Abstract
We have shown previously that interferon regulatory factor 7 (IRF7), a multifunctional protein intimately involved in latent Epstein-Barr virus (EBV) infection, is induced as well as activated by EBV latent membrane protein 1 (LMP1), the principal EBV oncoprotein. Since the LMP1 promoter (LMP1p) contains an interferon-stimulated response element (ISRE), we hypothesized that IRF7 might be able to regulate LMP1 expression and thus participate in a regulatory circuit between these two genes. In this study, IRF7 was shown first to activate LMP1p in transient transfection assays. Compared with EBV nuclear antigen 2 (EBNA2), the most potent viral transactivator of LMP1p, IRF7 has a lesser effect (approximately 10% that of EBNA2) on induction of LMP1p. Study with IRF7 deletion mutants showed that IRF7 functional domains have similar effects on both the beta interferon (IFN-β) and LMP1 promoters in BJAB and 293 cells, and study with IRF7 phosphomimetic mutants showed that IRF7 phosphorylation may be involved in the activation of these two promoters. Further, the ISRE in LMP1p responds to IRF7 induction and IRF7 binds to this element. In the EBV-positive cell line P3HR1, which lacks the complete EBNA2 and EBV-encoded leader protein genes and hence expresses low-level LMP1, IRF7 alone can notably increase the endogenous LMP1 mRNA and protein levels. These results indicate that LMP1 is regulated by this host cell gene in addition to the viral factor, EBNA2, and may help to explain how LMP1 is expressed in type II latency in the absence of EBNA2. Moreover, IRF7 can regulate a viral gene in addition to a host cellular gene such as the IFN-β gene. Together with the previous data that LMP1 can induce IRF7 expression and facilitate IRF7 phosphorylation and nuclear translocation, these results suggest a positive regulatory circuit between IRF7 and LMP1.
Interferons (IFNs) are a large family of multifunctional secreted proteins involved in antiviral defense, cell growth regulation, and immune activation (26, 44). The IFN regulatory factors (IRFs) are a growing family of virus- and IFN-inducible cellular proteins that now consists of 10 members, including the recently discovered IRF-10 (32) as well as four Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded viral IRFs (4, 26, 33). A hallmark of IRF proteins (but not including viral IRFs) is an N-terminal DNA-binding domain (DBD), which contains a five-tryptophan-residue repeat. This repeat forms a helix-turn-helix motif determining the characteristic DNA-binding site, 5′-(A/G)NGAAANNGAAACT-3′. Distinct properties and functions of IRF family members depend on their different C-terminal sequences and structures (26). IRFs are critical in the regulation of expression of type I IFNs (IFN-α and IFN-β), IFN-stimulated genes, and other cytokines and chemokines; three of them, IRF3, IRF5, and IRF7, are direct inducers of virus-mediated signaling (3, 4).
IRF7 was cloned and identified within the biologic context of Epstein-Barr virus (EBV) latency (52) and is expressed predominately in spleen, thymus, and peripheral blood leukocytes. IRF7 has been implicated in IFN gene activation in response to virus infection (1, 52). In addition to IFNs, IRF7 expression is also stimulated by other factors such as sodium butyrate (55), lipopolysaccharide, topoisomerase II inhibitors, 12-O-tetra-decanoylphorbol-13-acetate (TPA), and tumor necrosis factor alpha (TNF-α) (24). Cellular factors, including NF-κB, are involved in the induction of IRF7 by TNF-α (24). Interestingly, IRF7 also can be induced by the principal EBV oncoprotein, latent membrane protein 1 (LMP1) (53). IRF7 appears to be a multifunctional protein involved in multiple physiological processes during viral infection, including regulation of the host immune system, IFN gene expression, and EBV latency, as well as acting as a putative oncogene (L. Zhang, C. Der, and J. S. Pagano, unpublished results). IRF7 has also been reported to participate in monocyte differentiation (25).
EBV LMP1 is an integral membrane protein with six transmembrane N-terminal domains and a long C-terminal cytoplasmic tail. LMP1, as a key EBV oncogenic factor, is a constitutively active receptor-like molecule that does not need the binding of a ligand. Expression of LMP1 is essential for growth transformation of B lymphocytes (38, 40). Several studies have demonstrated that efficient LMP1 promoter activation is dependent upon virus-specific factors; some host cellular factors are also involved in these processes (11, 15, 19, 38-40, 45). EBV nuclear antigen 2 (EBNA2) is the best-characterized viral factor that transactivates the LMP1 promoter (LMP1p) (15, 38, 39, 45, 48). Also, EBV-encoded leader protein (EBNA-LP) can activate the LMP1p, especially in cooperation with EBNA2 (12, 29, 34). In addition to LMP1p, it is well established that EBNA2 can transactivate a number of other genes, including the cellular CD21, CD23, c-Bcl-2, and c-fgr and EBV LMP2A and LMP2B genes and the EBNA-1 C promoter (21, 39, 43). EBNA2 does not bind directly to DNA but exerts its function by interacting with the cellular protein, recombination signal-binding protein (RBP-Jκ), and PU.1 protein; other cellular transcriptional factors mediate the activation of LMP1 by EBNA2 (15, 19, 38, 40, 45). The latency-associated nuclear antigen encoded by KSHV can also activate LMP1p (11). LMP1 stimulates the expression of IRF7 (53), ICAM-1, LFA-3, CD40, EBI-3, the transporter associated with antigen processing 2 (Tap-2), Fas, and TNF receptor-associated factor 1 in EBV-negative Burkitt's lymphoma (BL) cells and of the epidermal growth receptor in epithelial cells (5, 8-10, 16, 47). LMP1 also induces or enhances expression of matrix metalloproteinase 1 (22), matrix metalloproteinase 9 (51), Bcl-2 (13), cyclooxygenase-2, and vascular endothelial growth factor (31) as well as fibroblast growth factor 2 (46) and hypoxia-inducible factor 1α (N. S. Wakisaka and J. S. Pagano, unpublished results), all of which are associated with tumor invasion and metastasis. Therefore, LMP1 is a key mediator for host viral defense as well as oncogenesis.
Because of a conserved DBD in the N-terminal region of all members of the IRF family, IRFs can bind to consensus or similar sequences: IFN-stimulated response element (ISRE), positive regulatory element, and IFN consensus sequence, all of which have conserved GAAA repeats. LMP1 has an ISRE within its promoter (Fig. 1A). In this paper, we show that IRF7 can bind to and regulate the LMP1 promoter and that IRF7 can stimulate expression of LMP1 mRNA and protein independently of as well as cooperatively with EBNA2.
