Abstract
The hepatitis C virus (HCV) pandemic affects the health of more than 170 million people and is the major indication for orthotopic liver transplantations. Although the human liver is the primary site for HCV replication, it is not known whether extrahepatic tissues are also infected by the virus and whether nonprimate cells are permissive for RNA replication. Because HCV exists as a quasispecies, it is conceivable that a viral population may include variants that can replicate in different cell types and in other species. We have tested this hypothesis and found that subgenomic HCV RNAs can replicate in mouse hepatoma and nonhepatic human epithelial cells. Replicons isolated from these cell lines carry new mutations that could be involved in the control of tropism of the virus. Our results demonstrated that translation and RNA-directed RNA replication of HCV do not depend on hepatocyte or primate-specific factors. Moreover, our results could open the path for the development of animal models for HCV infection.
Hepatitis C virus (HCV) is an enveloped, positive-stranded RNA virus that belongs to the Flaviviridae, a family that includes other human pathogens such as Yellow fever virus, Dengue virus, and West Nile virus (4). Although broad tissue and species tropisms are hallmarks of these viruses, HCV replication has so far been detected only in human and chimpanzee livers. Moreover, for reasons that are not yet understood, the amount of HCV RNA even in infected liver tissue is generally below one copy of RNA per hepatocyte on average. Hence, this viral RNA can be detected only with PCR, making it difficult to determine whether secondary sites for viral replication exist in the infected host (2, 11).
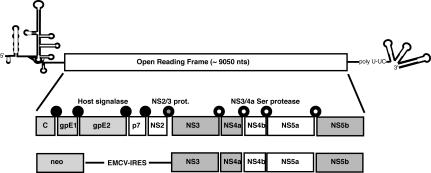
HCV encodes a single polyprotein that is processed proteolytically into 10 polypeptides (20). Three of these products are structural proteins required for capsid formation (core) and assembly into enveloped viral particles (E1 and E2). Four of the products are enzymes including cysteine and serine proteases (NS2 and NS3), an ATP-dependent helicase (NS3), and an RNA-directed RNA polymerase (NS5B). The functions of the remaining three polypeptides, p7, NS4B, and NS5A, are not yet known (Fig. 1).
FIG. 1.
Physical map of HCV. The open reading frame is flanked by the IRES and the 3′ untranslated region containing a polypyrimidine tract (poly U-UC), respectively. Subgenomes contain the neomycin phosphotransferase gene (neo) in lieu of the structural genes (C, core; gpE1 and gpE2, envelope proteins; p7) and the NS protein 2 (NS2 protease). Translation of the remaining NS proteins (NS3, protease; NS4A, cofactor for protease; NS4B; NS5A; NS5B, polymerase) is regulated by the IRES derived from EMCV. Proteins of unknown function for viral replication are marked with white rectangles. The cleavage sites for the three proteases required for processing of the polyprotein are indicated with circles.
For study of HCV replication in tissue culture cells, the structural proteins can be replaced with a selectable marker, such as the neomycin phosphotransferase (Fig. 1) (15). Replication of such subgenomic HCV replicons has so far been demonstrated only in the human hepatoma cell line Huh7. Although the apparent restriction of HCV replication to Huh7 cells would be consistent with the narrow host and tissue tropism of HCV infections, direct evidence for a role of hepatocyte-specific factors in HCV replication has so far been lacking. In fact, other members of the Flaviviridae whose RNA genomes are replicated in a manner very similar to that of HCV generally exhibit broad tissue and host tropism. Moreover, the efficient replication of HCV in Huh7 cells depends on adaptive mutations located in the nonstructural (NS) genes (1, 7, 14). Such considerations raise the possibility that additional mutations might allow HCV replication to occur in nonhepatic or nonhuman cells. This notion is supported by results described in this report. We show that amplification of HCV RNA can occur in HeLa and mouse hepatoma cells, indicating that host factors required for RNA replication are not hepatocyte specific and not restricted to cells of human origin.
MATERIALS AND METHODS
Cell culture.
Cells were purchased from the American Type Culture Collection (Table 1). The Huh7-derived cell lines GS4.1 and GS4.5 are subclones derived from cell lines FCA1 and FCA4, respectively (7). Cell line Bsp8 is a Huh7-derived cell line expressing HCV-N subgenomic replicon 1bneoΔS (7). All cultures were grown in Dulbecco's modified Eagle's medium (Gibco-Invitrogen) supplemented with 10% fetal bovine serum, l-glutamine, nonessential amino acids, penicillin, and streptomycin. The conditions used for the transfection of cells with total RNA were identical to those used for the transfection with in vitro-transcribed RNA (7). Colonies were selected with G418 at a concentration of 1 mg/ml.
TABLE 1.
