Abstract
A family of endogenous retroviruses (enJSRV) closely related to Jaagsiekte sheep retrovirus (JSRV) is ubiquitous in domestic and wild sheep and goats. Southern blot hybridization studies indicate that there is little active replication or movement of the enJSRV proviruses in these species. Two approaches were used to investigate the distribution of proviral loci in the sheep genome. Fluorescence in situ hybridization (FISH) to metaphase chromosome spreads using viral DNA probes was used to detect loci on chromosomes. Hybridization signals were reproducibly detected on seven sheep chromosomes and eight goat chromosomes in seven cell lines. In addition, a panel of 30 sheep-hamster hybrid cell lines, each of which carries one or more sheep chromosomes and which collectively contain the whole sheep genome, was examined for enJSRV sequences. DNA from each of the lines was used as a template for PCR with JSRV gag-specific primers. A PCR product was amplified from 27 of the hybrid lines, indicating that JSRV gag sequences are found on at least 15 of the 28 sheep chromosomes, including those identified by FISH. Thus, enJSRV proviruses are essentially randomly distributed among the chromosomes of sheep and goats. FISH and/or Southern blot hybridization on DNA from several of the sheep-hamster hybrid cell lines suggests that loci containing multiple copies of enJSRV are present on chromosomes 6 and 9. The origin and functional significance of these arrays is not known.
Jaagsiekte sheep retrovirus (JSRV) is an exogenous betaretrovirus, responsible for ovine pulmonary adenocarcinoma in sheep. The sheep genome contains a family of endogenous retroviral sequences (enJSRV) closely related to JSRV (12). Interestingly, the integration pattern, as assayed by Southern blot hybridization, does not vary much among a wide range of domestic sheep breeds and even among wild sheep, including bighorn sheep and mouflon sheep, the likely ancestor of domestic breeds. Thus, it appears that these viruses became fixed in the genome early in the development of the genus Ovis and have not shown much recent movement. Closely related endogenous viruses are also found in the genus Capra, which includes wild and domestic goats. Indeed related viruses were found in several other genera of the subfamily Caprinae of the family Bovidae (12).
Much of the interest in this family of viruses has been directed at developing ways to distinguish exogenous JSRV from enJSRV, and to this end restriction maps and partial sequences were derived for several of the enJSRV loci (2, 3, 15). This led to the identification of diagnostic restriction endonuclease cleavage sites and hybridization probes that allowed the isolation of two infectious clones of JSRV (17, 19). Recently, three complete enJSRV proviral loci have been sequenced (18). These were defective in producing virus particles and differed from infectious JSRV by numerous point mutations and deletions. Sequence differences between the long terminal repeats (LTRs) of JSRV and enJSRV seem to be particularly important in determining the differences in expression in different tissues. While JSRV LTRs appear to be most active in lung cells, the enJSRV LTRs seem to be most active in the uterus (16, 22). Further identification and characterization of the enJSRV loci will be necessary in determining their functional significance.
We have sought insight into the distribution of endogenous proviral loci in sheep chromosomes by using two approaches to mapping. In the first approach, fluorescence in situ hybridization (FISH) was used to localize the endogenous viral loci to sheep and goat chromosomes. The second approach made use of a recently developed panel of sheep-hamster hybrid cell lines. Each of the cell lines in this panel contains one or a few sheep chromosomes, and collectively the panel contains the entire sheep genome. This panel allows one to study individual sheep chromosomes in the absence of the rest of the genome, and it was used to detect enJSRV loci by PCR and Southern blotting. Using these approaches we report that the distribution of enJSRV proviruses on the genome is unusual because two chromosomes carry loci with several copies of the provirus.
MATERIALS AND METHODS
Ovine and caprine cell cultures.
Metaphase spreads were prepared from caprine and ovine cell lines: CAT2 (caprine testis-derived fibroblasts with a 6/15 translocation [11]), SGH2 (skin fibroblasts derived from a fertile ewe-goat hybrid [7]), MFP2 (fetal lung-derived fibroblasts) obtained from H. Hayes, TIGEF (T-immortalized goat embryonic fibroblast) developed in our laboratory (8), and IDO5 (sheep dermal fibroblasts), generously given by Chappuis, Institut Mérieux, Lyon, France. H921 fibroblasts were obtained from explants of cardiac muscle of a sheep with lung tumor and maintained in culture for six passages. OF686 fibroblasts emerged from epithelial culture of a nasal tumor from sheep. CAT2, SGH2, and MPF2 were cultured in complete minimal essential medium (MEM) (MEM [GIBCO BRL] supplemented with 2 mM l-glutamine [GIBCO BRL], 1% antibiotic-antimycotic solution with 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 250 ng of amphotericin B [GIBCO BRL[/ml, and 10% decomplemented fetal calf serum [GIBCO BRL]) at 37°C with 5% CO2. TIGEF, IDO5, OF686, and H921 were cultured in complete Dulbecco's modified Eagle medium (DMEM) medium (DMEM medium [GIBCO BRL], 2 mM l-glutamine, 10% decomplemented fetal calf serum, 10 U of penicillin/ml and 10 μg of streptomycine/ml).
