Abstract
Mutation in the prion gene, PRNP, accounts for approx. 10–15% of human prion diseases. However, little is known about the mechanisms by which a mutant prion protein (PrP) causes disease. We compared the biochemical properties of a wild-type human prion protein, rPrPC (recombinant wild-type PrP), which has five octapeptide-repeats, with two recombinant human prion proteins with insertion mutations, one with three more octapeptide repeats, rPrP8OR, and the other with five more octapeptide repeats, rPrP10OR. We found that the insertion mutant proteins are more prone to aggregate, and the degree and kinetics of aggregation are proportional to the number of inserts. The octapeptide-repeat and α-helix 1 regions are important in aggregate formation, because aggregation is inhibited with monoclonal antibodies that are specific for epitopes in these regions. We also showed that a small amount of mutant protein could enhance the formation of mixed aggregates that are composed of mutant protein and wild-type rPrPC. Accordingly, rPrP10OR is also more efficient in promoting the aggregation of rPrPC than rPrP8OR. These findings provide a biochemical explanation for the clinical observations that the severity of the disease in patients with insertion mutations is proportional to the number of inserts, and thus have implications for the pathogenesis of inherited human prion disease.
Keywords: aggregation, antibody blocking, insertion mutation, recombinant wild-type prion protein (rPrPC)
Abbreviations: AS-ELISA, aggregation-specific ELISA; GAG, glycosaminoglycans; GdmCl, guanidinium chloride; HRP, horseradish peroxidase; mAb, monoclonal antibody; PBS-T, PBS containing 0.05% Tween 20; PK, proteinase K; PrP, prion protein; PrPC, normal cellular PrP; PrPSc, scrapie PrP; rPrPC, recombinant wild-type PrPC; rPrP8OR, rPrPC with eight octapeptide repeats; rPrP10OR, rPrPC with ten octapeptide repeats; ThT, thioflavin T
INTRODUCTION
Misfolding and aggregation of cellular protein is thought to play a critical role in a growing number of human neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease and prion disease [1–4]. Among all neurodegenerative diseases, prion disease is unique because of its pathogenic mechanisms. All three forms of prion disease, sporadic, familial and infectious, are believed to share the same pathogenic mechanism, which is based on the conversion of PrPC (normal cellular prion protein) into the infectious and pathogenic PrPSc (scrapie PrP) [1,5].
Familial prion disease accounts for approx. 10–15% of human prion disease and is caused by mutations in the germ line prion gene, PRNP. More than 30 different pathogenic mutations in the human PRNP gene have been identified [6–9]. These are either insertion or point mutations. Insertion mutation occurs solely in the octapeptide-repeat region. Human PrPC has five octapeptide repeats. In pathogenic mutations, the numbers of additional octapeptide-repeats range from one to nine [10,11]. Patients with more octapeptide-repeat insertions have earlier disease onset and shorter disease duration [11]. Point mutations, however, can occur along the entire length of the molecule.
It is thought that the mutant PrP is inherently unstable, and prone to misfolding and aggregation, leading to the formation a structure which acts as a ‘seed’ to recruit additional mutant proteins, eventually leading to the formation of pathogenic and infectious PrPSc [12,13]. Although it is clear that mutation of PRNP causes human prion diseases, little is known about the mechanisms by which the mutant protein causes disease. PrPs with pathogenic mutations may cause disease because of gain of toxic functions, loss of normal physiological functions or both.
Bacterially produced rPrPC (recombinant PrPC) and rPrPC with pathogenic mutations have been used extensively as model systems for studying the mechanisms of inherited human prion diseases [14–21]. Thermostability studies suggest that the destabilization of PrP is not a general mechanism underlying the pathogenesis of the disease [17]. On the other hand, some of the mutant proteins exhibit biochemical and biophysical properties similar to infectious PrPSc [16,19,21,22]. Recently, we have shown that rPrP8OR (rPrPC with eight octapeptide repeats) has a more exposed N-terminus, binds better to GAGs (glycosaminoglycans), behaves differently after binding Cu2+ and is more susceptible to oxidative attack than wild-type rPrPC. Most importantly, the aberrant properties associated with rPrP8OR are also observed in another insertion mutant, rPrP10OR (rPrPC with ten octapeptide repeats), and the aberrations are even more profound in this protein [23]. Resistance to PK (proteinase K) digestion is a hallmark of PrPSc [1,5]. The number of octapeptide-repeat inserts has been reported to influence the rate of formation of PK-resistant PrP species [24,25]. Collectively these findings provide a biochemical explanation for the clinical observations that, for patients with insertion mutations, the onset and severity of the disease is proportional to the number of inserts [11].
In the present study, further work on the properties of these rPrPs in vitro was performed. We found that rPrPs with insertion mutations have the propensity to form aggregates in a manner that is proportional to the number of octapeptide-repeat inserts. We have also identified regions of the rPrP that are important in the aggregation process, and have investigated the ability of the insertion mutant proteins to enhance the formation of mixed aggregates composed of both mutant and wild-type rPrPC. Since aggregation is an essential step in the conversion of PrPC into PrPSc, the significance of these findings with respect to the pathogenesis of inherited human prion disease will be discussed.
EXPERIMENTAL
Plasmid construction and recombinant protein preparation
Cloning, generation and purification of human rPrPC, rPrP8OR and rPrP10OR were performed as described previously [23]. Protein solutions were concentrated using Centricon Plus-20 centrifugal filter devices (10 kDa nominal molecular-mass limit; Millipore) and stored in 20 mM sodium acetate (pH 5.5) at −80 °C. Protein concentration was determined with a Bio-Rad Protein Assay Kit. All the recombinant prion proteins were freshly purified.
Antibodies
The generation, purification and characterization of all the anti-PrPC murine mAbs (monoclonal antibodies) have been described in detail previously [26,27]. mAb 8B4 recognizes an epitope between residues 35 and 45; 5B2 recognizes residues 34–52; SAF32 and SAF34 react with residues 63–94, covering the octapeptide-repeat sequences [28]; 11G5 reacts with residues 115–130 spanning β-sheet 1; 7H6 recognizes residues 130–140; 7A12 interacts with helix 1 between residues 143 and 155; 2C2 reacts with residues 153–165 in β-sheet 2; 8H4 recognizes residues 175–185 of helix 2; 8F9 reacts with residues 220–231; and 5C3 and 6H3 react with residues 145–231. mAbs 8B4, 5B2, SAF32, SAF34, 7A12, 6H3, 8H4 and 8F9 are IgG1, whereas mAbs 11G5 and 7H6 are IgG2b, and 5C3 is IgG2a. All mAbs were affinity purified using Protein G chromatography. Biotinylation of mAbs was performed using the EZ-linked sulfo-N-hydroxysuccinimido-biotin kit (Pierce Endogen) according to the manufacturer's recommendation.
SDS/PAGE and immunoblotting
Samples were mixed with 2× SDS loading buffer [100 mM Tris/HCl, 4% (w/v) SDS, 20% (v/v) glycerol and 0.04% Bromphenol Blue, pH 6.8, without 2-mercaptoethanol] and heated for 5 min at 95 °C before being resolved by SDS/PAGE on 12% (v/v) gels. The gel was then transferred on to a nitrocellulose membrane and the PrPs detected by mAb 8H4. HRP (horseradish peroxidase)-conjugated goat anti-mouse IgG Fc-specific antibody was used as the secondary antibody and proteins were visualized using the chemiluminescence blotting system (Roche Applied Science).