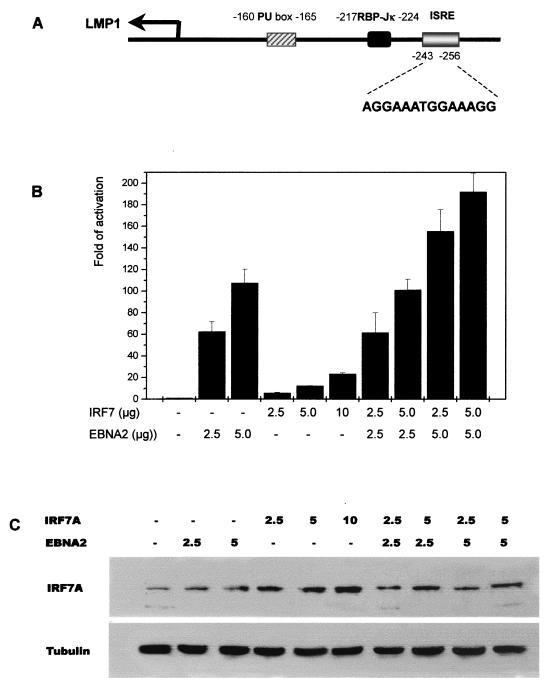
FIG. 1.
IRF7 upregulates the LMP1 promoter in BJAB cells. (A) Schematic illustration of the LMP1 promoter. Numbers indicate the nucleotide sites relative to the transcriptional start site. The ISRE sequence is shown. (B) Promoter activity was monitored by investigating luciferase activity. BJAB cells were transfected with pGL2(−512/+72)-luciferase plasmid (LMP1p) and IRF7 or with IRF7 plus EBNA2. Luciferase activity was analyzed at 48 h after transfection. RLU levels and severalfold activation relative to the basal level of reporter gene in the presence of the vector after normalization with β-Gal activity were measured. Equal amounts of cell lysates were assayed for luciferase activity. Each data point represents the average of seven to nine repeats in five independent experiments. Error bars represent means ± standard errors (SE). (C) Western blot analysis showing that IRF7 was expressed in transfected BJAB cells. Monoclonal IRF7 antibody was used at a dilution of 1:300. A total of 100 μg of protein was loaded in each lane. Tubulin was used as loading control.
MATERIALS AND METHODS
Plasmids.
The LMP1 promoter construct pGL2(−512/+72)-luciferase plasmid, which has two directly repeated copies of the LMP1 −512/+72 promoter element, was provided by Jeffery Lin and Elliott Kieff (19). The LMP1 promoter construct pgLRS(−259)-CAT and its mutations, pgLRS(−259)-ISRE−-CAT, pgLRS(−259)-Jκ−-CAT, and pgLRS(−259)-ISRE−/Jκ−-CAT, were gifts from Anna Sjoblom and Lars Rymo (39). The wild-type IRF7A expression plasmid pcDNA3/IRF7A was constructed in this laboratory (52). The IFN-β promoter (IFNβp) construct (pGL3/IFNβp-Luc) and a series of pCMV2-Flag plasmids containing wild-type IRF7A and its deletion or substitution mutations were described previously (20) and were gifts from Rongtuan Lin and John Hiscott. The pBS-LMP1 plasmid for RNase protection assays (RPA) was provided by Paul Farrell (42). The EBNA2-expressing plasmid construct was made by inserting a 2.1-kb MluI-AhaIII restriction fragment containing the EBNA2b coding region from the AG876 virus into the pGEM derivative pHD 101-3 (7). IRF7-DN, which lacks the DBD of the human IRF7A gene, was provided by Tom Maniatis.
Cell lines.
BJAB is an EBV-negative human BL cell line. P3HR-1, an EBV-positive cell line from which portions of the EBNA-LP and EBNA2 open reading frames have been deleted, expresses low levels of LMP1. Both BJAB and P3HR1 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively). The 293 cell line (derived from human kidney epithelial cells) was grown in Dulbecco's modified Eagle's medium with 10% FBS and penicillin-streptomycin (5 U/ml and 5 μg/ml, respectively).
Transfections.
BJAB and P3HR-1 cells were transfected with an electroporator at 210 V and 975 μF with 5-μg quantities of the various promoter constructs or vector and increasing concentrations of the IRF7 clones in 0.5 ml of RPMI 1640 medium containing 10% FBS. Vector DNA was added to equalize the total amount of DNA used in all transfections. After electroporation, cells were resuspended in 10 ml of complete medium and incubated for 48 h. Cells were harvested, washed twice with phosphate-buffered saline, and lysed in appropriate buffer.
Reporter assays.
For luciferase assays, the transfected cells were collected at 48 h posttransfection and resuspended in 200 μl of 1× reporter lysis buffer (Promega). The cells were placed in a dry ice-isopropanol bath for 2 min, thawed at 37°C, vortexed, and then centrifuged at 16,000 × g for 20 s. Cell lysates (20 μl each) were combined with luciferase assay reagent (Promega), and the relative light units (RLU) were measured in an Lmax luminometer (Molecular Devices Corp.). The pcDNA3 empty vector alone was used in these assays to determine the ability of IRF7 to activate the vector alone, and the transfection efficiencies were normalized by β-galactosidase (β-Gal) values. Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (18); results were analyzed by use of ImageQuant software (version 5; Molecular Dynamics). All reporter assay results presented are from single experiments representative of multiple independent trials or are averages derived from multiple independent repetitions.
Western blotting with enhanced chemiluminescence detection.
Cells were collected and washed with 5 ml of phosphate-buffered saline. The pellets were resuspended in 200 μl of cell lysis buffer and mixed with 200 μl of 2× sodium dodecyl sulfate loading buffer. Lysates were separated on sodium dodecyl sulfate-10% polyacrylamide electrophoresis gels for detection of IRF7 and LMP1 and 8% gels for EBNA2. The proteins were then transferred to nitrocellulose membranes. The membranes were blocked in 5% milk for 1 h at room temperature (RT), rinsed in Tris-buffered saline with Tween (TBST) three times for 5 min each time, and then incubated with the specific primary antibody at RT for 1 h or at 4°C overnight. After three rinses for 5 min each rinse in TBST, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences) (1:3,000) for 1 h at RT and subsequently washed with TBST three times for 5 min each time. Specific signals were detected by enhanced chemiluminescence (ECL) following the manufacturer's instruction (Amersham Pharmacia Biotech). IRF7 was detected with rabbit polyclonal antibody (Santa Cruz) (1:300), and EBNA2, LMP1, and Flag were detected with mouse monoclonal antibodies PE2 (DAKO) (1:500), CS1-4 (DAKO) (1:100), and M2 (Sigma) (1:3,000), respectively.
