffer, suspended in SDS sample buffer [20 mM Tris, pH 7.5, 2 mM EDTA, 5% (w/v) SDS, 0.02% (w/v) Bromophenol Blue, 20% (v/v) glycerol and 200 mM dithiothreitol] and boiled for 5 min. Proteins were subjected to SDS/PAGE (12% gels) and immunoblotting with anti-Myc antibody.
RESULTS
cDNA cloning of human NgalR variants
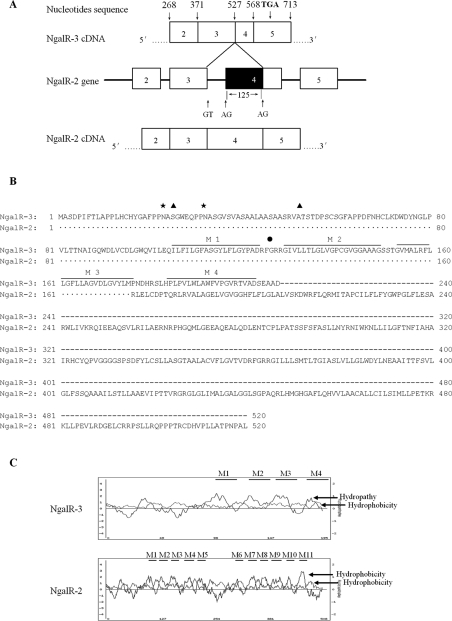
During the cloning of NgalR isoforms from EC18 cells by RT–PCR, we isolated two cDNA clones: the expected molecular mass NgalR-2 fragment and a shorter fragment. Since PCR was carried out by using specific primers complementary to sequences upstream to the 5′ translation start site and 3′-untranslated region of NgalR-2/1 cDNA (NgalR-2 cDNA presents sequences at the 5′ and 3′ termini identical with those of NgalR-1, but contains an internal deletion of 54 nucleotides at exon 7), we thought that a process of alternative splicing could account for the generation of this shorter fragment. Sequence analysis demonstrated that the large inserted cDNA fragment, of 1563 bp, was identical with the published NgalR-2 cDNA sequence (results not shown); however, the shorter cDNA fragment, of 1438 bp, designated NgalR-3 (GenBank® accession number DQ658848) was distinct from NgalR-2. Figure 1(A) shows a diagram of the NgalR-2 gene showing a possible mechanism for alternative splicing giving rise to NgalR-3. The NgalR-3 cDNA presents sequences at 5′ and 3′ termini identical with those of NgalR-2 cDNA. However, it contains an internal deletion of 125 nucleotides at exon 4 that produces a frameshift at amino acid 176 and encodes a predicted 207-amino-acid protein with a unique 32-amino-acid (residues 176–207) C-terminus. The alternative splicing at the junctions conforms to the GT–AG rule. This structure strongly suggests that the NgalR-3 cDNA molecule represents a new spliced variant. Figure 1(B) shows the deduced amino acid sequence of NgalR-3 and its alignment with NgalR-2. NgalR-3 showed an amino acid identity of 100% in the N-terminal 175 amino acids compared with NgalR-2. Based on a Kyte–Doolittle hydropathy analysis [27] and a PSORT II program developed by Horton and Nakai [28], the NgalR-3 polypeptide is predicted to be localized mainly at the membrane and encompass an extracellular N-terminus with two N-linked carbohydrate chains, four putative membrane-spanning domains, one amidation site and two potential CK2 phosphorylation sites (Figure 1C).
Figure 1. Identification of a novel NgalR alternatively spliced variant.
(A) Diagram of the NgalR-2 gene showing a possible mechanism for alternative splicing giving rise to NgalR-3 (GenBank® accession number DQ658848). Lines and boxes show the introns and exons respectively of the NgalR-2 gene. The sequence region which is present in NgalR-2 cDNA (bottom) but not in NgalR-3 cDNA (top) is shown by a solid box. (B) Residues conserved between the two sequences are indicated by dots. Two potential sites of N-linked glycosylation (23 and 32) are marked by ☆. Potential CK2 phosphorylation sites (25 and 53) and an amidation site (125) are marked by ▲ and ● respectively. Putative transmembrane domains are indicated by lines over the sequence with numbers (M1–M4). (C) Kyte–Doolittle hydropathy plots with a window of 13 amino acid residues. Hydropathy and hydrophobicity curves are indicated, and the bars indicate putative transmembrane domains.