Cell lines tested in this study
| Name | Description | Organism | Source |
|---|---|---|---|
| BHK | Kidney | Mesocricetus auratus (Syrian golden hamster) | ATCC CRL-1632 |
| Vero | Kidney epithelial | Cercopithecus aethiops (African green monkey) | ATCC CCL-81 |
| CV-1 | Kidney fibroblast | Cercopithecus aethiops (African green monkey) | ATCC CCL-70 |
| HT1080 | Fibrosarcoma | Homo sapiens (human) | ATCC CRL12012 |
| HeLa | Cervix carcinoma | Homo sapiens (human) | ATCC CCL2 |
| McA-RH7777 | Hepatoma | Rattus norvegicus (rat) | ATCC CRL-1601 |
| FTO2B | Hepatoma | Rattus norvegicus (rat) | Keith Fourniera |
| Hepa1-6 | Hepatoma | Mus musculus (mouse) | ATCC CRL-1830 |
| AML12 | Hepatocyte | Mus musculus (mouse) | ATCC CRL-2254 |
| FL83B | Hepatocyte | Mus musculus (mouse) | ATCC CRL-2390 |
Fred Hutchinson Cancer Center.
RNA transfection.
All the plasmids were linearized with ScaI, and RNA was synthesized with the MEGAscript kit (Ambion). In vitro-transcribed RNA was purified as previously described (7). Total cellular RNA was extracted with Trizol reagent (Invitrogen). The conditions used for the transfection of cells with total RNA were identical to those used for the transfection with in vitro-transcribed RNA (7). Colonies were selected with G418 at a concentration of 1 mg/ml.
RNA analysis.
Total cellular RNA was extracted with Trizol reagent. Five micrograms of total RNA was fractionated on 1% agarose gels containing 2.2 M formaldehyde and transferred onto a nylon membrane. Membranes were hybridized with riboprobes specific for plus-stranded HCV replicon RNA, human papillomavirus (HPV) E6, and mouse albumin mRNA as described previously (7). The HPV and mouse albumin probes spanned nucleotides 811 to 1491 (GenBank accession number M20325) and nucleotides 1501 to 1988 (GenBank accession number XM_132149), respectively.
Reverse transcription-PCR and DNA sequencing.
Nucleotide and amino acid numbers correspond to the HCV type 1b genome Con-1 (AJ238799). HCV replicons were isolated and cloned from established cell lines by PCR amplification of three fragments spanning the entire NS region from position 3420 to 9410. The untranslated regions at the 5′ and 3′ ends of HCV RNA were cloned separately for nucleotide sequence analysis. DNA synthesis was carried out with Superscript II reverse transcriptase provided in a cDNA synthesis kit (Gibco-Invitrogen). The DNA oligomers used as primers for the reverse transcription reaction mapped to positions 485 to 465, 5492 to 5473, 7256 to 7234, 9410 to 9388, and 9616 to 9597. The reaction mixtures were incubated for 1 h at 45°C. PCR was performed with an Advantage PCR kit (Clontech). One microliter of the cDNA reaction mixture was used for PCRs with 19- to 23-nucleotide-long primers that yielded fragments spanning positions 1 to 464, 1387E to 5082, 5016 to 7226, 7154 to 9387, and 9239 to 9616. Position 1387E refers to an oligomer specific for the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) element located upstream of NS3. The PCR products were cloned into plasmid pGEM-T Easy (Promega). Four clones of each fragment were sequenced with an ABI automatic DNA sequencer, and a consensus sequence was established with the help of a sequence assembly program (Genetics Computer Group).
Long reverse transcription-PCR was performed with an Advantage-GC kit (Clontech) with a pair of primers beginning at positions 1415E, upstream of NS3, and 7989 within NS5B. The PCR conditions were modified as follows: step 1, 95°C for 3 min; step 2, 5 cycles, 30 s at 95°C and 6 min at 72°C; step 3, 27 cycles, 30 s at 95°C and 6 min at 68°C; step 4, 68°C for 6 min. PCR products were gel purified and digested with HindIII and MfeI and replaced with the corresponding fragment in plasmid I377/NS3-3′.
Plasmid construction.
All plasmids (Table 4) were derived from the parental HCV Con-1 replicon I377/NS3-3′ (AJ242652). Subgenomes containing consensus mutations were constructed by replacing DNA restriction fragments with the corresponding fragments from the pGEM-T Easy cDNA libraries (see above). The resulting plasmids with the amino acid changes in the NS region are listed in Table 4. Sequence files for each plasmid are available upon request.
TABLE 4.