EMSA.
BJAB and 293 cells in 100-mm-diameter dishes were transfected with 5 μg of Flag-tagged IRF7A or its mutants. Cells were collected and lysed in binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 50 mM NaCl, 2 mM dithiothreitol [DTT], 5% glycerol, 0.5% NP-40, 10 μg of bovine serum albumin [BSA]/μl) with protease inhibitors at 48 h posttransfection. The whole-cell lysates were used for electrophoretic mobility shift assays (EMSA), as described previously (20). Poly(dI-dC) (Amersham) at a final concentration of 62.5 μg/ml was added to reduce nonspecific binding. The double-stranded sequence containing LMP1 ISRE is GATCCAACAGGAAATGGAAAGGCAGTG. Another sequence containing mutated LMP1 ISRE is GATCCAACAGGAggTGGAggGGCAGTG (the mutated nucleotides are indicated by lowercase letters). These double-stranded sequences with 5′-GATC adhesive ends were labeled (using Klenow fragments) with [α-32P]dCTP. For competitor assays, 100-fold excess cold probe or AP-1 sequence (Promega) was added to the binding mixture. For supershift assays, whole-cell lysates were incubated with 0.2 μg of Flag antibody M2 (Sigma) before probe was added. Protein-DNA complexes were separated on 5% 60:1 acrylamide gels.
RNA isolation and RPA.
P3HR1 cells were transfected with different amounts of IRF7 and EBNA2 expression plasmids together with CD4 expression plasmid. Using a QIAgen RNeasy mini kit, total RNAs were isolated from CD4-conjugated magnetic bead (Dynal)-selected cells. RPA were performed with total RNA using an RNase protection kit II (Ambion). The hybridization temperature was 37°C. The human GAPDH probe was supplied by US Biochemicals Inc. The LMP1 probe for RPA, which was from the pBS-LMP1 construct and labeled with [α-32P]UTP by in vitro transcription, corresponds to nucleotides 169033 to 169423 in the B95-8 EBV genome. The protected fragments were 220 and 90 bp (42).
RESULTS
IRF7 upregulates the LMP1 promoter.
The positive regulators of the LMP1 promoter (LMP1p) identified to date center mostly on the EBNA2-responsive enhancer region to which RBP-Jκ binds (43, 45, 48, 49). Here, we examined whether another region of LMP1p that contains an ISRE would be responsive to induction by IRF7. BJAB, an EBV-negative human BL cell line, was used for this study because it has little endogenous IRF7. The assayed LMP1p construct, pLMP1(−512/+72), has two directly repeated copies of the LMP1 −512/+72 promoter element (19) (Fig. 1A). Using luciferase assays, we examined the effect of the presence of IRF7 on the regulation of LMP1p after transient expression of IRF7. In cells transfected with IRF7A, LMP1p activity was increased about 10-fold compared with that produced by the vector pcDNA3 alone (Fig. 1B). The regulatory effects were clearly dose dependent, with increasing amounts of IRF7A resulting in elevated levels of luciferase activity (Fig. 1B). In this assay, EBNA2, which is known to transactivate LMP1p, was used as a positive control. Compared with IRF7, EBNA2 had a greater effect on transactivation of LMP1p (up to 100-fold with 5 μg of EBNA2 expression plasmid). However, the cotransfection of IRF7A and EBNA2 resulted in up to 200-fold activation of LMP1p; the combined effect was generally greater than the effect of IRF7 alone plus that of EBNA2 alone (Fig. 1B). Thus, LMP1p can respond to induction by IRF7 independently of the EBNA2 enhancer region, and the two can function together to activate the promoter. Western blot analyses show that exogenous IRF7 was expressed in the transfected BJAB cells (Fig. 1C) and that a low level of endogenous IRF7 is detected in these cells (Fig. 1C, first lane).
To elaborate on the significance of the results obtained with BJAB cells, we tested the effects of IRF7 on LMP1p in another human cell line, 293. These cells contain no detectable endogenous IRF7 and EBNA2, and LMP1p constructs cannot be activated by endogenous factors in 293 cells (data not shown). Luciferase assays show that IRF7 can activate LMP1p in 293 cells. However, the effect is less striking than in BJAB cells. IRF7 consistently activates LMP1p in 293 cells at levels two- to threefold higher than those seen with the vector-only control (Fig. 2B). The effect is approximately threefold less than that obtained with BJAB cells.
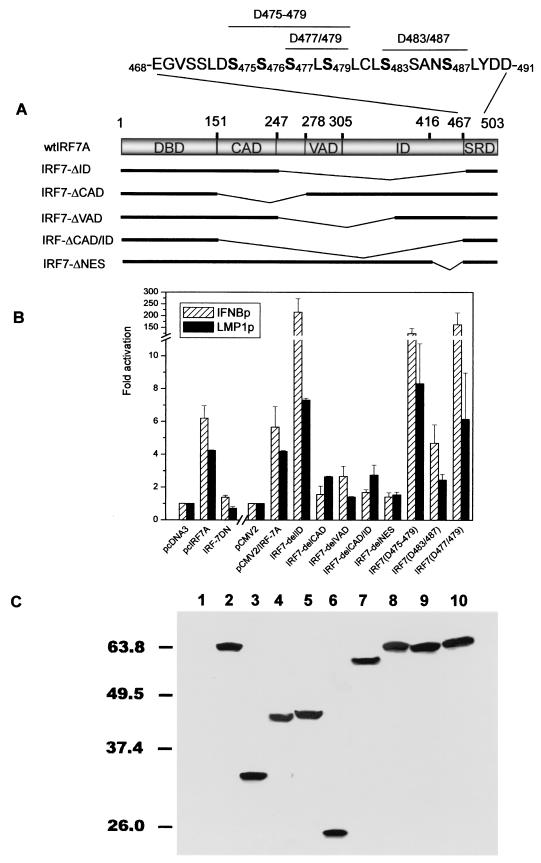
FIG. 2.