Expression of NgalR variant mRNAs in cell lines and normal tissues
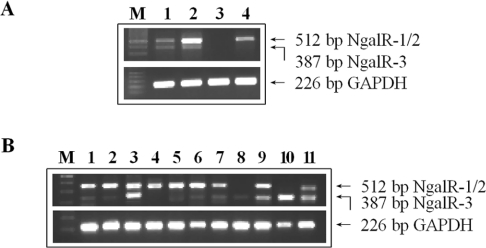
The relative expression of the NgalR-2/1 and NgalR-3 variants was analysed by RT–semi-quantitative PCR in different oesophageal carcinoma cell lines and several human normal tissues using primers flanking the spliced regions (primers 3 and 4). As observed in Figure 2(A), two different PCR fragments were obtained, including the longer fragment (512 bp) corresponding to the NgalR-2/1 variant and a shorter fragment (387 bp) corresponding to the NgalR-3 variant. Results demonstrated that there are some differences in the expression level of these isoforms depending on the cell line. In EC18 and EC8712 cells, the major NgalR form is NgalR-2/1. In EC171 cells, NgalR-3 and NgalR-2/1 isoforms are present at similar levels. No fragment was observed in EC109 cells. RT–PCR analysis from several human normal tissues revealed that both NgalR variants were widely expressed (Figure 2B).
Figure 2. Expression of NgalRs in cell lines and normal tissues by RT–PCR.
Total RNA was extracted from the various oesophageal carcinoma cell lines and human normal tissues and reverse-transcribed. The PCR was conducted with primers flanking the spliced regions. (A) Lane M, 100 bp ladder; lane 1, EC171; lane 2, EC18; lane 3, EC109; lane 4, EC8712. (B) Lane M, 100 bp ladder; lane 1, oesophagus; lane 2, stomach; lane 3, rectum; lane 4, colon; lane 5, cardia uraniscus; lane 6, thyroid; lane 7, ovary; lane 8, amnion; lane 9, cervix; lane 10, palate; lane 11, lung.
NgalR isoform mRNA levels in oesophageal carcinoma and adjacent normal mucosa tissues
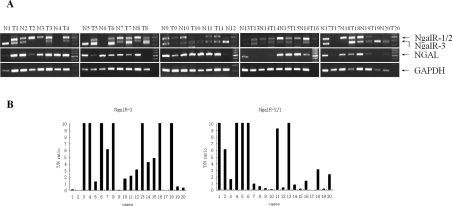
We performed an RT–semi-quantitative PCR amplification to identify the expression level of NgalRs and NGAL by using the frozen stored oesophageal carcinoma and the adjacent normal epithelial tissues. The intensity ratio of NgalRs to GAPDH was scored. Each specimen was evaluated by the ratio of the NgalR mRNA level in the tumour to that in the adjacent normal epithelial tissues (T/N ratio). As observed in Figure 3, the prevalence of the various NgalR isoforms and NGAL were found in oesophageal carcinoma and the adjacent normal tissues. Moreover, NgalR-3 expression was up-regulated in 70% of cases (14/20) of oesophageal carcinoma in comparison with the adjacent normal epithelium (T/N ratio>1.0), whereas in only 55% of specimens was NgalR-2/1 expression up-regulated.
Figure 3. NgalR and NGAL mRNA levels in oesophageal carcinoma and adjacent normal mucosa tissues.