Colony formation efficiency of mutant replicons in Huh7 cells
| Vector | Conserved mutation(s) | Mean (SD) CFE/μg of RNAg | NS protein(s) affected |
|---|---|---|---|
| I377/NS3-3′ | NAh | 2.3 (1.5) | NA |
| pZS10 | A1a | 3.3 (3.5) | NS3 |
| pZS1 | C1b | 1.4 × 103 (3.9 × 102) | NS5A |
| pZS20 | Bc | 1.7 (0.6) | NS4AB |
| pZS11 | A1 + C1 | 2.4 × 105 (7 × 104) | NS3, NS5A |
| pZS2 | B + C1 | 7.8 × 104 (1.7 × 104) | NS4AB, NS5A |
| pZS12 | A1 + B | 0.3 (0.6) | NS3, NS4AB |
| pZS5 | A1 + B + C1 | 15 | NS3, NS4AB, NS5A |
| pZS4 | B + C1 + A2d | 165 (21) | NS3, NS4AB, NS5A |
| pZS8 | A2 + B | 0 | NS3, NS4AB |
| pZS6 | A2 + B + C2e | 8 (4) | NS3, NS4AB, NS5A |
| pZS15 | C3f | 491 (183) | NS5A |
| pZS25 | B + C3 | 1.7 × 104 (2.6 × 103) | NS4AB, NS5A |
| pZS45 | A2 + B + C3 | 19 (2) | NS3, NS4AB, NS5A |
A1, mutation E1202G.
C1, mutations S2204I and D2254E.
B, mutations L1701F, Q1720R, Q1727H, and V1749A.
A2, mutations E1202G, S1128A, and S1323P.
C2, mutations S2204I, D2254E, R2290I, and I2324R.
C3, all the mutations of C2 plus the deletion 2371-2413.
CFE, colony formation efficiency. Values are derived from three independent transfections of each replicon RNA.
NA, not applicable.
Immunofluorescence.
Cells were plated on coverslips in six-well plates at least 16 h before treatment, washed with phosphate-buffered saline, and fixed with cold methanol-acetone (1:1) for 15 to 20 min. Next, the cells were blocked in phosphate-buffered saline containing 10% fetal bovine serum for 30 min at room temperature and then incubated with anti-NS5A antibodies (a gift from Chen Liu) and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin antibodies (Jackson Laboratories). In addition, cells were stained with the DNA-binding fluorochrome DAPI (4′,6′-diamidino-2-phenylindole). Coverslips were mounted with antifade agent (Molecular Probes), examined with a Nikon immunofluorescence microscope, and photographed with a charge-coupled device camera.
RESULTS
HCV replication in cells of nonhepatic origin.
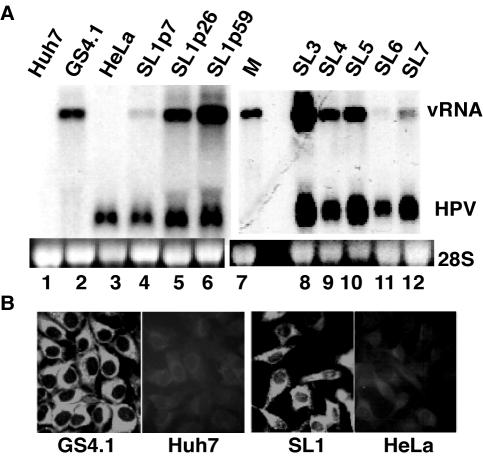
As HCV exhibits a very narrow host range and infects only humans and chimpanzees, we asked whether this limitation was due to determinants of RNA replication. Because efficient replication of subgenomes depends on genetic adaptations of the replicon (1, 7, 14), presumably to compensate for subtle variations in the cellular environments among cells from different tissues, we hypothesized that replication in cells of nonhepatic origin would require additional, cell-type-specific adaptive mutations. Transfection of several primate- and rodent-derived cell lines with subgenomic RNA transcribed from plasmid DNA carrying previously identified adaptive mutations in Huh7 cells did not yield cell lines expressing replicons (Table 1 and results not shown). To increase the chance for the selection of RNA subgenomes capable of replicating in cells of nonhepatic origin, we used subgenomic RNA isolated from Huh7 cell lines that replicate HCV RNA. Because of the high rate of nucleotide incorporation errors that occur during RNA-directed RNA synthesis, this population of viral subgenomes exhibited a much larger genetic heterogeneity than did RNA transcribed from a DNA template in vitro previously used for the transfection of Huh7 cells. Upon transfection of BHK, Vero, CV-1, HT1080, and HeLa cells with total RNA obtained from Huh7 cell lines GS4.1, GS4.5, and Bsp8, we obtained G418-resistant cell clones only with HeLa cells. The number of clones ranged from approximately 2 (Bsp8) to 50 (GS4.1) per 10 μg of total RNA depending on the origin of the RNA used for the transfections. Replicons in these three Huh7-derived cell lines contained different adaptive mutations and replicated two different HCV 1b genomes (7). Several HeLa-derived colonies obtained with total RNA from GS4.1 cells were subsequently expanded into seven stable cell lines (SL1 to SL7; Fig. 2A, lanes 4 and 8 to 12). The amounts of viral RNA present in early passages of these cell lines examined ranged from 0.05 to 7.5 ng/10 μg of total RNA, which corresponded to 20 to 3,000 copies of RNA per cell. In general, the amounts of RNA increased upon passage of cells and reached levels that were comparable to those obtained with the most productive Huh7-derived cell lines such as GS4.1 (lanes 2 and 4 to 6). As expected, expression of viral gene products could be confirmed by immunofluorescence with antibodies directed against NS5A (Fig. 2B). As with GS4.1 cells, more than 90% of SL1 cells expressed viral proteins. However, in contrast to Huh7 cell lines where the accumulation of HCV RNA declines approximately 100-fold when cells become confluent, viral replication in HeLa cells was not affected by the growth conditions of the cells, i.e., SL1 cells continued to produce high amounts of viral RNA even when they became confluent (results not shown) (7, 16).