Acidic native gel electrophoresis
Because of the basic pI (>7.0) and the auto-aggregation property of rPrP8OR and rPrP10OR at neutral pH (results not shown), native gel electrophoresis was performed under acidic conditions in combination with reversed polarity of the leads. Samples were mixed with 5× dissolving buffer [37.5% (v/v) glycerol, 60 mM potassium acetate, pH 5.5, and 0.05% Methyl Green] and loaded on to 8% polyacrylamide gels and electrophoresis was performed in a running buffer containing 350 mM β-alanine and 140 mM acetic acid (pH 4.3) at 150 V for 120 min at 4 °C followed by Coomassie Blue staining.
Far-UV CD
The far-UV CD spectra were recorded on a Jasco J-810 spectropolarimeter (Jasco) at room temperature (25 °C). To improve the signal-to-noise ratio three spectra were averaged and the background buffer spectra were subtracted. The measurements were performed in a 0.1-cm-pathlength cylindrical cuvette for 20 μM rPrPC, rPrP8OR or rPrP10OR in 20 mM sodium acetate (pH 5.5). Wavelength scans were collected every 0.5 nm from 200–250 nm (5 nm bandwidth). The scanning speed was 100 nm/min.
Turbidity measurement
The aggregation reactions were performed according to Frankenfield et al. [29] with minor modifications. The assays were performed at 37 °C in a volume of 200 μl in 96-well plates. rPrPC, rPrP8OR and rPrP10OR at various concentrations were suspended in 50 mM sodium acetate and 150 mM NaCl (pH 4.0). The solutions were pre-incubated at 37 °C before mixing. GdmCl (guanidinium chloride; 0.5 M final concentration) was added to initiate the aggregation reactions. After addition of GdmCl, the turbidity of the samples was monitored within 15 s by reading the attenuance at 405 nm in a Beckman Coulter AD340 micro-ELISA plate reader, using a kinetic photometric model (interval time 50 s, 60 cycles with 1 s shaking before every cycle). Each experiment was repeated at least three times with freshly prepared rPrPs.
ThT (thioflavin T)-binding assay
The assays were performed at room temperature in a volume of 100 μl in a 0.1 cm path-length cuvette. ThT (50 μM) was mixed with 10 μM rPrP in 50 mM sodium acetate and 150 mM NaCl (pH 4.0). After the addition of 0.5 M GdmCl, the fluorescence of ThT was monitored within 10 s in a Quanta Master spectrofluorimeter (Photon Technology International) with excitation at 450 nm and emission at 482 nm. The kinetic assay was performed (1800 s duration with 1 point/s) without agitation. The background values for ThT fluorescence were subtracted from the results.
AS-ELISA (aggregation-specific ELISA)
We have described the rationale for AS-ELISA previously [30]. mAb 11G5 was used to coat 96-well Costar plates (Corning) at 0.5 μg/well for 3 h at room temperature. Then the wells were blocked with 3% (w/v) BSA (Sigma) at 4 °C overnight. Aggregates were formed using 0, 1, 3, 5 and 7 μM of rPrPC, rPrP8OR and rPrP10OR in 50 mM sodium acetate and 150 mM NaCl (pH 4.0) in the presence of 0.5 M GdmCl at room temperature for 1 h. Then the rPrPs (including the monomer and aggregates) were diluted 500-fold with 20 mM sodium acetate, pH 5.5 and added into wells pre-coated with 11G5. After incubation at room temperature for 2 h, the plates were washed three times with PBS-T (PBS containing 0.05% Tween 20). An appropriate dilution of biotinylated 11G5 was added into the wells and incubated for 2 h. After washing again with PBS-T, HRP-conjugated streptavidin (Chemicon) was added and incubated for 1 h. The plates were washed three times with PBS-T before the addition of 100 μl ABTS [2,2′-azinobis-(3-ethylbenzthiazoline-6-sulphonic acid); Roche Diagnostics]. After 15–30 min, the absorbance was read at 405 nm on a Beckman Coulter AD340 micro-ELISA plate reader. Each experiment was repeated at least three times with freshly prepared rPrPs.
Blocking of aggregation of rPrPs with mAbs
Appropriate concentrations of different mAbs were added to 10 μM rPrP10OR in 50 mM sodium acetate and 150mM NaCl (pH 4.0). After incubation at 37 °C for 10 min, 0.5 M GdmCl was added and turbidity measurements were performed as described above. BSA and an irrelevant mAb 9C1 [anti-(brain-derived neurotrophic factor)] were used as negative controls. Each experiment was repeated at least three times with freshly prepared rPrPs.
Effects of rPrP8OR and rPrP10OR on the aggregation of rPrPC
Various concentrations of rPrP10OR were mixed with 5 μM rPrPC in 50 mM sodium acetate and 150 mM NaCl (pH 4.0), and then 0.5 M GdmCl was added to the reaction to initiate the aggregation at 37 °C. Turbidity measurements were performed at 405 nm following the kinetic model (interval time 50 s, 60 cycles with 1 s shaking before every cycle) on the ELISA plate reader. BSA was used as a control. We also compared the promoting efficiencies of rPrPC, rPrP8OR and rPrP10OR by mixing respectively with 5 μM rPrPC.
To study the contents of the aggregates generated in this experiment, the aggregates at 90 min were centrifuged at 13000 g for 10 min at 4 °C and the supernatants were removed. The pellets were washed twice with 50 mM sodium acetate, 150 mM NaCl (pH 4.0) and 0.5 M GdmCl, and then washed twice with 50 mM sodium acetate and 150 mM NaCl (pH 4.0). The pellets were dissolved in 1× SDS loading buffer [100 mM Tris/HCl, 2% (w/v) SDS, 10% (v/v) glycerol, 0.04% Bromphenol Blue, pH 6.8, and 100 mM 2-mercaptoethanol] and heated at 95 °C for 10 min. The samples were resolved by SDS/PAGE (12% gels), followed by silver staining. Density measurements were performed with a BiospectrumAC Imaging System (UVP). Each experiment was repeated at least three times with freshly prepared rPrPs.
RESULTS
Characterization of rPrPC, rPrP8OR and rPrP10OR
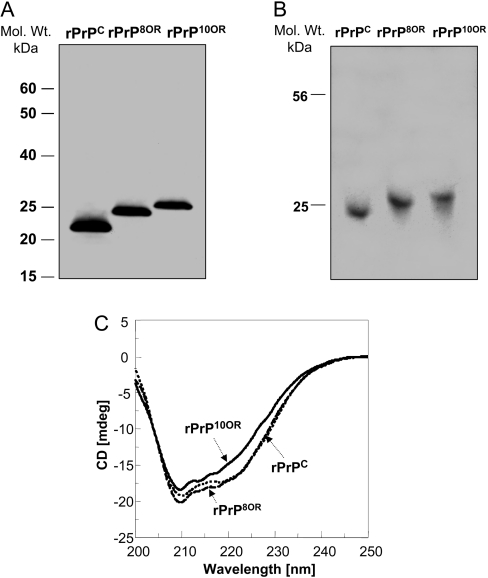
We analysed three freshly prepared recombinant proteins by SDS/PAGE under non-reducing conditions, followed by immunoblotting with anti-PrP mAb 8H4 (Figure 1A). rPrPC appears as a single band, with a molecular mass of approx. 23 kDa, which is the expected molecular mass of full-length rPrPC protein. rPrP8OR also appears as a single band, but has slower migration, with a molecular mass of 25 kDa, reflecting the addition of 24 amino acids in the octapeptide-repeat region. rPrP10OR has a slower migration than rPrP8OR with a molecular mass of approx. 26.5 kDa. We did not observe any immunoreactivity at the higher molecular mass regions, even with over-exposure of the film (results not shown). We also ran the samples on a native gel with non-reducing conditions and stained with Coomassie Blue (Figure 1B). We did not detect any larger molecular aggregate using this condition. Therefore these rPrPs exist as monomers.