Expression of NgalRs and NGAL were determined by RT–semi-quantitative PCR in 20 samples of oesophageal carcinoma (T) and adjacent normal mucosa tissues (N), and the amplification of a 226 bp fragment of GAPDH is shown as control (A). The levels of NgalR isoform expression were evaluated by calculating the T/N ratio (B). The distribution of NgalR-3 mRNA expression among the 20 patients was as follows: 14 cases had a T/N ratio >1.0, and six cases had a T/N ratio <1.0. NgalR-2/1 mRNA expression was up-regulated in 11 cases.
NgalR-3 and NGAL interact in vivo and co-localize mainly at the cell membrane in COS-7 cells
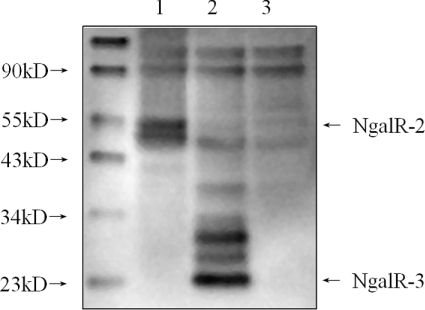
To evaluate the potential translated products of alternatively spliced NgalR-2 and NgalR-3 mRNA, we first transfected Myc-tagged NgalR-2 and Myc-tagged NgalR-3 respectively into COS-7 cells. The proteins were subjected to Western blotting (Figure 4). Both Myc-tagged NgalR-2 (approx. 55 kDa) and Myc-tagged NgalR-3 proteins (approx. 24 kDa) were detected by Western blot analysis using anti-Myc antibody, whereas no band was seen in the control tests. This result indicates that alternatively spliced variants of NgalR-2 and NgalR-3 can be translated into proteins and that the two expression protein bands have the predicted molecular mass.
Figure 4. Expression of NgalR-2 and NgalR-3 in COS-7 cells determined by Western blot.
There was a prominent band at 55 kDa, corresponding to the expected molecular mass of Myc–NgalR-2 in lane 1, and a band at 24 kDa, corresponding to the expected molecular mass of Myc–NgalR-3 in lane 2. There was no band in the control cell lysates (lane 3). kD, kDa.
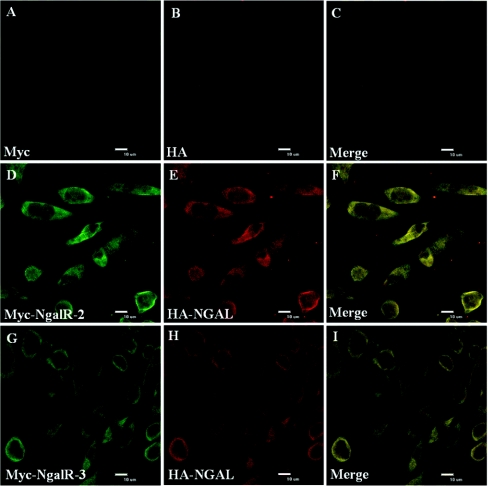
Subsequently, we performed experiments to see whether we could detect any evidence of co-localization of NgalRs and NGAL. We examined the subcellular localization patterns of NgalR-2 and NgalR-3 with NGAL respectively in COS-7 cells. The subcellular patterns of distribution of co-expressed Myc-tagged forms of NgalR-2 or Myc-tagged forms of NgalR-3 with HA-tagged NGAL are shown in Figure 5. Staining with an anti-HA or an anti-Myc antibody revealed a mainly cell membrane localization and a partial cytoplasm localization for the NgalR-3 and NGAL protein, and a nearly complete co-localization of the two proteins was observed after summation of the two staining patterns (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/403/bj4030297add.htm). Identical subcellular localization was also observed between the NgalR-2 and NGAL protein by immunofluorescence experiments.
Figure 5. Confocal fluorescence microscopy analysis of cellular localization of NgalR-2 or NgalR-3 with NGAL.