Effect of IRF7 mutants on LMP1 and IFN-b promoters. (A) Schematic representation of the IRF7 mutants used in this study. The amino acid sequence from 468 to 491 is shown, serines replaced with phosphomimetic aspartic acids are in bold, and the sites are indicated as subscripts. SRD, signal response domain (20). (B) Comparison of the effects of IRF7 mutants on the IFN-b and LMP1 promoters in 293 cells. 293 cells were transfected with pGL3/IFNb-Luc or pGL2(2512/172)-Luc and a series of expression plasmids encoding IRF7 or IRF7 mutants as indicated. RLU levels and severalfold activation relative to the basal level of reporter gene in the presence of the vector after normalization with b-Gal activity were measured. Equal amounts of cell lysates were assayed for luciferase activity. Each data point represents the average of seven to nine repeats in five independent experiments. Error bars represent means 6 SE. (C) Western blot analysis of transfected IRF7 mutants. Monoclonal Flag antibody M2 was used at a dilution of 1:3,000. Lanes: 1, pCMV2-Flag; 2, IRF7A; 3, IRF7-DID; 4, IRF7-DCAD; 5, IRF7-DVAD; 6, IRF7-DCAD/ID; 7, IRF7-DNES; 8, IRF7(D475-479); 9, IRF7(D483/487); 10, IRF7(D477/479).
IRF7 functional domains and phosphorylation in LMP1p activation.
Functional domains of IRF7 protein have been defined based on effects of mutated IRF7 on the IFNA4, IFN-β, and RANTES promoters (20). This study showed that the C terminus of IRF7 protein contains a constitutive activation domain (CAD), a signal response domain, a virus-inducible activity domain (VAD), and an inhibition domain (ID), as shown in Fig. 2A. Also, a nuclear export sequence (NES) was identified between amino acids 416 and 467 which overlaps with the ID (20) (Fig. 2A). To check whether these functional domains have similar functions for LMP1p, we transfected IRF7 domain deletion mutants (Fig. 2A) IRF7(Δ247-467) (designated as IRF-ΔID), IRF7(Δ151-278) (IRF7-ΔCAD), IRF7(Δ247-372) (IRF7-ΔVAD), IRF7(Δ151-467) (IRF7-ΔCAD/ID), and IRF7(Δ416-467) (IRF7-ΔNES), together with the LMP1p construct pLMP1 (−512/+72) or the IFNβp construct pGL3/IFNβp-Luc, in 293 cells. Interestingly, these IRF7 functional domains have similar functional effects on two different promoters, IFNβp and LMP1p, in the same cell line, 293 (Fig. 2B). IRF-ΔID has a higher level of activation ability than wild-type IRF7 for both IFNβp and LMP1p, although the level of activation is much higher for IFNβp than for LMP1p. Although deletion of the VAD (IRF7-ΔVAD) impaired transactivation greatly, the VAD is not necessary for retention of IRF7 activity in the absence of virus infection, since IRF7-ΔID (Δ247-467) without the VAD also has very high constitutive activity. In contrast, the CAD is necessary for constitutive IRF7 activity. All the other deletion mutants have lower activation capabilities than wild-type IRF7, and IRF7-ΔNES almost abolished activation of LMP1p (Fig. 2B). The dominant-negative mutant, IRF7-DN, which lacks the DBD, cannot activate LMP1p (Fig. 2B), indicating that direct or indirect binding of IRF7 to LMP1p is necessary for LMP1p activation. Similar results were obtained with BJAB cells (data not shown).
The sequence and phenotype of each of the IRF7A point mutants with serines replaced by phosphomimetic aspartic acids, IRF7(D483/487), IRF7(D475-479), and IRF7(D477/479), are shown in Fig. 2A (20). These substitution mutants show that serines at different sites have different functional capacities in LMP1p induction. IRF7(D483/487) produced much less induction than wild-type IRF7A; however, IRF7(D477/479) has much higher inducing ability. Substitution of two more serines at serine-475 and serine-476 (D475-479) did not result in a higher level of activity. The results suggest that only phosphorylation of serine-477 and serine-479 is necessary for LMP1p activation in the absence of virus infection (Fig. 2B).
Western blot analysis shows that IRF7 and its mutants are all expressed in the transfected cells (Fig. 2C). The bulk of each mutated protein was located in the nucleus, except for IRF7-ΔID and IRF7-ΔNES, both of which were located only in the nucleus with none of either detected in the cytoplasmic extracts (data not shown).
The ISRE of LMP1p responds to IRF7.
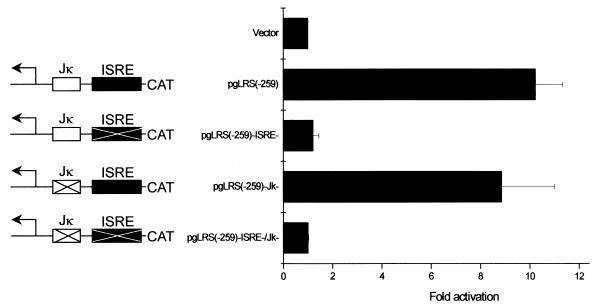
The LMP1 regulatory sequence (LRS) is defined as nucleotides 169477 to 170151 of the B95-8 genome, which corresponds to −634 to +40 relative to the LMP1 transcription start site. Because IRF7 can bind to ISRE-like elements, we tested whether the ISRE element in LMP1p is responsive to IRF7. BJAB cells were transfected with pcDNA3/IRF7A expression plasmid together with pgLRS(−259)-CAT LMP1p constructs and its mutants. As expected, both pgLRS(−259)-CAT (wild type) and the pgLRS(−259)-RBP-Jκ−-CAT mutant have similar activities after IRF7 induction; however, both the pgLRS(−259)-ISRE−-CAT mutant and the pgLRS(−259)-ISRE−/RBP-Jκ−-CAT mutant could not be activated by IRF7 (Fig. 3). From these data, we can conclude that the ISRE, not the RBP-Jκ binding site in LMP1p, is the target for IRF7.
FIG. 3.
The ISRE in LMP1p responds to IRF7. IRF7 and pLRS(−259)-CAT or its mutants as indicated were transfected into BJAB cells. Transfected cells were collected after 48 h, and CAT assays were performed (18). Equal amounts of cell lysates were used. The CAT signals were quantitated by Molecular Dynamics PhosphorImager and normalized with β-Gal activities. Each data point represents the average of eight repeats in two independent experiments. Error bars represent means ± SE.
IRF7 binds to the ISRE of LMP1p.