FIG. 2.
Replication of HCV subgenomic replicons in HeLa cells. (A) Detection of HCV viral RNA. Total RNA (5 μg) was isolated from HeLa cell lines that were established from G418-resistant cell colonies and analyzed by Northern blot analysis. Blots were hybridized with radiolabeled RNA probes corresponding to the HCV NS5 region to detect viral RNA (vRNA) and the ΔE1 region of HPV present in HeLa cells. In vitro-transcribed HCV RNA (1 ng plus 5 μg of total RNA from Huh7 cells, lane 7) served as a marker (M) and control for the hybridization reaction, and 28S rRNA served as a control for the amount of RNA present in each sample analyzed. GS4.1 is a Huh7-derived cell line expressing HCV subgenomic replicons. RNA in SL1 cells was analyzed from cells harvested at the indicated passage (p). SL3 to SL7 were analyzed at passage 3. (B) Immunohistochemical analysis of HCV replication in HeLa cells. Expression of NS5A in GS4.1 and SL1 cells (passage 26) was detected with a monoclonal antibody bound to fluorescein isothiocyanate-conjugated antibody. Parental Huh7 and HeLa cells served as controls.
Adaptation of HCV replicons.
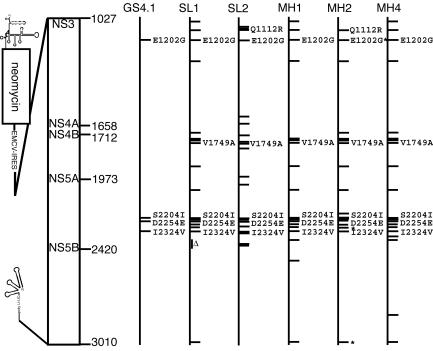
To determine whether HCV replication in HeLa cells led to the selection of subgenomes with cell-type-specific adaptive mutations, we compared the efficiency by which G418-resistant colonies formed in Huh7 and HeLa cells transfected with total RNA isolated from GS4.1 and SL1 cells. Total RNA from GS4.1 cells led to the selection of approximately 166 G418-resistant colonies per ng of viral RNA in Huh7 cells compared with only 4 colonies in HeLa cells (Table 2). In contrast, total RNA from SL1 cells yielded 160 colonies in HeLa cells compared with about 20 in Huh7 cells. These results indicated that replication in HeLa cells led to the selection of variants with cell-type-specific adaptive mutations that were responsible for the 40-fold increase in colony formation efficiency between amplified RNA in GS4.1 and SL1 cells. Nucleotide sequence analysis of HCV cDNA clones obtained from the SL1 and SL2 cell lines confirmed this view. These data showed that replicons in the two HeLa cell lines maintained the previously identified adaptive mutations in GS4.1 cells and acquired several additional mutations that resulted in amino acid changes in the NS region (Fig. 3 and Table 3). Notably, some of the new mutations formed clusters in the NS4B and NS5A regions. In the case of SL1 cells, we observed a deletion of 43 amino acids near the C terminus of NS5A. Of particular interest were mutations in the amino-terminal region of NS4B, because they have so far not been found in cDNAs from replicons in Huh7 cells and hence could have been responsible for the observed adaptation of replicating RNA (1, 7, 10, 14). Moreover, one mutation at position 1749 was present in both SL1 and SL2 cells. In contrast to the results obtained with the NS regions, we could not detect any mutations in the 5′ and 3′ untranslated regions of replicons expressed in SL1 and SL2 cells.
TABLE 2.
Colony formation efficiency of total cellular RNAa
| Cell | Viral RNA (ng/10 μg) | No. of colonies in transfected cells
|
|||||
|---|---|---|---|---|---|---|---|
| Huh7
|
HeLa
|
Hepa1-6
|
|||||
| Mean (SD) | Colonies/ng of viral RNA | Mean (SD) | Colonies/ng of viral RNA | Mean (SD) | Colonies/ng of viral RNA | ||
| GS4.1 | 5 | 834 (64) | 166 | 22 (4) | 4 | 0 | <1 |
| SL1 | 5 | 100 (53) | 20 | 803 (81) | 160 | 1.3 (1.5) | <1 |
| MH1 | 0.5 | 20 (2) | 40 | 66 (9) | 132 | 1.7 (0.6) | 3 |
Results from three independent transfection experiments. Total RNA was extracted from GS4.1, SL1, and MH1 cells at passages 21, 26, and 4, respectively.