COS-7 cells were co-transfected with Myc–NaglR-2 (D) or Myc–NgalR-3 (G) with the HA–NGAL (E and H) respectively. Cells were stained with an anti-Myc antibody or an anti-HA antibody and developed with a fluorescein-conjugated goat anti-mouse IgG or Alexa Fluor® 594-conjugated donkey anti-rabbit IgG. Co-localization of the signals can be seen in the Merge panel (F and I) as a flavo-green signal. COS-7 cells co-transfected with Myc (A) and HA (B) empty vectors are shown as control.
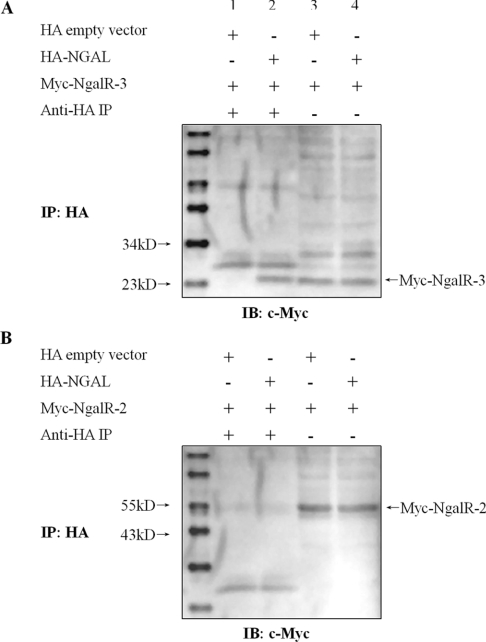
In view of our evidence demonstrating co-localization between NgalR-3 protein and NGAL in mammalian cells, we performed co-immunoprecipitation assays to determine whether a physical interaction between NGAL and NgalR-3 could occur in mammalian cells. Full-length NgalR-3 cDNA was co-expressed with either HA-tagged NGAL or HA-tagged empty vector in COS-7 cells. Cell lysates were subjected to immunoprecipitation with anti-HA affinity matrix followed by immunoblotting with anti-Myc antibody. As shown in Figure 6(A), a clear band was seen in cells where Myc–NgalR-3 was co-transfected with HA–NGAL, but not in cells where the empty vector was co-transfected with Myc–NgalR-3. The starting cell lysates contained equivalent amounts of Myc–NgalR-3 protein. However, we could not detect an in vivo interaction between NGAL and NgalR-2 protein (Figure 6B).
Figure 6. Interaction between NgalR-3 and NGAL by co-immunoprecipitation in vivo.
(A) A clear band was seen in cells where Myc–NgalR-3 was co-transfected with HA–NGAL (lane 2) but not in cells where the empty vector was co-transfected with Myc–NgalR-3 (lane 1). The starting cell lysates contained equivalent amounts of Myc–NgalR-3 protein (lanes 3 and 4). (B) In contrast, Myc–NgalR-2 was co-transfected with HA–NGAL into COS-7 cells. However, no co-immunoprecipitation evidence indicated any interaction between NgalR-2 and NGAL.
DISCUSSION
NGAL was originally purified from human neutrophils [4]. Up-regulation of NGAL expression was observed in several types of cancer [15–18]. According to previous studies by our group, NGAL is involved in the transformation and development of oesophageal carcinoma [23]. However, a clear explanation for the overexpression of NGAL as a result of malignancy has been lacking.
In the present study, we have demonstrated that NgalR-2, a receptor isoform of NGAL, was expressed in oesophageal carcinoma EC18 cells, which also expressed NGAL, according to our previous study [21]. However, this was not the only NgalR isoform found in oesophageal carcinoma cells, as we have also identified a new NgalR isoform designated NgalR-3 that results from alternative splicing. This new isoform encodes a predicted 207-amino-acid protein which presents amino acid sequences in the N-terminal 175 amino acids identical with those of NgalR-2 and contains a unique 32-amino-acid (residues 176–207) C-terminus. To identify the NgalR-3 isoform, we performed an RT–PCR analysis. Our results indicate that this new variant was widely expressed in human normal tissues and oesophageal carcinoma cell lines, and the relative expressed level of NgalR isoforms depended on the origin of the tissues and cells. Interestingly, it is noteworthy that NgalR-3 expression was up-regulated in 70% of cases of oesophageal carcinoma in comparison with the adjacent normal epithelium, whereas in only 55% of specimens was NgalR-2/1 expression up-regulated. This result could imply that this new NgalR-3 variant might play a more important role in oesophageal carcinoma.