Figure 1. Characterization of rPrPC, rPrP8OR and rPrP10OR.
(A) Freshly prepared rPrPC, rPrP8OR and rPrP10OR proteins were separated by SDS/PAGE under non-reducing conditions and then immunoblotted with mAb 8H4. Molecular-mass (Mol. Wt.) markers are indicated in kDa to the left-hand side of the panel. (B) Acidic native gel electrophoresis of rPrPC, rPrP8OR and rPrP10OR. The gel was stained with Coomassie Blue. All three recombinant proteins exist as monomers under these conditions. (C) Far-UV CD spectra of rPrPC, rPrP8OR and rPrP10OR. The spectra were obtained using 20 μM of rPrPC (dotted line), rPrP8OR (broken line) and rPrP10OR (solid line) in 20 mM sodium acetate (pH 5.5). While rPrPC and rPrP8OR have rather similar secondary structures, rPrP10OR has a small decrease in the negativity of the spectra. Therefore insertion mutations at the N-terminus are able to influence the secondary structure of the molecule, depending on the number of octapeptide-repeat inserts.
We next compared the secondary structures of the three rPrPs by far-UV CD spectroscopy at pH 5.5 without GdmCl. Under these conditions, the three rPrPs do not aggregate and exist mostly as monomers. The spectra of rPrPC and rPrP8OR are similar, showing characteristics of a typical α-helical protein with two minima at 208 and 220 nm in the far-UV region. In contrast, the spectrum of rPrP10OR differs slightly, with a small decrease in negativity (Figure 1C). These results provide evidence that the addition of three octapeptide-repeats does not significantly alter the overall secondary structure of the protein. However, longer inserts do have an impact on the secondary structure. Hence it is likely that the longer the insert, the greater the influence on the overall structural conformation.
The propensity of rPrPs to aggregate is proportional to the number of octapeptide inserts
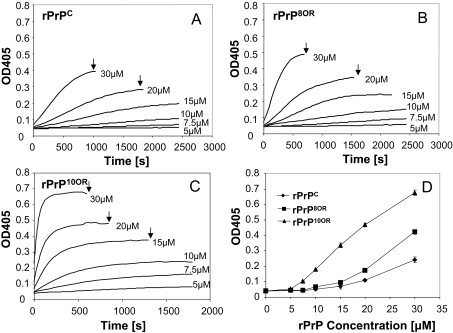
We used the turbidity assay to investigate whether rPrPs with insertion mutations are more prone to aggregate. Protein aggregation is pH- and denaturing agent concentration-dependent [29,31,32]. Therefore we first determined the optimal conditions for aggregation of our rPrPs. The optimal pH for aggregation is between pH 3.5 and pH 4.5 for all three rPrPs. The optimal GdmCl concentration is between 0.5 and 0.75 M for all three rPrPs (results not shown). At pH 4 and 0.5 M GdmCl, rPrPC, rPrP8OR and rPrP10OR aggregate in a protein concentration-dependent manner (Figures 2A–2C). However, rPrP10OR aggregates the most with the fastest kinetics, followed by rPrP8OR then rPrPC (Figure 2D). Therefore the degree and speed of aggregation is proportional to the number of octapeptide-repeat inserts.
Figure 2. Aggregation of rPrPC, rPrP8OR and rPrP10OR, quantified using the turbidity assay.
GdmCl (0.5 M) was added to various concentrations of rPrPC (A), rPrP8OR (B) and rPrP10OR (C) in 50 mM sodium acetate and 150 mM NaCl (pH 4.0), and the aggregation kinetics were monitored by the increase in A405 (OD405) at 37 °C. Concentrations of rPrPs are indicated to the right-hand side of the traces. (D) A comparison of the aggregations of rPrPC, rPrP8OR and rPrP10OR by recording the A405 (OD405) 500 s after the addition of GdmCl. The results presented in (D) are means±S.E.M. for at least three experiments. The degree and kinetics of aggregation are proportional to the number of inserts. (A–C) Arrows indicate when the readings were terminated owing to the precipitation of the proteins.
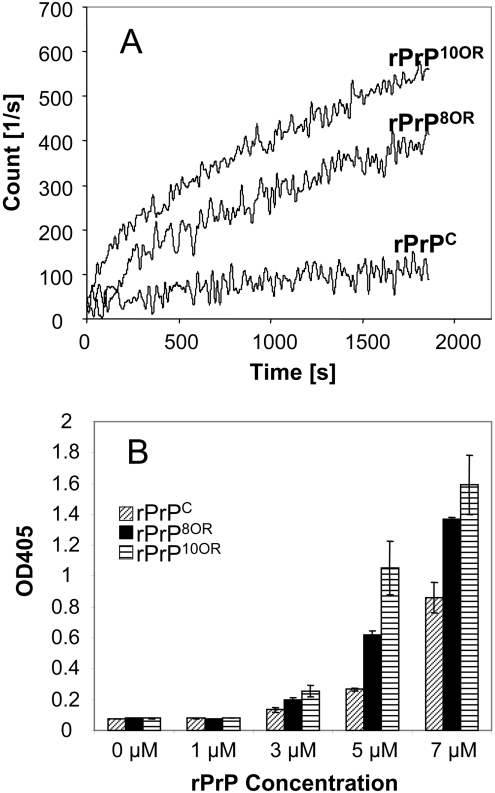
We also verified the aggregation of rPrPC, rPrP8OR and rPrP10OR using two additional techniques, namely ThT-binding fluorescence and an AS-ELISA [30]. Results from ThT-binding fluorescence (Figure 3A) and AS-ELISA (Figure 3B) are in good accord with the turbidity assay, showing that rPrP10OR forms the most aggregates, followed by rPrP8OR and then rPrPC.
Figure 3. rPrPs aggregation monitored by ThT-binding assay and AS-ELISA.
(A) Aggregation kinetics of rPrPC, rPrP8OR and rPrP10OR followed by ThT-binding assay. rPrPC, rPrP8OR and rPrP10OR (10 μM of each) were mixed with 50 μM ThT in 50 mM sodium acetate and 150 mM NaCl (pH 4.0), and then 0.5 M GdmCl was added to initiate aggregation. The experiments were performed at least three times and the background ThT fluorescence in the buffer was subtracted. (B) Aggregation of rPrPC, rPrP8OR and rPrP10OR monitored by AS-ELISA. ELISA plates were coated with mAb 11G5 at 0.5 μg/well, and prepared aggregates from different concentrations of rPrPC, rPrP8OR and rPrP10OR were added to the wells. After washing, bound rPrPs were detected by measuring the A405 (OD405) using biotinylated 11G5 followed by HRP-conjugated streptavidin. The results presented in (B) are the means±S.E.M for at least three experiments. Results from both assays are in good agreement with the results of the turbidity assay.