FIG. 3.
Sequence analysis of HCV subgenomes in HeLa and mouse hepatoma Hepa1-6 cells. (Left) Physical map of HCV subgenomic RNA including the positions of the first amino acid NS3 and the last residue of the polyprotein. The IRES for the translation of the NS genes is indicated (EMCV-IRES). (Right) Mutations causing amino acid changes identified in cDNAs isolated from subgenomic replicons present in GS4.1 (passage 21) and the indicated HeLa (SL1, passage 26; SL2, passage 5) and Hepa1-6 (MH1 [passage 12], MH2 [passage 4], and MH4 [passage 4]) cell lines are depicted with horizontal bars. Four independent clones were sequenced from each PCR fragment that was amplified from cDNAs obtained from total RNA purified from the indicated cell lines. Mutations present in more than one cell line are indicated with their amino acid positions. Mutations that occurred in 50% of the clones analyzed are indicated with an asterisk. Mutations that occurred in only one of four clones analyzed are not included in the figure. A deletion identified in cDNA clones obtained from SL1 cells spanning amino acids 2371 to 2413 is indicated (Δ).
TABLE 3.
Consensus mutations in replicons isolated from HeLa and mouse hepatoma cell clones
| Cell clone | Conserved mutation(s) | NS protein |
|---|---|---|
| GS4.1 | E1202G | NS3 |
| S2204I, D2254E, I2324V | NS5A | |
| SL1 | Q1067R, S1128A, E1202G, S1323P, S1560Ga | NS3 |
| L1701F | NS4A | |
| Q1720R, Q1727H, V1749A, V1893L | NS4B | |
| T2035A, S2204I, I2252V, D2254E, I2274V, R2290L, I2324V, del.2371-2413b | NS5A | |
| W2990R | NS5B | |
| SL2 | I1097V, Q1112R, P1115L, V1593M, M1647I | NS3 |
| L1715P, Q1737R, V1749A, I1797V, N1965Y | NS4B | |
| Q2012L, S2204I, E2247G, D2254E, K2302R, I2324V, S2336P, L2400S, E2411G, A2412V | NS5A | |
| MH1 | Q1067R, S1128A, E1202G, S1323P, S1560G | NS3 |
| Q1720R, Q1727H, V1749A, V1893L | NS4B | |
| T2035A, S2204I, I2252V, D2254E, I2274V, R2290L, I2324V, M2388T, T2496A | NS5A | |
| W2990R | NS5B | |
| MH2 | Q1112R, E1202G,a S1323P, S1560G | NS3 |
| L1701F | NS4A | |
| Q1720R, Q1727H, V1749A, V1893L | NS4B | |
| T2035A, T2185A, S2204I, I2252V, D2254E, I2274V, R2290,a I2324Va | NS5A | |
| W2990Ra | NS5B | |
| MH4 | Q1067R, S1128A, E1202G, S1323P, S1560G | NS3 |
| L1701F | NS4A | |
| Q1720R, Q1727H, V1749A, V1893L, A1841T | NS4B | |
| T2035A, S2204I, I2252V, D2254E, I2274V, R2290L, I2324V, T2364M, L2391R | NS5A | |
| I2843V, W2990R | NS5B |
Mutation that occurred in 50% of the clones analyzed.
del., deletion.
Mouse hepatoma cells can support HCV RNA replication.
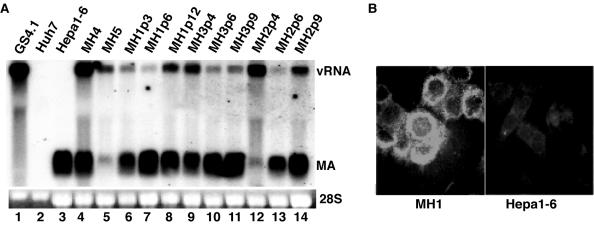
The discovery of several additional mutations in cDNA clones obtained from SL1 and SL2 cells prompted us to examine whether total RNA from these cell lines could yield colonies in cells that did not appear to be permissive for HCV replication after transfection with subgenomic RNA or total RNA from Huh7-derived cell lines. In addition to the five cell lines that we examined initially with Huh7-derived RNA, we examined five additional hepatoma and hepatocyte-derived cell lines (Table 1). G418-resistant colonies were obtained only with the mouse hepatoma cell line Hepa1-6 after transfection with total RNA from SL1 cells (Fig. 4A, lanes 4 to 6, 9, and 12). As with HeLa cells, the amounts of RNA ranged from 300 to 1,000 copies of RNA per cell and a large fraction of the cells expressed viral proteins (Fig. 4B). In contrast to Huh7 and HeLa cells, the amount of HCV RNA in the mouse cell lines appeared to vary between cell passages (Fig. 4A, lanes 6 to 14). Interestingly, total RNA isolated from one of the mouse cell lines, MH1, did not produce significantly more colonies in Hepa1-6 cells than did total RNA from SL1 cells, suggesting that the subgenomes present in SL1 cells were already adapted for replication in the mouse cells (Table 2). In support of this interpretation, nucleotide sequence analysis of viral cDNAs cloned from three mouse cell lines showed that the majority of the mutations identified in SL1 cells were maintained (Fig. 3). Surprisingly, the deletion in NS5A identified in four of four clones sequenced from SL1 cells was not present in replicons isolated from mouse cells, indicating that a subpopulation of replicons without the deletion was still present in these (SL1) cells.