Structure prediction analysis indicates that the deduced protein from this new spliced variant includes four putative membrane-spanning domains and features an extracellular N-terminal domain containing two potential N-linked glycosylation sites that is important for proper protein folding in the secretory pathway. The result indicates that this novel spliced variant might localize to the membrane similar to other NgalR isoforms and was expected to retain its bioactivity. In agreement with this prediction, as expressed in a heterologous system (COS-7 cells), this novel isoform conferred the expected functionality of co-localization with NGAL protein mainly at the cell membrane. We also found cytoplasmic localization for the isoform. Combined with results from a previous study that 24p3 enters the cell through an endosome recycling pathway and the 24p3R could come back to the cytomembrane circularly following endocytosis [26], we suppose that NGAL, a highly conserved human homologue of 24p3, might have a similar endocytosed pathway to that of 24p3. NGAL–NgalR complexes might be formed in the cytoplasm following internalization of NGAL through NgalR-3. Further research has demonstrated that a physical interaction between NGAL and NgalR-3 could occur in mammalian cells. All of these results suggest that NgalR-3 acts as a potential NgalR in oesophageal epithelial cells. We also found a similar co-localization between NGAL and the NgalR-2 protein. However, we could not detect an in vivo interaction between them. Structure prediction indicates that the NgalR-2 protein includes 11 membrane-spanning domains. The large hydrophobicity may make the NgalR-2 protein unable to interact with NGAL when the protein is ectopically expressed in the cytoplasm. Furthermore, many membrane proteins exert their biological function depending on their localization on the cell membrane. More experiments must be carried out to investigate the actual interaction on the cytomembrane between NGAL and NgalRs.
Alternative splicing is widespread in mammalian gene expression and is a biologically important mechanism for generating related molecules with a novel or complementary function. NgalR is a member of the organic cation transporter family. Molecular identification in the last decade of several genes encoding organic cation transporters has shown that most of them give rise to alternatively spliced isoforms, resulting in a variety of physiological consequences. Recent studies suggest that NGAL is a novel iron transporter with functions distinct from those of transferrin and mediates a new iron-delivery pathway [29,30]. Yang et al. [13] and Gwira et al. [14] have found that NGAL can bind iron specifically through the catecholate-type ferric siderophores and facilitate intracellular iron delivery by a specific cellular receptor, and thus play a regulatory role in inducing epithelial cell differentiation in kidney development. In the present study, we have identified a novel NgalR spliced variant. According to the study, we suppose that the NgalR-2 isoform would exhibit bioactivity as a bilateral iron-transporter mechanism, whereas the novel NgalR-3 isoform would mediate a unilateral intracellular iron-delivery pathway which adds to the growth potential of the tumour combined with a grave free radical reaction and gene mutation, thus promoting malignancy in oesophageal carcinoma. However, how NgalR acts in NGAL-mediated iron-delivery pathways in oesophageal carcinoma is as yet unknown, and will require extensive study to determine.
In summary, we have isolated a cDNA clone in oesophageal carcinoma cells that is a novel spliced variant of the NgalR gene and demonstrated that the protein produced from this cDNA exhibits the biochemical characteristics of interaction and co-localization in vivo with NGAL protein. This new finding suggests that NgalR-3 may act as a potential NgalR receptor and play a role in NGAL-mediated iron transport in oesophageal carcinoma. The isolation of this novel spliced variant will enable further studies of NgalR structure and function, as well as permitting studies of the factors that control alternative splicing of the gene in different cell types.