Blocking of rPrP10OR aggregation with anti-PrP mAbs
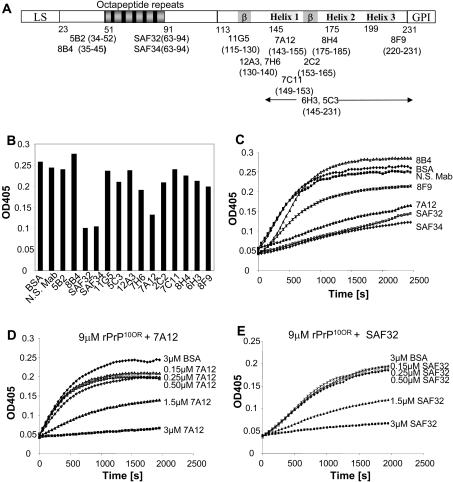
We next sought to identify which region(s) of the PrP is important in the aggregation process. Using a panel of mAbs which react with epitopes along the entire PrP molecule (Figure 4A) [26,27], we investigated whether any of these mAbs could block aggregation of rPrP10OR. Of the 14 mAbs tested, only mAbs SAF32, SAF34 and 7A12 blocked aggregation of rPrP10OR (Figures 4B and 4C). Only mAbs specific for the octapeptide-repeat region and the α-helix 1 region blocked aggregation.
Figure 4. Blocking of rPrP10OR aggregation by anti-PrP mAbs.
(A) The location of mAb-binding epitopes along the length of the PrP. LS, leader sequence; GPI, glycosylphosphatidylinositol. (B) The effects of all 14 mAbs on the aggregation of rPrP10OR. Anti-PrP mAbs (2 μM) were mixed with 10 μM rPrP10OR in 50 mM sodium acetate and 150 mM NaCl (pH 4.0), and incubated at 37 °C for 10 min. GdmCl (0.5 M) was added and the A405 (OD405) was recorded after 1500 s. (C) The kinetics of the aggregation of 10 μM rPrP10OR in the presence of anti-PrP mAbs (2 μM). Anti-PrP mAbs (2 μM) were mixed with 10 μM rPrP10OR in 50 mM sodium acetate and 150 mM NaCl (pH 4.0), and incubated at 37 °C for 10 min. GdmCl (0.5 M) was added and turbidity assay was performed as described in the Experimental section. N. S. Mab, non-specific mAb. Various concentrations of mAb 7A12 (D) or SAF32 (E) were mixed with 9 μM rPrP10OR and incubated at 37 °C for 10 min, then 0.5 M GdmCl was added and turbidity assay was performed.
Blocking is mAb-concentration-dependent. At a molar ratio of 1:3 (mAb/rPrP10OR), mAb 7A12 (Figure 4D) and SAF32 (Figure 4E) inhibited aggregation by approx. 100%. One may argue that the reason the other mAbs were unable to block aggregation was that these mAbs were unable to bind rPrP10OR at pH 4.0 and in the presence of 0.5 M GdmCl, the conditions used for the aggregation assay. To examine this possibility, we performed ELISA at pH 4.0, with 0.5 M GdmCl and demonstrated that mAbs such as 8B4 and 8F9, which were unable to block aggregation, reacted with rPrP at pH 4.0 and 0.5 M GdmCl (results not shown). Therefore the inability of these mAbs to block aggregation is not due to their inability to bind rPrPs. Since only mAbs that are specific for epitopes in the octapeptide-repeat and α-helix 1 regions blocked aggregation, then these regions can be considered as important for aggregate formation.
rPrP10OR and rPrP8OR enhance rPrPC aggregation
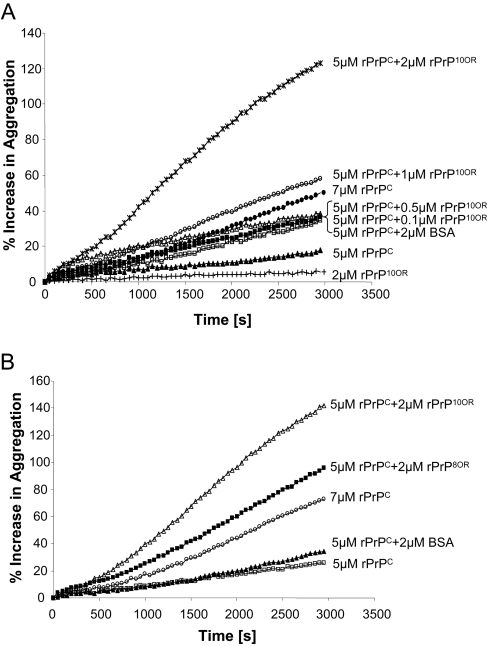
We next investigated whether a small amount of rPrP10OR could recruit rPrPC to form mixed aggregates. Titration experiments revealed that 5 μM rPrPC, which does not aggregate by itself, would aggregate in the presence of rPrP10OR, in an rPrP10OR-concentration-dependent manner (Figure 5A). Similar results were obtained using rPrP8OR (Figure 5B); however, the efficiency of rPrP8OR to promote aggregation of rPrPC was consistently less than rPrP10OR.
Figure 5. Enhancement of aggregation of rPrPC in the presence of rPrP10OR and rPrP8OR.
(A) rPrPC (5 μM) was mixed with 0.1, 0.5, 1 or 2 μM rPrP10OR in 50 mM sodium acetate and 150 mM NaCl (pH 4.0), followed by the addition of 0.5M GdmCl at 37 °C, and the turbidity assay was performed. The enhanced aggregation is given as an increased percentage of starting turbidity (formula: P=(T/T0-1)×100; P, percentage increase; T, turbidity; T0, starting turbidity). BSA and rPrPC (both at 2 μM) were used as controls. (B) rPrP10OR is more efficient than rPrP8OR in promoting the aggregation of rPrPC. Each experiment was performed at least three times.
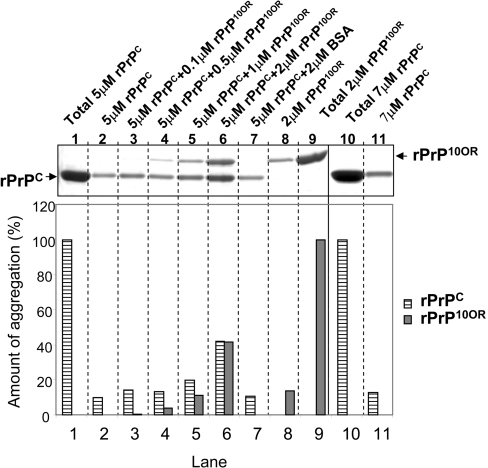
We next sought to verify that the aggregates indeed consisted of rPrP10OR and rPrPC. We first removed monomeric rPrPC and rPrP10OR from the mixture by repeat centrifugation at 13000 g for 10 min, and washing the pellets. We then resolved the proteins by SDS/PAGE under reducing conditions. Since rPrP10OR has a lower rate of migration than rPrPC, we were able to distinguish these two proteins in a mixture. As the concentration of rPrP10OR increased, the amount of rPrPC in the aggregates increased accordingly (Figure 6, top panel). Densitometric analysis of the bands in each sample is shown in the bottom panel of Figure 6. The total amount of rPrPC by itself (5 μM) was arbitrarily considered to be 100% (Figure 6, top panel, lane 1). Approx. 6% of this rPrPC precipitates spontaneously (Figure 6, top panel, lane 2). The total amount of rPrP10OR by itself (2 μM) was arbitrarily considered to be 100% (Figure 6, top panel, lane 9). Approx. 10% of this rPrP10OR precipitates spontaneously (Figure 6, top panel, lane 8). However, when 5 μM of rPrPC was mixed with increasing amounts of rPrP10OR, the amount of rPrPC in the mixed aggregate precipitates increased proportionally to the amount of rPrP10OR added. In samples containing 5 μM of rPrPC and 2 μM of rPrP10OR, close to 40% of the total rPrPC and 40% of the total rPrP10OR were detected in the aggregate precipitate (Figure 6, top panel, lane 6). This increase in aggregation is not due to non-specific protein ‘crowding’, because addition of the same concentration of BSA did not alter the aggregation of either rPrPC or rPrP10OR. When 7 μM rPrPC was studied under identical conditions, <10% of the total rPrPC (Figure 6, top panel, lane 10) was present as aggregates in the pellet (Figure 6, top panel, lane 11). Collectively these results provide strong evidence that small amounts of rPrP10OR can enhance the formation of mixed aggregates of normal rPrPC and mutant rPrP10OR.