FIG. 4.
Replication of HCV subgenomes in mouse hepatoma cells. (A) Detection of HCV viral RNA. RNA in Hepa1-6 cell lines (MH1 to MH5) that were established from G418-resistant cell colonies was analyzed as described in the legend to Fig. 2 except that a radiolabeled probe specific to the mouse albumin cDNA was used in lieu of the probe against HPV. MH4 and MH5 were analyzed at passage 4. (B) Immunohistochemical analysis of HCV replication in MH1 cells (passage 3). Expression of NS5A was detected as described in the legend to Fig. 2. Hepa1-6 cells served as a negative control.
Cell-derived HCV RNA is more efficient than in vitro-transcribed RNA in initiating replication in HeLa and mouse hepatoma cells.
So far, our results showed that replication of HCV subgenomes in HeLa and mouse cells led to the selection of replicons with several novel mutations. The majority of these mutations were located in the NS3, NS4B, and NS5A regions. Moreover, the results showed that cell-derived RNA carrying some or all of these mutations was much more efficient in establishing G418-resistant colonies in HeLa cells than was RNA derived from Huh7 cells (Table 2). Based on these observations, we surmised that introduction of these mutations into available subgenomic replicons should alter or expand their tissue and host tropism. To test this hypothesis, we constructed 13 subgenomic replicons that carried mutations in NS3, NS4B, and NS5A alone or in combination with each other as described in Table 4. Of the 13 constructs examined, only two, pZS2 and pZS25, yielded a small number of G418-resistant colonies in HeLa cells (Table 5). Viral RNA replication was confirmed by Northern blot analysis of total RNA isolated from six cell lines derived from those colonies. None of the variants yielded colonies in Hepa1-6 cells. Moreover, negative-control experiments with in vitro-transcribed RNA derived from a variant containing a frameshift mutation in NS5B did not yield any colonies that could be expanded into cell lines. Notably, save for one, all replicons were permissive for replication in Huh7 cells, albeit with significantly different efficiencies (Table 4). Interestingly, both pZS2 and pZS25 carried mutations in NS4B that were conserved in replicons from two independent HeLa cell lines, SL1 and SL2. In addition, these replicons had the S2204I mutation in NS5A that was previously found to be one of the most potent adaptive mutations for HCV replication in Huh7 cells. Because both replicons replicated very efficiently in Huh7 cells, the results suggested that the NS4B mutations could have contributed to the observed expansion of the tissue tropism of HCV replicons. In support of this hypothesis, the subgenome with the highest efficiency in Huh7 cells, pZS11 lacking mutations in NS4B (Table 4), did not yield any colonies in HeLa cells. However, the number of colonies obtained with in vitro transcripts was too low to draw firm conclusions (Table 5).
TABLE 5.
Colony formation efficiency of in vitro-transcribed RNAa
| Source of cDNA library (cell line or plasmid) | No. of colonies in transfected cells
|
||
|---|---|---|---|
| Huh7 | HeLa | Hepa1-6 | |
| GS4.1 | >10,000 | 0, 0, 0 | 0, 1, 0 |
| SL1 | >10,000 | 0, 3, 2 | 0, 1, 0 |
| MH4 | >10,000 | 3, 4, 0 | 17, 0, 0 |
| pZS2 | >10,000 | 2, 3, 0 | 0, 0, 0 |
| pZS25 | >10,000 | 0, 2, 1 | 0, 0, 0 |
Results from transfection experiments with in vitro-transcribed RNA from pooled clones isolated from the indicated cell line and from in vitro-transcribed RNA from pZS2 and pZS25 (Table 4).