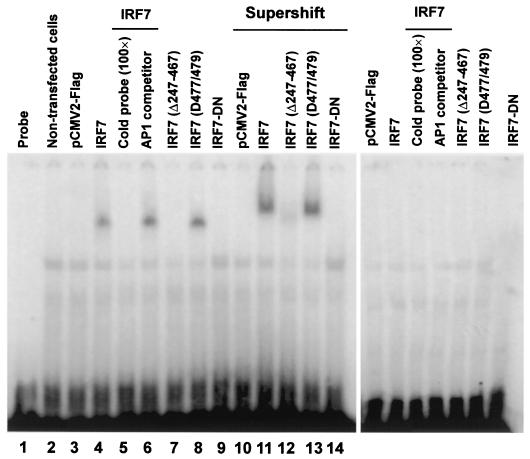
Further, whether IRF7 binds to the ISRE of LMP1p was investigated by EMSA. BJAB and 293 cells were transfected with IRF7 or its mutants, and whole-cell lysates were prepared for EMSA after 48 h. The ISRE probe that contains LMP1p ISRE and its flanking sequences was synthesized and labeled by 32P. As indicated in Fig. 4, the Flag-tagged wild-type IRF7A and IRF7(D477/479) can bind to the ISRE plus flanking sequences of LMP1p (see details in Materials and Methods) specifically. However, binding of IRF7-ΔID (Δ247-467), which can activate both LMP1p and IFNβp in both BJAB and 293 cells, was not detected (Fig. 4, lane 7) but did produce a weak signal in the supershift assay (Fig. 4, lane 12). So it seems that this IRF7 mutant has very weak DNA-binding activity. Consistent with this result, this mutant could activate the IFNA4 promoter but it could not be detected by EMSA as binding to the PRDI-III sequences in this promoter (20). Recently, Yang et al. (50) showed that this IRF7 mutant could activate transcription in insect cells but that it had very weak DNA-binding activity. Actually, some IRF7 protein-DNA complexes are not readily detected by EMSA (54). IRF7-DN, which lacks the DBD, cannot bind to the ISRE in LMP1p (Fig. 4, lane 9). When the ISRE sequence in the probe was mutated (see details in Materials and Methods), no specific band was detected with cell lysates from cells transfected with wild-type IRF7 or any IRF7 mutant (Fig. 4, right panel). Supershift assays with monoclonal Flag antibody confirmed that the protein-DNA complex contains IRF7 (Fig. 4, lanes 10 to 14).
FIG. 4.
IRF7 binds to the ISRE in LMP1p. BJAB cells were transfected with 5 μg of Flag-tagged IRF7A or its mutants as indicated. Cells were collected and lysed in binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 50 mM NaCl, 2 mM DTT, 5% glycerol, 0.5% NP-40, 10 μg of BSA/μl) with protease inhibitors at 48 h posttransfection. The binding reaction mixture contained 20 μg of total proteins, 62.5 μg of poly(dI-dC)/ml, 2 mM DTT, 10 μg of BSA/μl, and 40,000 cpm of 32P-labeled probe. (Left panel) EMSA with the double-stranded sequence containing LMP1p ISRE (GATCCAACAGGAAATGGAAAGGCAGTG). Lanes 10 to 14 show supershift with Flag antibody. (Right panel) EMSA with the mutated LMP1p ISRE (GATCCAACAGGAggTGGAggGGCAGTG [the mutated nucleotides are indicated by lowercase letters]). For competitor assays, 100-fold excess cold probe or AP-1 sequences were added to the binding mixture. For supershift assays, 0.2 μg of Flag antibody was added to each sample before the probe was added. Protein-DNA complexes were separated on 5% 60:1 acrylamide gels.
Exogenously expressed IRF7 induces expression of LMP1 in P3HR1 cells.
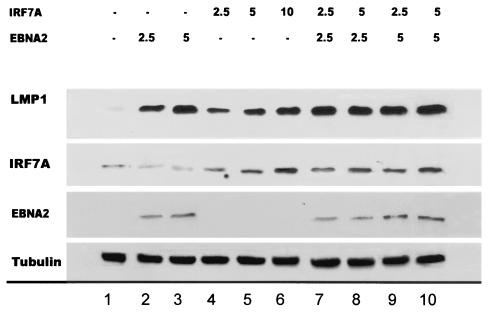
To determine whether the levels of endogenous LMP1 actually change when exogenous IRF7 is expressed, we transfected an EBV-positive cell line exhibiting low levels of LMP1 to determine whether increased expression of LMP1 would occur when the endogenous promoter was activated by IRF7. The P3HR1 genome is deleted from the EBNA2 gene, which transactivates LMP1p, and the cells express very low levels of LMP1. P3HR1 cells were transfected with increasing amounts of IRF7 and cotransfected with EBNA2 and IRF7A. The Western blotting results demonstrated that LMP1 protein levels increased with increasing amounts of transfected IRF7 (Fig. 5). LMP1 could be barely detected in the vector-alone control, although endogenous IRF7 protein was detected in these P3HR1 cells (Fig. 5, lane 1). The endogenous IRF7 may be inactive and therefore does not detectably induce LMP1. Transiently transfected IRF7A or EBNA2 or cotransfection with IRF7 and EBNA2 each resulted in elevated LMP1 protein levels. These results suggest that IRF7A can upregulate LMP1 protein levels in the context of the EBV genomic, native promoter elements.
FIG. 5.
Expression of LMP1 protein is upregulated in P3HR1 cells transiently transfected with IRF7 alone or together with EBNA2. P3HR1 cells contain an EBV genome from which EBNA2 and the C terminus of EBNA-LP have been deleted. Lanes 4 to 6 contain lysates from P3HR1 cells transfected with increasing amounts of IRF7, and lanes 7 to 10 contain lysates from P3HR1 cells transfected with different combinations of IRF7 and EBNA2. LMP1, EBNA2, and IRF7 were probed with anti-LMP1 (CS1-4), anti-EBNA2 (PE2), and anti-IRF7 (H-246) at dilutions of 1:100, 1:500, and 1:300, respectively. Equal amounts of proteins were loaded in each lane, as verified by tubulin blotting.