Figure 6. Demonstration of rPrP10OR and rPrPC aggregates by SDS/PAGE.
rPrPC (5 μM) was mixed with 0.1, 0.5, 1 or 2 μM rPrP10OR in 50 mM sodium acetate and 150 mM NaCl (pH 4.0), followed by the addition of 0.5 M GdmCl at 37 °C. After 90 min, the aggregates were collected by centrifugation and washed as described in the Experimental section. The pellets were dissolved by heating in 1× SDS loading buffer and the samples were separated by SDS/PAGE under reducing conditions (top panel). The amount of rPrPC and rPrP10OR in each precipitate was quantified by densitometric analysis (lanes 2–8 and 11) (bottom panel). The experiment was performed at least three times.
An identical experiment using rPrP8OR to recruit rPrPC was also carried out. Under identical conditions, rPrP8OR was less efficient in recruiting rPrPC. Instead of having 40% of the rPrPC in the aggregate, only approx. 20% of the rPrPC was recruited to the aggregates by rPrP8OR (results not shown). In multiple experiments, rPrP10OR was approx. 2–3-fold more efficient than rPrP8OR in promoting rPrPC aggregation.
DISCUSSION
We have reported previously [23] that full-length rPrPC with insertion mutations, such as rPrP8OR and rPrP10OR, share common aberrant features that are proportional to the number of inserts. Insertion mutants have a more exposed N-terminus, bind more GAG and are more susceptible to oxidative attack. In the present study, we show further characterization of rPrP8OR and rPrP10OR. (i) rPrP8OR and rPrP10OR are prone to aggregate in proportion to the number of inserts; (ii) the octapeptide-repeat and α-helix 1 regions of PrP are important in aggregate formation; (iii) a small amount of rPrP10OR can enhance the formation of mixed aggregates composed of rPrP10OR and rPrPC; and (iv) rPrP10OR is more effective in promoting mixed aggregate formation than rPrP8OR. Since aggregation of PrPC is an essential step in PrPC to PrPSc conversion, our findings may have implications for the pathogenesis of inherited human prion diseases.
Far-UV CD spectroscopic studies at pH 5.5 showed that rPrPC and rPrP8OR have almost identical secondary structures. These results are consistent with earlier findings [16,29,33–35]. On the other hand, there is a small decrease in the amount of α-helical content in rPrP10OR. Therefore mutation at the N-terminus does have an impact on the secondary structure of the C-terminal globular domain, although the effect is rather small. The impact is probably proportional to the number of octapeptide-repeat insertions.
Many groups have studied aggregation and fibril formation using rPrPs; fibril formation is protein-, denaturing-agent-concentration and pH-dependent [14,15,29,31,34,36–42]. However, in most of these studies, rPrP fragments, either C-terminal fragments or N-terminal fragments were used [13,14,22,31,34,36,38–41,43]. All three recombinant full-length proteins, rPrPC, rPrP8OR and rPrP10OR, also aggregate in a protein- and denaturing-agent-concentration- and pH-dependent manner. However, insertion mutant proteins are more prone to aggregate, and the degree and kinetics of aggregation are proportional to the number of inserts. Under identical conditions, rPrP10OR requires the lowest amount of protein to induce aggregation of the wild-type protein, forms the most aggregate and has the fastest aggregation kinetics, followed by rPrP8OR and then rPrPC. All three assays used to monitor protein aggregation (turbidity, ThT-binding fluorescence and AS-ELISA) revealed similar results. Interestingly, using atomic-force microscopy we did not observe any fibril in our rPrP preparations (results not shown). The reason we failed to observe fibril formation is probably because we used a much lower concentration of rPrPs (20 μM) and a shorter incubation time (<30 min). In some of earlier studies rPrP as great as 400 μM was used [13,22], and the incubation time was in either hours or days [13,14,31,37,39,41].
One shortcoming of bacterially produced rPrPs is their lack of N-linked glycans. Mammalian cell lines transfected to express rPrPC with insertion mutations [25,44] and a transgenic mouse line that expresses an insertion mutant form of the protein have been generated [45–48]. The insertion mutant PrPs in these models are also prone to aggregate and are more protease-resistant than wild-type PrPC [25]. Therefore our observations with rPrP8OR and rPrP10OR are consistent with the earlier findings. Furthermore, transgenic mice that are homozygous for the insertion mutation also spontaneously develop a fatal neurodegenerative disorder [45,48] and have a reduced incubation time when inoculated with PrPSc [46–48].
Our finding that aggregation of rPrP10OR and rPrP8OR (rPrP8OR results not shown) can be blocked with mAbs that are specific for either the octapeptide-repeat or the helix 1 region, suggests that these two regions are important in rPrPC aggregation. Using synthetic peptides, it was reported that the octapeptide-repeat region is an important domain in PrP aggregation [49]. The helix 1 region, which includes residues 145–155, has been suggested to be important in the formation of a hydrophilic core and in the seeding of PrP aggregates [50]. Biophysical studies have also provided strong evidence that PrPC transformation into PrPSc involves the conversion of the helix 1 to a β-sheet structure [51,52]. These mAbs probably block aggregation by preventing rPrP10OR molecules from binding to each other via either the octapeptide-repeat region or the α-helix 1 region. We have recently shown [53] that, in cell models, mAb 7A12, which recognizes the α-helix 1 region, can inhibit the replication of PrPSc and cure already infected cells. Therefore the α-helix 1 region is also important in the infection process. The ability of mAb SAF32 to inhibit PrPSc replication in cell models has not yet been examined. We were unable to demonstrate a synergistic effect when both mAbs SAF32 and 7A12 were added together to inhibit aggregation. Therefore it is likely that these two regions are equally important in aggregation (results not shown).
The biophysical properties that render the mutant proteins prone to aggregation are unclear. One possibility is that the three rPrPs may have different secondary structures prior to aggregate formation. However, preliminary experiments using far-UV CD spectroscopy revealed that the three rPrPs have comparable secondary structures at pH 5.5. The spectra of rPrPC and rPrP8OR are almost identical. It is now generally accepted that the octapeptide-repeat region binds divalent cations, such as Cu2+ and Zn2+ [54,55]. Addition of Cu2+ has been reported to promote the aggregation of rPrPs and render them partially PK-resistant [18,35,56]. However, under our experimental condition, at pH 4.0, Cu2+ did not alter the aggregation of the three rPrPs. This is most probably due to the destruction of the Cu2+-binding sites at this pH. It has been reported that the optimal pH for the binding of Cu2+ to rPrP is at pH 6.5 [57–59].