Online data
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (numbers 30672376, 30570849 and 30370641), Specialized Research Fund for the Doctoral Program of Higher Education of China (numbers 20050560002 and 20050560003), Guangdong Scientific Fund Key Items (numbers 37788 and 05104541), Natural Science Foundation of Guangdong Province (number 5301047) and China Postdoctoral Science Foundation (number 2004036491). We thank Professor S. H. He and Professor J. L. Lin for their kind assistance in confocal fluorescence microscopy analysis.
References
- 1.Flower D. R. The lipocalin protein family: structure and function. Biochem. J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flower D. R., North A. C., Sansom C. E. The lipocalin protein family: structural and sequence overview. Biochim. Biophys. Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 3.Flower D. R. Beyond the superfamily: the lipocalin receptors. Biochim. Biophys. Acta. 2000;1482:327–336. doi: 10.1016/s0167-4838(00)00169-2. [DOI] [PubMed] [Google Scholar]
- 4.Kjeldsen L., Johnsen A. H., Sengelov H., Borregaard N. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J. Biol. Chem. 1993;268:10425–10432. [PubMed] [Google Scholar]
- 5.Nielsen B. S., Borregaard N., Bundgaard J. R., Timshel S., Sehested M., Kjeldsen L. Induction of NGAL synthesis in epithelial cells of human colorectal neoplasia and inflammatory bowel diseases. Gut. 1996;38:414–420. doi: 10.1136/gut.38.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedl A., Stoesz S. P., Buckley P., Gould M. N. Neutrophil gelatinase-associated lipocalin in normal and neoplastic human tissues: cell type-specific pattern of expression. Histochem. J. 1999;31:433–441. doi: 10.1023/a:1003708808934. [DOI] [PubMed] [Google Scholar]
- 7.Yan L., Borregaard N., Kjeldsen L., Moses M. A. The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL): modulation of MMP-9 activity by NGAL. J. Biol. Chem. 2001;276:37258–37265. doi: 10.1074/jbc.M106089200. [DOI] [PubMed] [Google Scholar]
- 8.Tschesche H., Zolzer V., Triebel S., Bartsch S. The human neutrophil lipocalin supports the allosteric activation of matrix metalloproteinases. Eur. J. Biochem. 2001;268:1918–1928. doi: 10.1046/j.1432-1327.2001.02066.x. [DOI] [PubMed] [Google Scholar]
- 9.Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 10.Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 11.Mori K., Lee H. T., Rapoport D., Drexler I. R., Foster K., Yang J., Schmidt-Ott K. M., Chen X., Li J. Y., Weiss S., et al. Endocytic delivery of lipocalin–siderophore–iron complex rescues the kidney from ischemia-reperfusion injury. J. Clin. Invest. 2005;115:610–621. doi: 10.1172/JCI23056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong Z., Wu X., Ovcharenko D., Zhu J., Chen C. S., Kehrer J. P. Neutrophil gelatinase-associated lipocalin as a survival factor. Biochem. J. 2005;391:441–448. doi: 10.1042/BJ20051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., Mori K., Li J. Y., Barasch J. Iron, lipocalin, and kidney epithelia. Am. J. Physiol. Renal. Physiol. 2003;285:F9–F18. doi: 10.1152/ajprenal.00008.2003. [DOI] [PubMed] [Google Scholar]
- 14.Gwira J. A., Wei F., Ishibe S., Ueland J. M., Barasch J., Cantley L. G. Expression of neutrophil gelatinase-associated lipocalin regulates epithelial morphogenesis in vitro. J. Biol. Chem. 2005;280:7875–7882. doi: 10.1074/jbc.M413192200. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen B. S., Borregaard N., Bundgaard J. R., Timshel S., Sehested M., Kjeldsen L. Induction of NGAL synthesis in epithelial cells of human colorectal neoplasia and inflammatory bowel diseases. Gut. 1996;38:414–420. doi: 10.1136/gut.38.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartsch S., Tschesche H. Cloning and expression of human neutrophil lipocalin cDNA derived from bone marrow and ovarian cancer cells. FEBS Lett. 1995;357:255–259. doi: 10.1016/0014-5793(94)01303-i. [DOI] [PubMed] [Google Scholar]