To further explore the basis for the observed low colony formation efficiency of in vitro-transcribed RNA in HeLa cells, we examined whether replication in HeLa cells led to the selection of adaptive mutations that were not discovered previously when we sequenced cDNA clones from SL1 and SL2 cells. For this purpose, cDNA clones were isolated from total RNA obtained with pZS2- and pZS25-derived cell lines, respectively. Nucleotide sequence analysis of both cDNA clones did not reveal any additional consensus mutations, suggesting that the two subgenomes were sufficiently adapted for replication in HeLa cells (results not shown). However, as mentioned above, we could not exclude the possibility that a minor population of subgenomic replicons with additional mutations were present in these cell lines. To overcome this problem, we developed a method for the isolation and cloning of cDNAs spanning the NS3 to NS5B region (see Materials and Methods). We produced replicon cDNA libraries from GS4.1, SL1, and MH4 cells. Approximately 2,000 cDNA clones were pooled and subsequently used for in vitro transcription of subgenomic RNA. With Huh7 cells, the colony formation efficiency of the pooled clones was comparable to that of the most efficient subgenomes, such as pZS2 or pZS25, and did not vary significantly with the origin of the total RNA used for cDNA cloning (Table 5). Consistent with previous results, colony formation in HeLa and mouse cells was origin dependent, i.e., save for one case, colonies were observed only with clones derived from SL1 and MH4 cell lines. Notably, with this strategy we were able for the first time to obtain G418-resistant colonies with Hepa1-6 cells by using in vitro-transcribed RNA. To confirm the presence of viral RNA, we expanded 11 colonies and performed Northern blot analysis with total RNA. All 11 RNA samples analyzed contained viral RNA ranging from approximately 0.1 to 1 ng/5 μg of total RNA (results not shown).
Taken together, the results supported the hypothesis that mutations identified in subgenomic replicons expressed in HeLa and mouse cells play a role in adaptation of the replicons to certain cell-type-specific conditions. Importantly, the results invoked the possibility that differences exist between amplified RNA in cells and in vitro-transcribed RNA that influence the efficiency by which HCV subgenomes initiate replication in HeLa and mouse hepatoma cells.
DISCUSSION
HCV is known as a species- and tissue-specific virus. This report now shows that replication of HCV can occur in cells derived from tissues other than liver, indicating that cellular factors required for RNA replication are expressed in cell types other than hepatocytes. One interpretation of this result is that the apparent tropism of HCV for hepatocytes is determined primarily at the level of virus entry or assembly or, alternatively, that HCV can infect many other tissues but has escaped detection due to very low amounts of RNA replication or accumulation. Extrahepatic tissues could serve as reservoirs for HCV that, as with human immunodeficiency virus, could provide a source of viruses that are refractory to antiviral therapy and, importantly, can be responsible for infection of liver grafts following orthotopic liver transplantation (5, 12). Such a scenario would have profound implications for antiviral therapy. For example, the targeting of drugs to secondary sites of viral replication and the analysis of drug metabolism in cells other than hepatocytes would become important factors for the development of successful antiviral therapies. Although proof for HCV replication in cells of nonhepatic origin is still lacking, there is ample evidence for the presence of viral RNA in lymphocytes and other tissues (3, 18, 19, 21). The results of this study will encourage further investigations that might provide convincing evidence for HCV replication in extrahepatic tissues.
It is conceivable that HCV quasispecies in hepatocytes and other tissues exhibit differences in their composition due to the selection of variants with cell-type-specific adaptations. As shown in this report, replication of subgenomes in HeLa cells led to the accumulation of clusters of mutations in the NS3, NS4B, and NS5A regions including a deletion in NS5A (Fig. 3). Mutations and deletions in NS5A have been found previously in genomes that replicated in Huh7 cells, which could suggest that expression of the natural form of this protein in cell culture somehow interferes with RNA replication (1, 7, 9, 13, 14). However, mutations in the amino terminus of NS4B have previously not been observed. Notably, in both SL1 and SL2 cells, the mutations changed two or one glutamine residues, respectively, to one of the two basic amino acids arginine and histidine. Moreover, the mutation V1749A was present in all five cell lines examined (Table 3 and Fig. 3). Thus far, our results showed that these mutations appeared to be required for replication in HeLa cells, because only replicons pZS2 and pZS25 carrying these mutations yielded colonies after transfection with in vitro-transcribed RNA (Tables 4 and 5). However, due to the low efficiency in colony formation obtained with in vitro-transcribed RNA, our results did not yet provide definitive proof for such a conclusion (see below). The amino terminus of NS4B is predicted to reside on the cytoplasmic side of endoplasmic reticulum membranes and may interact with other host or viral proteins required for RNA replication (8). As an integral endoplasmic reticulum membrane protein, NS4B might provide a scaffold for the assembly of replication complexes and act as a regulator for RNA replication. More importantly, a recent study revealed that NS4B can induce particular membrane structures, called membranous webs, proposed to be the site for HCV replication (6). Interestingly, genetic analyses with an HCV-related pestivirus identified the amino-terminal region of NS4B as a determinant for cytotoxicity caused by high levels of virus replication (17). Although the exact mechanism by which NS4B exerts this activity is still unknown, it might interact with cell-type-specific factors and cause the selection of variants with adaptive mutations as shown in this study.