Chromosomal localization of enJSRV. (i) Preparation of metaphase spreads.
Metaphase spreads were obtained by a double thymidine synchronization as previously described (9). Briefly, on day 1, cells were seeded in complete medium and incubated at 37°C, with 5% CO2. At 30 to 40% confluence, cells were incubated in complete medium supplemented with 0.8 mg of thymidine (Sigma)/ml for 14 to 15 h. On day 2, cells were extensively washed three times for 20 min with MEM and incubated for 8 h with complete medium. A second thymidine synchronization was performed using the same conditions. On day 3, cells were washed with MEM medium and incubated in complete medium supplemented with 10 μg of bromodeoxy-uridine (Sigma)/ml to induce R-banding. After an hour of incubation, 0.5 μg of fluorodeoxyuridine (Sigma)/ml was added to the culture. Mitotic cells appeared as round cells at the surface of the culture 6 to 8 h after bromodeoxy-uridine treatment and were mechanically detached from the plastic. Supernatants were harvested and mitotic cells were collected by a 10-min centrifugation at 430 × g. The pellets were washed once in phosphate-buffered saline (PBS) without calcium or magnesium. Approximately 1.5 × 106 cells were incubated for 13 min at 37°C in 10 ml of a 0.22 μm-pore-size-filtered hypotonic solution containing 16% decomplemented fetal calf serum and 2 μg of EDTA/ml in pure sterile water. The mitotic nuclei were prefixed for 5 min at room temperature by adding 1 ml of 3:1 methanol-acetic acid solution and were centrifuged for 10 min at 430 × g. The pellets were washed three times after a 10-min incubation at room temperature in 10 ml of 3:1 methanol-acetic acid solution and then kept overnight at 4°C. Finally, nuclei were resuspended in 200 to 500 μl of 3:1 methanol-acetic acid solution, spread on cold slides, and stored at −20°C. Culture flasks were vigorously shaken, and the cells not attached to the plastic were recovered by centrifugation.
(ii) FISH.
The JSRV plasmid (23), containing the full-length genome of the South-African strain of JSRV, generously provided by Gilles Quérat (Marseille, France), was used to generate the probe. The genomic biotinylated 7.5-kb JSRV probe was obtained by random priming as previously described (10); briefly, 200 ng of denatured JSRV DNA was incubated for 2 h at 37°C in 50 μl of OLBF buffer (50 mM tris-HCl [pH 8], 5 mM MgCl2, 200 mM HEPES [pH 6.6], 5.4 U of Pd(N6) hexanucleotides (Boehringer, Meylan, France)/ml, 10 mM 2-B-mercaptoethenol), containing 40 μM (each) dATP, dCTP, and dGTP (Pharmacia), 20 μM biotin 11-dUTP (Sigma), 652 μg of bovine serum albumin (Sigma)/ml, and 2.5 U of Klenow fragment (Eurogentech). After a 2-h incubation at 37°C, the reaction was stopped by addition of 42 mM EDTA. The JSRV probe was coprecipitated with 4 μg of sonicated salmon sperm DNA and 20 μg of sonicated sheep competitor DNA, dissolved in 20 μl of hybridization buffer (50% formamide, 10% dextran sulfate in SSCP buffer [pH 7] [40 mM NaH2PO4-Na2HPO4, 240 mM NaCl, 30 mM Na3C6H5O7 · 2H2O]), and heat denatured. Hybridization was performed as previously described (1) with minor modifications. Slides were washed an hour at room temperature in 2× SSC (300 mM NaCl, 30 mM Na3C6H5O7 [pH 7]) and treated with 100 μg of RNase A (Sigma)/ml in 2× SSC (Sigma) for 1 h at 37°C. After a 15-min wash in 2× SSC at room temperature, the slides were dehydrated in an ethanol series (70, 80, and 90%). The metaphase spreads were denatured by a 2-min incubation at 72°C in 70% formamide in 2× SSC and dehydrated in an ethanol series at −20°C. After a 2-h incubation at 37°C in hybridization buffer (50% formamide, 10% dextran sulfate in SSCP buffer), the denatured JSRV probe was added to the slides and they were covered with “steam-cooking” paper. Slides were hybridized overnight at 37°C in a moist chamber and then successively washed at room temperature as follows: 15 min in 2× SSC; three times for 3 min in 50% formamide-2× SSC; four times for 2 min in 2× SSC; and four times for 2 min in 1× BN buffer (0.5 M NaHCO3, 25 ml of IGEPAL (Sigma)/liter, and 0.1 mg of sodium azide [pH 8]/ml). Slides were then incubated for 10 min at room temperature in 1× BN supplemented with 5% nonfat dry milk, followed by 30 min at 37°C in a dark, moist chamber with 50 μl of a 1/400 dilution of avidin-FITC (Vector) in 1× BN supplemented with 5% nonfat dry milk, followed by three washes for 2 min in 1× BN at 42°C. After a 10-min incubation, signal amplification was performed with 50 μl per slide of a 1/20 dilution of goat serum (Vector) in 1× BN. Each slide was incubated for 30 min in the dark at 37°C with 50 μl of a 1/20 dilution of biotinylated antiavidin antibodies (Vector) in 1× BN and washed three times for 2 min at 42°C in 1× BN. After 10 min at room temperature in 1× BN supplemented with 5% nonfat dry milk, a second treatment with avidin-FITC was done under the same conditions. Slides were washed in PBS and stained in the dark with 60 μl of 0.5% propidium iodide in PBS for 10 min. R-banding was revealed by addition of 10 μl of antifade mixture (90% glycerol, 0.1% p-phenylene diamine dihydrochloride σ [pH 11] in PBS) (14). The slides were examined with a Zeiss MCD80DX microscope coupled with a charge-coupled device camera, and metaphases were analyzed with the CytoVision System (Applied Imaging). Chromosomes were identified according to the new International System for Chromosome Nomenclature of Domestic Bovids (6).