Most human PRNP pathogenic mutations are heterozygous, producing both wild-type and mutant PrPC. A previous study showed that in Creutzfeldt–Jakob disease patients with five or six octapeptide-repeat insertions, both mutant and wild-type PrPs were converted into detergent-insoluble and protease-resistant isoforms [60]. A recent study also found that both the mutant and the wild-type PrPC could contribute to pathogenesis [61]. Therefore our finding that rPrP10OR can enhance the formation of mixed aggregates composed of both rPrP10OR and rPrPC are in good accordance with these findings, and may have in vivo relevance. A small amount of mutant PrP in vivo, which by itself is unable to form aggregates, will be able to recruit PrPC to form larger aggregates with pathogenic properties. Again, rPrP10OR is more effective than rPrP8OR in recruiting PrPC. Using an in vitro conversion assay, it has been reported that full-length, glycosylated, hamster PrP with 15 octapeptide-repeat insertions is converted to a PK-resistant PrP species more rapidly than wild-type hamster PrPC [24]. Therefore PrP with an insertion mutation can not only promote mixed aggregate formation, but it is also a better substrate for exogenous PrPSc.
Whether the aggregates we have detected in the present study have any relevance to the pathogenesis of prion disease is unknown. Although most of the structure studies using rPrP tend to focus on the ability of the protein to form large fibrils in vitro, a recent report [62] revealed that the most infectious PrPSc molecules were spherical or ellipsoidal, 20–25 nm in diameter, with a molecular mass of approx. 300–600 kDa, which corresponds with 14–28 PrP molecules. Resistance to PK digestion is a fundamental feature of infectious PrPSc [5,63]. Others have reported that rPrP aggregates or fibrils generated in vitro are relatively more PK-resistant than monomeric rPrP [15,18,24,34,38,40,64,65]. The number of octapeptide-repeat inserts has also been reported to influence the rate of formation of PK-resistant PrP species [24]. We found that aggregated rPrPs tend to be more resistant to PK degradation than monomeric rPrPs. However, the degree of resistance is rather marginal and is much lower than in vivo generated PrPSc (results not shown).
On the basis of the findings of the present study, we hypothesize that an increase in the number of octapeptide repeats renders the PrP molecule more prone to aggregation. Both the octapeptide-repeat region and the α-helix 1 region are important in aggregate formation. Furthermore, mutant PrP can enhance the formation of larger mixed aggregates, composed of wild-type and mutant proteins. This enhancement model should provide a useful in vitro model for further investigation of the conversion process in the absence of other cellular components. Since all these aberrant features are proportional to the number of insertions, our earlier and current findings also provide a cogent biochemical explanation for the observation that patients with more octapeptide-repeat insertions have earlier disease onset and shorter disease duration [11].
Acknowledgments
We would like to thank Dr Michael Lamm (Department of Pathology, Case Western Reserve University, Cleveland, OH, U.S.A.) for careful reading of the manuscript and suggestions before submission, and Dr Jacques Grassi (Atomic Energy Commission, Saclay, France) for his gift of mAbs SAF32 and SAF34. We also thank Dr Witold Surewicz (Department of Biophysics, Case Western Reserve University, Cleveland, OH, U.S.A.) for performing a preliminary experiment on the CD spectrum of rPrPC and rPrP8OR, and Dr Jim De Yoreo (BioSecurity and Nanosciences Laboratory, Lawrence Livermore National Laboratory, Livermore, CA, U.S.A.) for performing the atomic-force microscopy experiment. This work was supported in part by NIH (National Institutes of Health) grant NS-045981-01 and an award/contract from the U.S. Department of the Army, DAMD17-03-1-0286 (to M.S.S.).
References
- 1.Prusiner S. B. Prions. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen F. E. Protein misfolding and prion diseases. J. Mol. Biol. 1999;293:313–320. doi: 10.1006/jmbi.1999.2990. [DOI] [PubMed] [Google Scholar]
- 3.Dobson C. M. Protein misfolding, evolution and disease. Trends. Biochem. Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 4.Ross C. A., Poirier M. A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 5.Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 6.Collinge J. Human prion diseases and bovine spongiform encephalopathy (BSE) Hum. Mol. Genet. 1997;6:1699–1705. doi: 10.1093/hmg/6.10.1699. [DOI] [PubMed] [Google Scholar]
- 7.Kovacs G. G., Trabattoni G., Hainfellner J. A., Ironside J. W., Knight R. S., Budka H. Mutations of the prion protein gene phenotypic spectrum. J. Neurol. 2002;249:1567–1582. doi: 10.1007/s00415-002-0896-9. [DOI] [PubMed] [Google Scholar]
- 8.Gambetti P., Kong Q., Zou W., Parchi P., Chen S. G. Sporadic and familial CJD: classification and characterisation. Br. Med. Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 9.Mead S. Prion disease genetics. Eur. J. Hum. Genet. 2006;14:273–281. doi: 10.1038/sj.ejhg.5201544. [DOI] [PubMed] [Google Scholar]
- 10.Owen F., Poulter M., Lofthouse R., Collinge J., Crow T. J., Risby D., Baker H. F., Ridley R. M., Hsiao K., Prusiner S. B. Insertion in prion protein gene in familial Creutzfeldt–Jakob disease. Lancet. 1989;1:51–52. doi: 10.1016/s0140-6736(89)91713-3. [DOI] [PubMed] [Google Scholar]
- 11.Croes E. A., Theuns J., Houwing-Duistermaat J. J., Dermaut B., Sleegers K., Roks G., Van den Broeck M., van Harten B., van Swieten J. C., Cruts M., et al. Octapeptide repeat insertions in the prion protein gene and early onset dementia. J. Neurol. Neurosurg. Psychiatry. 2004;75:1166–1170. doi: 10.1136/jnnp.2003.020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen F. E., Pan K. M., Huang Z., Baldwin M., Fletterick R. J., Prusiner S. B. Structural clues to prion replication. Science. 1994;264:530–531. doi: 10.1126/science.7909169. [DOI] [PubMed] [Google Scholar]
- 13.Jones E. M., Surewicz K., Surewicz W. K. Role of N-terminal familial mutations in prion protein fibrillization and prion amyloid propagation in vitro. J. Biol. Chem. 2006;281:8190–8196. doi: 10.1074/jbc.M513417200. [DOI] [PubMed] [Google Scholar]
- 14.Baskakov I. V., Legname G., Baldwin M. A., Prusiner S. B., Cohen F. E. Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 2002;277:21140–21148. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 15.Bocharova O. V., Breydo L., Parfenov A. S., Salnikov V. V., Baskakov I. V. In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrPSc. J. Mol. Biol. 2005;346:645–659. doi: 10.1016/j.jmb.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 16.Cappai R., Stewart L., Jobling M. F., Thyer J. M., White A. R., Beyreuther K., Collins S. J., Masters C. L., Barrow C. J. Familial prion disease mutation alters the secondary structure of recombinant mouse prion protein: implications for the mechanism of prion formation. Biochemistry. 1999;38:3280–3284. doi: 10.1021/bi982328z. [DOI] [PubMed] [Google Scholar]