- 17.Furutani M., Arii S., Mizumoto M., Kato M., Imamura M. Identification of a neutrophil gelatinase-associated lipocalin mRNA in human pancreatic cancers using a modified signal sequence trap method. Cancer Lett. 1998;122:209–214. doi: 10.1016/s0304-3835(97)00391-1. [DOI] [PubMed] [Google Scholar]
- 18.Stoesz S. P., Friedl A., Haag J. D., Lindstrom M. J., Clark G. M., Gould M. N. Heterogeneous expression of the lipocalin NGAL in primary breast cancers. Int. J. Cancer. 1998;79:565–572. doi: 10.1002/(sici)1097-0215(19981218)79:6<565::aid-ijc3>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez C. A., Yan L., Louis G., Yang J., Kutok J. L., Moses M. A. The matrix metalloproteinase-9/neutrophil gelatinase-associated lipocalin complex plays a role in breast tumor growth and is present in the urine of breast cancer patients. Clin. Cancer Res. 2005;11:5390–5395. doi: 10.1158/1078-0432.CCR-04-2391. [DOI] [PubMed] [Google Scholar]
- 20.Xu L. Y., Li E. M., Xiong H. Q., Cai W. J., Shen Z. Y. Study of neutrophil gelatinase-associated lipocalin (NGAL) gene overexpression in the progress of malignant transformation of human immortalized esophageal epithelial cell. Prog. Biochem. Biophys. 2001;28:839–843. [Google Scholar]
- 21.Li E. M., Xu L. Y., Cai W. J., Xiong H. Q., Shen Z. Y., Zeng Y. Functions of neutrophil gelatinase-associated lipocalin in the esophageal carcinoma cell line SHEEC. Acta Biochim. Biophys. Sin. 2003;35:247–254. [PubMed] [Google Scholar]
- 22.Lin J. L., Xu L. Y., Li E. M., Cai W. J., Niu Y. D., Fang K. Y., Xiong H. Q., Shen Z. Y., Zeng Y. Antisense blocking of NGAL gene expression affects the microfilament cytoskeleton in SHEEC esophageal cancer cells. Prog. Biochem. Biophys. 2004;31:409–415. [Google Scholar]
- 23.Zhang H. H., Xu L. Y., Xiao D. W., Xie J. J., Zeng H. M., Wang Z. Y., Zhang X. L., Niu Y. D., Shen Z. Y., Shen J. H., et al. Up-regulation of neutrophil gelatinase-associated lipocalin in esophageal squamous cell carcinoma: significantly correlated with cell differentiation and tumor invasion. J. Clin. Pathol. 2006 doi: 10.1136/jcp.2006.039297. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L. Y., Li E. M., Niu Y. D., Cai W. J., Yuan H. M., Chang J. X., Shen Z. Y., Zeng Y. There are TPA response elements in −152∼−60 position of NGAL gene 5′ flanking region in the esophageal cancer cells EC109. Prog. Biochem. Biophys. 2006;33:140–148. [Google Scholar]
- 25.Hvidberg V., Jacobsen C., Strong R. K., Cowland J. B., Moestrup S. K., Borregaard N. The endocytic receptor megalin binds the iron transporting neutrophil-gelatinase-associated lipocalin with high affinity and mediates its cellular uptake. FEBS Lett. 2005;579:773–777. doi: 10.1016/j.febslet.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 26.Devireddy L. R., Gazin C., Zhu X., Green M. R. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 2005;123:1293–1305. doi: 10.1016/j.cell.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 27.Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 28.Horton P., Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 29.Yang J., Goetz D., Li J. Y., Wang W., Mori K., Setlik D., Du T., Erdjument-Bromage H., Tempst P., Strong R., Barasch J. An iron delivery pathway mediated by a lipocalin. Mol. Cell. 2002;10:1045–1056. doi: 10.1016/s1097-2765(02)00710-4. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan J. Mechanisms of cellular iron acquisition: another iron in the fire. Cell. 2002;111:603–606. doi: 10.1016/s0092-8674(02)01164-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.