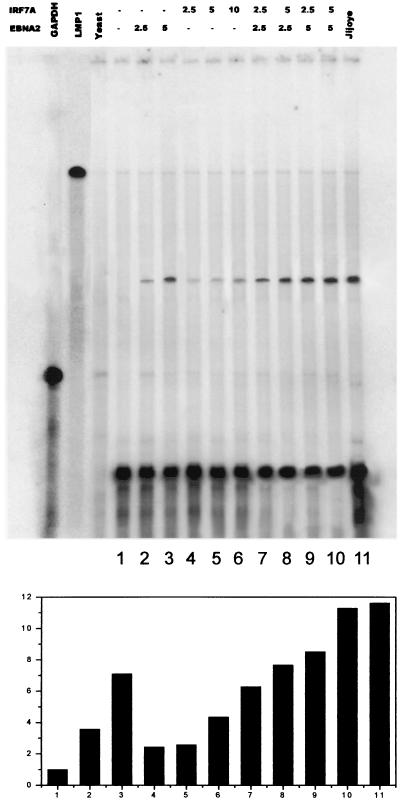
To confirm this interesting result with RPA, we also assayed LMP1 RNA levels in P3HR1 cells after transfection. The RPA results (Fig. 6) show a pattern of increase in LMP1 RNA levels corresponding to that of LMP1 protein (shown in Fig. 5). In the vector-alone control, endogenous LMP1 mRNA could not be detected in P3HR1 cells. With increasing amounts of IRF7 or EBNA2 or the combination of IRF7 and EBNA2, the LMP1 mRNA level is elevated gradually. By titrating in IRF7 with EBNA2, we could essentially reconstitute normal LMP1 levels as expressed in the parental JiJoye cell line (Fig. 6). Thus, IRF7 can activate the endogenous LMP1 promoter independently or together with EBNA2 in cell culture and the results confirm those obtained with the promoter-reporter assays.
FIG. 6.
IRF7 upregulates LMP1 mRNA in P3HR1 cells. P3HR1 cells were transfected with IRF7 or EBNA2 or combinations of different amounts of IRF7 and EBNA2. Transfected cells were selected using anti-CD4 magnetic beads, and total RNAs were isolated. LMP1 and GAPDH probes were labeled with [α-32P]UTP and used for RPA. Saccharomyces cerevisiae RNA was used as the negative control, and RNA from JiJoye cells was used as the positive control. Equal amounts of RNAs were loaded in each lane, as verified by GAPDH signals. A histogram quantifying LMP1 mRNA signals by normalization with GAPDH mRNA signals is shown at the bottom.
DISCUSSION
Our results clearly show that IRF7 can activate the LMP1 promoter by binding to its ISRE and that IRF7 can directly induce LMP1 expression. Furthermore, the study with IRF7 deletion mutants showed that IRF7 functional domains have similar functions for two different promoters, IFNβp and LMP1p, in both 293 and BJAB cells. Our results with IFNβp confirm those of Lin et al. (20). From our results together with those from the study of Lin et al. (20), it can be concluded that IRF7 functional domains are constitutive and independent of its targets and cell lines. It has been reported that NF-κB plays an important role in the activation of IFNβp independently of the IRF family (35), but no NF-κB binding site has been identified in LMP1p. It is unknown whether IRF7 can activate transcription without involvement of another transcription factor. IRF3 has been shown to associate with IRF7 in IFN gene activation (2, 30, 37). In this study, we used a constitutively active form of IRF7, IRF7(Δ247-467), which cannot dimerize with either IRF3 or IRF7. However, this mutant has very high activity on both the promoters, indicating that dimerization is not necessary for IRF7 transactivation ability, even though it has been reported that artificially induced IRF7 dimers can bind DNA and induce target genes independently of phosphorylation or other downstream effects of viral infection (28).
IRF7 is modified by phosphorylation after exposure to various stimuli, including viral infection, double-stranded RNA (14), chemotherapeutic DNA-damaging drugs (17), and expression of specific viral gene products such as LMP1 (53). This phosphorylation is thought to regulate IRF7 activity through elements in the C terminus which dictate nuclear localization, dimer formation, and promoter targeting (2, 20, 28). Two groups reported different results in investigations of the identity of phosphorylated sites needed for IRF7 transactivation activity. Marie et al. showed that murine IRF7 activity is dependent on phosphorylation of serine-425/426, which correspond to human IRF7A serine-471/472 (27). Hiscott and colleagues showed that human IRF7A activity depends on the phosphorylation of serine-477/479. Our results at least support the latter, but we did not test for the effect of serine-471/472.
Several studies have demonstrated that LMP1 is regulated by virus-specific factors, especially EBNA2 and EBNA-LP, mediated by some host cellular factors that tether to the LMP1 promoter (11, 15, 19, 38-40, 40, 45, 48). Actually, EBNA2 is not the only viral activator for LMP1. In KSHV-infected cells as well as in cells coinfected with both KSHV and EBV, LMP1 is activated by the KSHV antigen latency-associated nuclear antigen in the absence of EBNA2 (11). To our knowledge, no report has shown that LMP1 is regulated directly by a host cellular factor. In this study, we demonstrated that LMP1 can be activated by cellular IRF7 without the involvement of any viral factor. This interesting finding may account for a puzzling phenomenon in EBV biology: LMP1 is expressed in type II latency but EBNA2 is absent in type II cells (6, 16, 36). Type II latency is exemplified by nasopharyngeal carcinoma (NPC), and in fact, Zhang and Pagano have detected high levels of expression of IRF7 in most NPC tissues tested as well as in LMP1-positive NPC lines passaged in nude mice (L. Zhang and J. S. Pagano, unpublished results). So the LMP1 levels detected in type II cells may be the consequence of IRF7 expressed in type II cells. Therefore, like EBNA2, which is the key viral antigen for maintaining type III latency, IRF7 may play an equally important role in maintaining type II latency through regulation of LMP1 expression.
IRF7 functions as a multifunctional protein. First, IRF7 was cloned on the basis of its ability to bind to and repress EBNA-1 Qp in EBV type III latency (52). Second, IRF7, like other IRFs, plays a primary role in regulating the expression of type I IFN genes in virus-infected cells (3, 41). Zhang and Pagano showed that IRF7 was involved in the activation of Tap-2 by LMP1 in B lymphoma cells (54); Tap-2 mutations are involved in diseases associated with the immune system. Moreover, IRF7 has properties consistent with a putative oncogene (Zhang et al., unpublished). Here, we found that IRF7 can regulate expression of a viral gene in addition to host cellular genes such as IFNA genes.