- 17.Liemann S., Glockshuber R. Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry. 1999;38:3258–3267. doi: 10.1021/bi982714g. [DOI] [PubMed] [Google Scholar]
- 18.Qin K., Yang D. S., Yang Y., Chishti M. A., Meng L. J., Kretzschmar H. A., Yip C. M., Fraser P. E., Westaway D. Copper(II)-induced conformational changes and protease resistance in recombinant and cellular PrP: effect of protein age and deamidation. J. Biol. Chem. 2000;275:19121–19131. doi: 10.1074/jbc.275.25.19121. [DOI] [PubMed] [Google Scholar]
- 19.Vanik D. L., Surewicz W. K. Disease-associated F198S mutation increases the propensity of the recombinant prion protein for conformational conversion to scrapie-like form. J. Biol. Chem. 2002;277:49065–49070. doi: 10.1074/jbc.M207511200. [DOI] [PubMed] [Google Scholar]
- 20.Lewis P. A., Tattum M. H., Jones S., Bhelt D., Batchelor M., Clarke A. R., Collinge J., Jackson G. S. Codon 129 polymorphism of the human prion protein influences the kinetics of amyloid formation. J. Gen. Virol. 2006;87:2443–2449. doi: 10.1099/vir.0.81630-0. [DOI] [PubMed] [Google Scholar]
- 21.Swietnicki W., Petersen R. B., Gambetti P., Surewicz W. K. Familial mutations and the thermodynamic stability of the recombinant human prion protein. J. Biol. Chem. 1998;273:31048–31052. doi: 10.1074/jbc.273.47.31048. [DOI] [PubMed] [Google Scholar]
- 22.Jones E. M., Surewicz W. K. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 23.Yin S., Yu S., Li C., Wong P., Chang B., Xiao F., Kang S. C., Yan H., Xiao G., Grassi J., et al. Prion proteins with insertion mutations have altered N-terminal conformation and increased ligand binding activity and are more susceptible to oxidative attack. J. Biol. Chem. 2006;281:10698–10705. doi: 10.1074/jbc.M511819200. [DOI] [PubMed] [Google Scholar]
- 24.Moore R. A., Herzog C., Errett J., Kocisko D. A., Arnold K. M., Hayes S. F., Priola S. A. Octapeptide repeat insertions increase the rate of protease-resistant prion protein formation. Protein Sci. 2006;15:609–619. doi: 10.1110/ps.051822606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priola S. A., Chesebro B. Abnormal properties of prion protein with insertional mutations in different cell types. J. Biol. Chem. 1998;273:11980–11985. doi: 10.1074/jbc.273.19.11980. [DOI] [PubMed] [Google Scholar]
- 26.Li R., Liu T., Wong B. S., Pan T., Morillas M., Swietnicki W., O'Rourke K., Gambetti P., Surewicz W. K., Sy M. S. Identification of an epitope in the C-terminus of normal prion protein whose expression is modulated by binding events in the N-terminus. J. Mol. Biol. 2000;301:567–573. doi: 10.1006/jmbi.2000.3986. [DOI] [PubMed] [Google Scholar]
- 27.Zanusso G., Liu D., Ferrari S., Hegyi I., Yin X., Aguzzi A., Hornemann S., Liemann S., Glockshuber R., Manson J. C., et al. Prion protein expression in different species: analysis with a panel of new mAbs. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8812–8816. doi: 10.1073/pnas.95.15.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feraudet C., Morel N., Simon S., Volland H., Frobert Y., Creminon C., Vilette D., Lehmann S., Grassi J. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J. Biol. Chem. 2005;280:11247–11258. doi: 10.1074/jbc.M407006200. [DOI] [PubMed] [Google Scholar]
- 29.Frankenfield K. N., Powers E. T., Kelly J. W. Influence of the N-terminal domain on the aggregation properties of the prion protein. Protein Sci. 2005;14:2154–2166. doi: 10.1110/ps.051434005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan T., Chang B., Wong P., Li C., Li R., Kang S. C., Robinson J. D., Thompsett A. R., Tein P., Yin S., et al. An aggregation-specific enzyme-linked immunosorbent assay: detection of conformational differences between recombinant PrP protein dimers and PrPSc aggregates. J. Virol. 2005;79:12355–12364. doi: 10.1128/JVI.79.19.12355-12364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokolowski F., Modler A. J., Masuch R., Zirwer D., Baier M., Lutsch G., Moss D. A., Gast K., Naumann D. Formation of critical oligomers is a key event during conformational transition of recombinant syrian hamster prion protein. J. Biol. Chem. 2003;278:40481–40492. doi: 10.1074/jbc.M304391200. [DOI] [PubMed] [Google Scholar]
- 32.Swietnicki W., Petersen R., Gambetti P., Surewicz W. K. pH-dependent stability and conformation of the recombinant human prion protein PrP(90–231) J. Biol. Chem. 1997;272:27517–27520. doi: 10.1074/jbc.272.44.27517. [DOI] [PubMed] [Google Scholar]
- 33.Cordeiro Y., Machado F., Juliano L., Juliano M. A., Brentani R. R., Foguel D., Silva J. L. DNA converts cellular prion protein into the β-sheet conformation and inhibits prion peptide aggregation. J. Biol. Chem. 2001;276:49400–49409. doi: 10.1074/jbc.M106707200. [DOI] [PubMed] [Google Scholar]
- 34.Swietnicki W., Morillas M., Chen S. G., Gambetti P., Surewicz W. K. Aggregation and fibrillization of the recombinant human prion protein huPrP90–231. Biochemistry. 2000;39:424–431. doi: 10.1021/bi991967m. [DOI] [PubMed] [Google Scholar]
- 35.Wong E., Thackray A. M., Bujdoso R. Copper induces increased β-sheet content in the scrapie-susceptible ovine prion protein PrPVRQ compared with the resistant allelic variant PrPARR. Biochem. J. 2004;380:273–282. doi: 10.1042/BJ20031767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson G. S., Hosszu L. L., Power A., Hill A. F., Kenney J., Saibil H., Craven C. J., Waltho J. P., Clarke A. R., Collinge J. Reversible conversion of monomeric human prion protein between native and fibrilogenic conformations. Science. 1999;283:1935–1937. doi: 10.1126/science.283.5409.1935. [DOI] [PubMed] [Google Scholar]
- 37.Cordeiro Y., Kraineva J., Gomes M. P., Lopes M. H., Martins V. R., Lima L. M., Foguel D., Winter R., Silva J. L. The N-terminal PrP domain is crucial to modulate prion misfolding and aggregation. Biophys. J. 2005;89:2667–2676. doi: 10.1529/biophysj.105.067603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martins S. M., Frosoni D. J., Martinez A. M., De Felice F. G., Ferreira S. T. Formation of soluble oligomers and amyloid fibrils with physical properties of the scrapie isoform of the prion protein from the C-terminal domain of recombinant murine prion protein mPrP-(121–231) J. Biol. Chem. 2006;281:26121–26128. doi: 10.1074/jbc.M605367200. [DOI] [PubMed] [Google Scholar]
- 39.Ricchelli F., Buggio R., Drago D., Salmona M., Forloni G., Negro A., Tognon G., Zatta P. Aggregation/fibrillogenesis of recombinant human prion protein and Gerstmann–Straussler–Scheinker disease peptides in the presence of metal ions. Biochemistry. 2006;45:6724–6732. doi: 10.1021/bi0601454. [DOI] [PubMed] [Google Scholar]