For reasons that we do not yet understand, we could not yet obtain subgenomes that replicated with high efficiency, transiently or permanently, in HeLa or mouse cells (Tables 4 and 5). Although it is conceivable that we missed a critical mutation, because it was for some reason underrepresented in our cDNA clones, it is puzzling that it did not arise following the transfection of HeLa cells with RNA. Based on the experience with Huh7 cells, we would have expected that such an event would have occurred and eventually led to the identification of the critical adaptive mutation(s). Nevertheless, our results indicated that the mutations in NS4B and NS5A in replicons pZS2 and pZS25 were sufficient to establish replication in a small number of HeLa cells, because, based on our sequence analysis of cDNA clones, cell lines obtained with these subgenomes did not contain any additional mutations (results not shown). Moreover, transfection of HeLa and mouse cells with heterogeneous populations of subgenomes that should have represented the populations of amplified RNA in cells, with one exception, did not yield more colonies than in vitro-transcribed RNA (Table 5). Hence, based on our results we were speculating that in vitro-transcribed RNA exhibited some toxicity in HeLa or mouse cells. However, this is an unlikely scenario, because we found that in vitro-transcribed RNA did not alter the colony formation efficiency in HeLa cells when added to total RNA isolated from HCV-containing cell lines, such as GS4.1 and SL1 (results not shown). Similarly, addition of small amounts of in vitro-transcribed RNA to total RNA from normal Huh7 or HeLa cells did not yield any G418-resistant colonies. These results also indicated that cellular mRNAs did not influence the colony formation efficiency. In addition, we did not observe an increase in colony formation efficiency when we used cured HeLa cells that were obtained through the treatment of subgenome-expressing cells with an HCV polymerase inhibitor. It is conceivable that establishment of HCV replicons in HeLa cells requires certain adaptive mutations that are not required for the maintenance of replicons during the expansion of G418-resistant colonies. Because the amount of viral RNA in total cellular RNA is too low to permit detection of transient replication, we have not been able to directly test this hypothesis. Finally, our results could indicate that amplified viral RNA isolated from cells exhibits physical differences from RNA that is transcribed in vitro, such as methylation of certain residues or other, so far unrecognized modifications at the termini of viral RNA that are required for initiation of replication in HeLa cells.
In summary, we have shown in this report that HCV RNA replication is not restricted to the human hepatoma cell line Huh7 but instead can occur in HeLa cells and hepatoma cells derived from mice. These findings suggest that it may be possible to develop a mouse model for HCV infection. Establishment of such a model will depend on the isolation of HCV variants that can infect mouse hepatocytes or transgenic mice that express the still elusive HCV receptor.
Acknowledgments
We thank Rich Katz, Luis Sigal, Ann Skalka, and Ken Zaret for their helpful comments on the manuscript and acknowledge services provided by the Fox Chase Cancer Center nucleotide sequencing, tissue culture, and imaging facilities.
This work was supported by grants from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 2.Boisvert, J., X. S. He, R. Cheung, E. B. Keeffe, T. Wright, and H. B. Greenberg. 2001. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J. Infect. Dis. 184:827-835. [DOI] [PubMed] [Google Scholar]
- 3.Chang, T. T., K. C. Young, Y. J. Yang, H. Y. Lei, and H. L. Wu. 1996. Hepatitis C virus RNA in peripheral blood mononuclear cells: comparing acute and chronic hepatitis C virus infection. Hepatology 23:977-981. [DOI] [PubMed] [Google Scholar]
- 4.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 5.Chun, T. W., J. S. Justement, P. Pandya, C. W. Hallahan, M. McLaughlin, S. Liu, L. A. Ehler, C. Kovacs, and A. S. Fauci. 2002. Relationship between the size of the human immunodeficiency virus type 1 (HIV-1) reservoir in peripheral blood CD4+ T cells and CD4+:CD8+ T cell ratios in aviremic HIV-1-infected individuals receiving long-term highly active antiretroviral therapy. J. Infect. Dis. 185:1672-1676. [DOI] [PubMed] [Google Scholar]
- 6.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanford, R. E., D. Chavez, F. V. Chisari, and C. Sureau. 1995. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J. Virol. 69:8079-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laskus, T., M. Radkowski, J. Wilkinson, H. Vargas, and J. Rakela. 2002. The origin of hepatitis C virus reinfecting transplanted livers: serum-derived versus peripheral blood mononuclear cell-derived virus. J. Infect. Dis. 185:417-421. [DOI] [PubMed] [Google Scholar]
- 13.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 16.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu, L., L. K. McMullan, and C. M. Rice. 2001. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in viral cytopathogenicity. J. Virol. 75:10651-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radkowski, M., J. Kubicka, E. Kisiel, J. Cianciara, M. Nowicki, J. Rakela, and T. Laskus. 2000. Detection of active hepatitis C virus and hepatitis G virus/GB virus C replication in bone marrow in human subjects. Blood 95:3986-3989. [PubMed] [Google Scholar]
- 19.Radkowski, M., J. Wilkinson, M. Nowicki, D. Adair, H. Vargas, C. Ingui, J. Rakela, and T. Laskus. 2002. Search for hepatitis C virus negative-strand RNA sequences and analysis of viral sequences in the central nervous system: evidence of replication. J. Virol. 76:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 21.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]