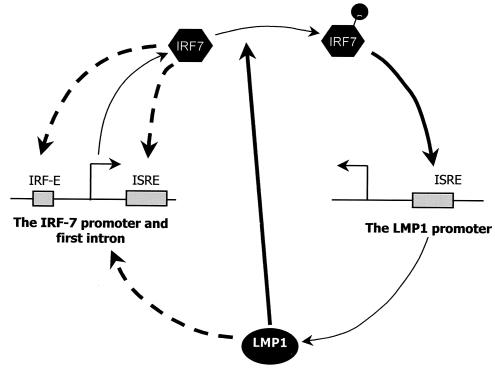
Expression of IRF7 mRNA and protein is clearly inducible by LMP1 in EBV-infected cells (53). Since LMP1 can activate NF-κB and since there is an NF-κB binding site in the IRF7 promoter, it is likely that IRF7 induction by LMP1 is mediated by NF-κB, at least in part. In fact, Zhang et al. have found that a suppression of IκB can block the induction of IRF7 by LMP1. Furthermore, overexpression of NF-κB can induce IRF7 expression in DG75, the EBV-negative BL cell line (55). However, the results of a promoter-reporter assay indicated that LMP1 could not activate IRF7 promoter constructs in HeLa cells, suggesting that IRF7 induction by LMP1 is cell type specific and that NF-κB activation needs cooperation with other transcription factors (23). In addition to induction of IRF7 expression, LMP1 also regulates IRF7 protein activity: LMP1 augments the phosphorylation status and facilitates the nuclear localization of IRF7 (54). Based on our results, a regulatory circuit between LMP1 and IRF7 is outlined (Fig. 7): IRF7 activates the LMP1 promoter by binding to its ISRE, which results in LMP1 protein expression. In turn, the LMP1 expression can induce IRF7 expression and regulate its protein activity. In addition, IRF7 may be autoregulated through binding of IRF7 protein to the ISRE in the first intron or the IRF-E in the IRF7 promoter since IRF7 is capable of binding to these elements; preliminary results indicate that IRF7 can induce the IRF7 promoter in transient transfection assays (S. Ning and J. S. Pagano, unpublished results).
FIG. 7.
A scheme for the proposed regulatory circuit between IRF7 and LMP1. IRF7 protein can activate the LMP1 promoter, as demonstrated in this study. The activated LMP1 promoter results in elevated LMP1 protein levels, which in turn induce IRF7 mRNA and protein and regulate IRF7 protein activity through phosphorylation as indicated (53). Thick, solid arrows indicate identified regulatory pathways, and broken arrows indicate pathways which are proposed but still under study.
P3HR1 cells are defective for EBNA2 expression (53); therefore, little if any LMP1 is detected. The absence of LMP1 leads to low levels of IRF7. We show here that IRF7 can induce a low level of LMP1 in these cells. However, since EBNA2 is totally absent and since the IRF7 and EBNA2 inducing effects are independent and EBNA2 is the stronger inducer and normally necessary for LMP1 induction, the level of LMP1 remains subnormal in P3HR1 cells.
Our results suggest that IRF7 potentiates the oncogenic effect of LMP1. Since LMP1 can induce IRF7, the net effect should be augmented expression of this EBV oncoprotein. In other work, Zhang et al. have found that IRF7 by itself has oncogenic properties in 3T3 cells, as determined by focus formation and colony formation as well as production of tumors, and that the combination of LMP1 and IRF7 produces at least additive effects (Zhang et al., unpublished). Whether IRF7 is essential for effects of LMP1 including oncogenesis remains to be determined.
Acknowledgments
We thank R. Lin and J. Hiscott (Lady Davis Institute for Medical Research, McGill University, Montreal, Quebec, Canada) for pCMV2/IRF7A-Flag plasmid and a series of its mutants and the pGL3/IFNβp-Luc plasmid; A. Sjoblom and L. Rymo (Goteborg University, Goteborg, Sweden) for pgLRS(−259)-CAT plasmid and its mutants; J. Lin and E. Kieff (Brigham and Women's Hospital and Harvard Medical School, Boston, Mass.) for pGL2(−512/+72)LMP1p-Luc plasmid; L. C. Spender and P. J. Farrell (Ludwig Institute for Cancer Research, London, United Kingdom) for pBS-LMP1 plasmid, and T. Maniatis (Harvard University, Boston, Mass.) for IRF7-DN construct. We also thank Luwen Zhang (University of Nebraska, Lincoln) for valuable input and discussion.
This work was supported by grants from the National Institute of Allergy and Infectious Disease (AI 42372-01) and from the National Cancer Institute (CA 19014). L. E. Huye is supported by an NCI training grant (T32-CA09156-27).
REFERENCES
- 1.Au, W.-C., P. A. Moore, D. W. LaFleur, B. Tombal, and P. M. Pitha. 1998. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J. Biol. Chem. 273:29210-29217. [DOI] [PubMed] [Google Scholar]
- 2.Au, W.-C., W.-S. Yeow, and P. M. Pitha. 2001. Analysis of functional domains of interferon regulatory factor 7 and its association with IRF-3. Virology 280:273-282. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, B. J., A. E. Field, and P. M. Pitha. 2003. Virus-induced heterodimer formation between IRF-5 and IRF-7 modulates assembly of the IFNA enhanceosome in vivo and transcriptional activity of IFNA genes. J. Biol. Chem. 278:16630-16641. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, B. J., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 5.Busch, L. K., and G. A. Bishop. 2001. Multiple carboxyl-terminal regions of the EBV oncoprotein, latent membrane protein 1, cooperatively regulate signaling to B lymphocytes via TNF receptor-associated factor (TRAF)-dependent and TRAF-independent mechanisms. J. Immunol. 167:5805-5813. [DOI] [PubMed] [Google Scholar]
- 6.Contreras-Brodin, B. A., M. Anvret, S. Imreh, E. Altiok, G. Klein, and M. G. Masucci. 1991. B cell phenotype-dependent expression of the Epstein-Barr virus nuclear antigens EBNA-2 to EBNA-6: studies with somatic cell hybrids. J. Gen. Virol. 72:3025-3033. [DOI] [PubMed] [Google Scholar]
- 7.Davis, M., S. Kenney, J. Kamine, J. S. Pagano, and E. S. Huang. 1987. Immediate-early gene region of human cytomegalovirus transactivates the promoter of human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 84:8642-8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devergne, O., E. C. McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliopoulos, A. G., E. R. Waites, S. M. S. Blake, C. Davies, P. Murray, and L. S. Young. 2003. TRAF1 is a critical regulator of JNK signaling by the TRAF-binding domain of the Epstein-Barr virus-encoded latent infection membrane protein 1 but not CD40. J. Virol. 77:1316-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 16:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 75:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, S., M. Rowe, C. Gregory, D. Croom-Carter, F. Wang, R. Longnecker, E. Kieff, and A. Rickinson. 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65:1107-1115. [DOI] [PubMed] [Google Scholar]
- 14.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 15.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff, E., and A. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2574. In D. M. Knipe and P. M. Howley (ed.), Virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 17.Kim, T. K., T. Kim, T. Y. Kim, W. G. Lee, and J. Yim. 2000. Chemotherapeutic DNA-damaging drugs activate interferon regulatory factor-7 by the mitogen-activated protein kinase kinase-4-c-Jun NH2-terminal kinase pathway. Cancer Res. 60:1153-1156. [PubMed] [Google Scholar]