- 40.Torrent J., Alvarez-Martinez M. T., Harricane M. C., Heitz F., Liautard J. P., Balny C., Lange R. High pressure induces scrapie-like prion protein misfolding and amyloid fibril formation. Biochemistry. 2004;43:7162–7170. doi: 10.1021/bi049939d. [DOI] [PubMed] [Google Scholar]
- 41.Tahiri-Alaoui A., James W. Rapid formation of amyloid from α-monomeric recombinant human PrP in vitro. Protein Sci. 2005;14:942–947. doi: 10.1110/ps.041000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S., Eisenberg D. Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process. Nat. Struct. Biol. 2003;10:725–730. doi: 10.1038/nsb961. [DOI] [PubMed] [Google Scholar]
- 43.Tahiri-Alaoui A., Gill A. C., Disterer P., James W. Methionine 129 variant of human prion protein oligomerizes more rapidly than the valine 129 variant: implications for disease susceptibility to Creutzfeldt–Jakob disease. J. Biol. Chem. 2004;279:31390–31397. doi: 10.1074/jbc.M401754200. [DOI] [PubMed] [Google Scholar]
- 44.Lehmann S., Harris D. A. Mutant and infectious prion proteins display common biochemical properties in cultured cells. J. Biol. Chem. 1996;271:1633–1637. doi: 10.1074/jbc.271.3.1633. [DOI] [PubMed] [Google Scholar]
- 45.Chiesa R., Piccardo P., Ghetti B., Harris D. A. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- 46.Chiesa R., Piccardo P., Quaglio E., Drisaldi B., Si-Hoe S. L., Takao M., Ghetti B., Harris D. A. Molecular distinction between pathogenic and infectious properties of the prion protein. J. Virol. 2003;77:7611–7622. doi: 10.1128/JVI.77.13.7611-7622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castilla J., Gutierrez-Adan A., Brun A., Pintado B., Parra B., Ramirez M. A., Salguero F. J., Diaz San Segundo F., Rabano A., Cano M. J., Torres J. M. Different behavior toward bovine spongiform encephalopathy infection of bovine prion protein transgenic mice with one extra repeat octapeptide insert mutation. J. Neurosci. 2004;24:2156–2164. doi: 10.1523/JNEUROSCI.3811-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castilla J., Gutierrez-Adan A., Brun A., Pintado B., Salguero F. J., Parra B., Segundo F. D., Ramirez M. A., Rabano A., Cano M. J., Torres J. M. Transgenic mice expressing bovine PrP with a four extra repeat octapeptide insert mutation show a spontaneous, non-transmissible, neurodegenerative disease and an expedited course of BSE infection. FEBS Lett. 2005;579:6237–6246. doi: 10.1016/j.febslet.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 49.Zahn R. The octapeptide repeats in mammalian prion protein constitute a pH-dependent folding and aggregation site. J. Mol. Biol. 2003;334:477–488. doi: 10.1016/j.jmb.2003.09.048. [DOI] [PubMed] [Google Scholar]
- 50.Morrissey M. P., Shakhnovich E. I. Evidence for the role of PrPC helix 1 in the hydrophilic seeding of prion aggregates. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11293–11298. doi: 10.1073/pnas.96.20.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Safar J., Roller P. P., Gajdusek D. C., Gibbs C. J., Jr Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J. Biol. Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 52.Speare J. O., Rush T. S., 3rd, Bloom M. E., Caughey B. The role of helix 1 aspartates and salt bridges in the stability and conversion of prion protein. J. Biol. Chem. 2003;278:12522–12529. doi: 10.1074/jbc.M211599200. [DOI] [PubMed] [Google Scholar]
- 53.Pankiewicz J., Prelli F., Sy M. S., Kascsak R. J., Kascsak R. B., Spinner D. S., Carp R. I., Meeker H. C., Sadowski M., Wisniewski T. Clearance and prevention of prion infection in cell culture by anti-PrP antibodies. Eur. J. Neurosci. 2006;23:2635–2647. doi: 10.1111/j.1460-9568.2006.04805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chattopadhyay M., Walter E. D., Newell D. J., Jackson P. J., Aronoff-Spencer E., Peisach J., Gerfen G. J., Bennett B., Antholine W. E., Millhauser G. L. The octarepeat domain of the prion protein binds Cu(II) with three distinct co-ordination modes at pH 7.4. J. Am. Chem. Soc. 2005;127:12647–12656. doi: 10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown L. R., Harris D. A. Copper and zinc cause delivery of the prion protein from the plasma membrane to a subset of early endosomes and the Golgi. J. Neurochem. 2003;87:353–363. doi: 10.1046/j.1471-4159.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- 56.Tsiroulnikov K., Rezaei H., Dalgalarrondo M., Chobert J. M., Grosclaude J., Haertle T. Cu(II) induces small-size aggregates with amyloid characteristics in two alleles of recombinant ovine prion proteins. Biochim. Biophys. Acta. 2006;1764:1218–1226. doi: 10.1016/j.bbapap.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 57.Miura T., Sasaki S., Toyama A., Takeuchi H. Copper reduction by the octapeptide repeat region of prion protein: pH dependence and implications in cellular copper uptake. Biochemistry. 2005;44:8712–8720. doi: 10.1021/bi0501784. [DOI] [PubMed] [Google Scholar]
- 58.Miura T., Hori-i A., Mototani H., Takeuchi H. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–11569. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- 59.Wells M. A., Jackson G. S., Jones S., Hosszu L. L., Craven C. J., Clarke A. R., Collinge J., Waltho J. P. A reassessment of copper(II) binding in the full-length prion protein. Biochem. J. 2006;399:435–444. doi: 10.1042/BJ20060458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S. G., Parchi P., Brown P., Capellari S., Zou W., Cochran E. J., Vnencak-Jones C. L., Julien J., Vital C., Mikol J., et al. Allelic origin of the abnormal prion protein isoform in familial prion diseases. Nat. Med. 1997;3:1009–1015. doi: 10.1038/nm0997-1009. [DOI] [PubMed] [Google Scholar]
- 61.Wadsworth J. D., Joiner S., Linehan J. M., Cooper S., Powell C., Mallinson G., Buckell J., Gowland I., Asante E. A., Budka H., et al. Phenotypic heterogeneity in inherited prion disease (P102L) is associated with differential propagation of protease-resistant wild-type and mutant prion protein. Brain. 2006;129:1557–1569. doi: 10.1093/brain/awl076. [DOI] [PubMed] [Google Scholar]
- 62.Silveira J. R., Raymond G. J., Hughson A. G., Race R. E., Sim V. L., Hayes S. F., Caughey B. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prusiner S. B., McKinley M. P., Bowman K. A., Bolton D. C., Bendheim P. E., Groth D. F., Glenner G. G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 64.Luhrs T., Zahn R., Wuthrich K. Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle solutions. J. Mol. Biol. 2006;357:833–841. doi: 10.1016/j.jmb.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 65.Baskakov I. V., Aagaard C., Mehlhorn I., Wille H., Groth D., Baldwin M. A., Prusiner S. B., Cohen F. E. Self-assembly of recombinant prion protein of 106 residues. Biochemistry. 2000;39:2792–2804. doi: 10.1021/bi9923353. [DOI] [PubMed] [Google Scholar]