- 18.Kingston, R. E. 1988. Introduction of DNA into mammalian cells. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 19.Lin, J., E. Johannsen, E. Robertson, and E. Kieff. 2002. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J. Virol. 76:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275:34320-34327. [DOI] [PubMed] [Google Scholar]
- 21.Ling, P. D., J. J. Hsieh, I. K. Ruf, D. R. Rawlins, and S. D. Hayward. 1994. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J. Virol. 68:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, J., H. H. Chua, S. Y. Chen, J. Y. Chen, and C. H. Tsai. 2003. Regulation of matrix metalloproteinase-1 by Epstein-Barr virus proteins. Cancer Res. 63:256-262. [PubMed] [Google Scholar]
- 23.Lu, R., W.-C. Au, W.-S. Yeow, N. Hageman, and P. M. Pitha. 2000. Regulation of the promoter activity of interferon regulatory factor-7 gene. Activation by interferon and silencing by hypermethylation. J. Biol. Chem. 275:31805-31812. [DOI] [PubMed] [Google Scholar]
- 24.Lu, R., P. A. Moore, and P. M. Pitha. 2002. Stimulation of IRF7 gene expression by TNF alpha: requirement for NF-kB transcription factor and gene accessibility. J. Biol. Chem. 277:16592-16598. [DOI] [PubMed] [Google Scholar]
- 25.Lu, R., and P. M. Pitha. 2001. Monocyte differentiation to macrophage requires interferon regulatory factor 7. J. Biol. Chem. 276:45491-45496. [DOI] [PubMed] [Google Scholar]
- 26.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. J. Servant, C. LePage, C. Deluca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 27.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marie, I., E. Smith, A. Prakash, and D. E. Levy. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20:8803-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCann, E. M., G. L. Kelly, A. B. Rickinson, and A. I. Bell. 2001. Genetic analysis of the Epstein-Barr virus-coded leader protein EBNA-LP as a co-activator of EBNA2 function. J. Gen. Virol. 82:3067-3079. [DOI] [PubMed] [Google Scholar]
- 30.Morin, P., J. Braganca, M. T. Bandu, R. Lin, J. Hiscott, J. Doly, and A. Civas. 2002. Preferential binding sites for interferon regulatory factors 3 and 7 involved in interferon-α gene transcription. J. Mol. Biol. 316:1009-1022. [DOI] [PubMed] [Google Scholar]
- 31.Murono, S., H. Inoue, T. Tanabe, I. Joab, T. Yoshizaki, M. Furukawa, and J. S. Pagano. 2001. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc. Natl. Acad. Sci. USA 98:6905-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nehyba, J., R. Hrdlicková, J. Burnside, and H. R. Bose, Jr. 2002. A novel interferon regulatory factor (IRF), IRF-10, has a unique role in immune defense and is induced by the v-Rel oncoprotein. Mol. Cell. Biol. 22:3942-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, H., J. Hiscott, and P. M. Pitha. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293-312. [DOI] [PubMed] [Google Scholar]
- 34.Peng, R., A. V. Gordadze, E. M. F. Pananá, F. Wang, J. Zong, G. S. Hayward, J. Tan, and P. D. Ling. 2000. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J. Virol. 74:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitha, P. M., W. C. Au, W. Lowther, Y. T. Juang, S. L. Schafer, L. Burysek, J. Hiscott, and P. A. Moore. 1998. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie 80:651-658. [DOI] [PubMed] [Google Scholar]
- 36.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 37.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunology 13:539-548. [DOI] [PubMed] [Google Scholar]
- 38.Sjoblom, A., A. Jansson, W. Yang, S. Lain, T. Nilsson, and L. Rymo. 1995. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J. Gen. Virol. 76:2679-2692. [DOI] [PubMed] [Google Scholar]
- 39.Sjoblom, A., A. Nerstedt, A. Jansson, and L. Rymo. 1995. Domains of the Epstein-Barr virus nuclear antigen 2 (EBNA2) involved in the transactivation of the latent membrane protein 1 and the EBNA Cp promoters. J. Gen. Virol. 76:2669-2678. [DOI] [PubMed] [Google Scholar]
- 40.Sjoblom, A., W. Yang, L. Palmqvist, A. Jansson, and L. Rymo. 1998. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J. Virol. 72:1365-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Iκ B kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 42.Spender, L. C., G. H. Cornish, B. Rowland, B. Kempkes, and P. J. Farrell. 2001. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J. Virol. 75:3537-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung, N. S., S. Kenney, D. Gutsch, and J. S. Pagano. 1991. EBNA2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J. Virol. 65:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniguchi, T., and A. Takaoka. 2002. The interferon-α/β system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 45.Voss, M. D., A. Hille, S. Barth, A. Spurk, F. Hennrich, D. Holzer, N. Mueller-Lantzsch, E. Kremmer, and F. A. Grässer. 2001. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter. J. Virol. 75:11781-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakisaka, N., S. Murono, T. Yoshizaki, M. Furukawa, and J. S. Pagano. 2002. Epstein-Barr virus latent membrane protein 1 induces and causes release of fibroblast growth factor-2. Cancer Res. 62:6337-6344. [PubMed] [Google Scholar]
- 47.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, F., S.-F. Tsang, M. G. Kurilla, J. I. Cohen, and E. Kieff. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, H., C. H. Lin, G. Ma, M. O. Baffi, and M. G. Wathelet. 2003. Interferon regulatory factor (IRF)-7 synergizes with other transcription factors through multiple interactions with p300/CBP coactivators. J. Biol. Chem. 278:15495-15504. [DOI] [PubMed] [Google Scholar]
- 51.Yoshizaki, T., H. Sato, M. Furukawa, and J. S. Pagano. 1998. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad. Sci. USA 95:3621-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, L., and J. S. Pagano. 2000. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J. Virol. 74:1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., and J. S. Pagano. 2001. Interferon regulatory factor 7 mediates activation of Tap-2 by Epstein-Barr virus latent membrane protein 1. J. Virol. 75:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, L., L. Wu, K. Hong, and J. S. Pagano. 2001. Intracellular signaling molecules activated by Epstein-Barr virus for induction of interferon regulatory factor 7. J. Virol. 75:12393-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]