Sheep-hamster somatic cell hybrid lines.
A panel of 30 sheep-hamster cell lines was obtained from the Eleanor Roosevelt Institute for Cancer Research, Denver, Colo. A full description of the construction and characterization of these lines is found in the work of Burkin et al. (4). Chinese hamster ovary cells and the hybrid cell lines were grown at 37°C in Ham's F-12 media supplemented with 5% fetal bovine serum to generate enough cells for cryopreservation and DNA isolation.
DNA isolation.
Each cell line was grown to confluency in a roller bottle, trypsinized, and then incubated at 50°C for 3 h in 10 ml of extraction buffer containing 10 mM Tris-Cl (pH 8.0), 0.1 mM EDTA (pH 8.0), 20 μg of pancreatic RNase/ml, 0.5% sodium dodecyl sulfate (SDS), and 100 μg of proteinase K/ml. DNA from lysed cells was extracted twice in equilibrated phenol (pH 8.0), precipitated by addition of 0.7 vol of isopropanol, spooled with a glass rod, and resuspended in Tris-EDTA. DNA was also isolated from a normal sheep lung (98AO1) for use as a control.
gag PCR.
Each 50-μl reaction consisted of 35 ng of genomic DNA, standard 1× PCR buffer, 200 μM deoxynucleoside triphosphates, 20 pmol (each) of gag primer (15) (sense [P1], 5′-GCTGCTTTRAGACCTTATCGAAA-3′, and antisense [P2], 5′-ATACTGCAGCYCGATGGCCAG-3′) and 1.5 U of Taq polymerase. The thermocycler was programmed for an initial cycle of 94°C for 3 min and then 30 cycles of 94°C for 30 s, 58°C for 30s, and 72°C for 30 s, with a final extension of 72°C for 5 min. PCR products were analyzed by electrophoresis in 1.5% agarose in 1× Tris-borate EDTA.
Southern blot hybridization.
Ten micrograms of each genomic DNA was cleaved with EcoRI, XbaI, or BamHI, and fragments were separated on a 0.7% agarose gel and transferred by a PosiBlot 30-30 pressure blotter (Stratagene) to a nylon membrane (MSI, Westborough, Mass.) as described previously (21). The DNA probe was generated by PCR with P1 and P2 primers for JSRV capsid from gel-purified fragments with the addition of [α32-P]dCTP to the reaction. Each membrane was hybridized with 32P-labeled capsid probe with a specific activity of 106 cpm of probe/ml of UltraHyb (Ambion) at 42°C. Following hybridization, the membrane was washed at 42°C twice for 5 min (2× SSC, 0.1% SDS) and then twice for 15 min (0.1% SSC, 0.1% SDS) followed by exposure to a PhosphorImager screen for either 5 h or 2 weeks by autoradiography.
RESULTS
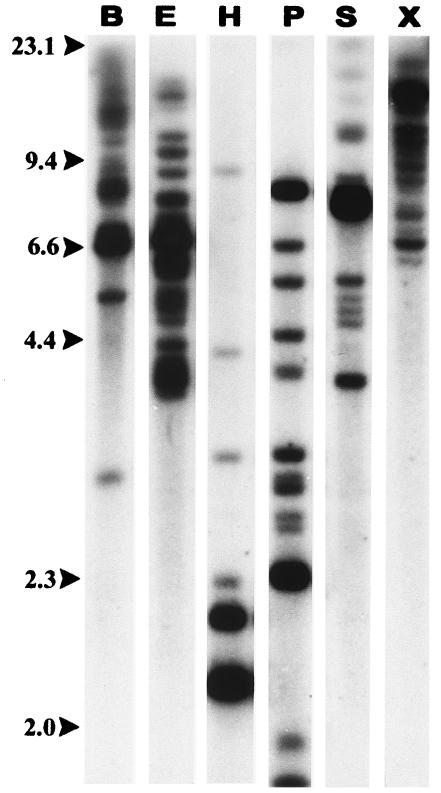
Southern blot hybridization on sheep genomic DNA cut with any of several restriction enzymes generally shows multiple bands of widely varying intensity when probed with capsid or envelope probes (Fig. 1) The dark bands presumably represent dropout fragments conserved in several endogenous proviruses. Southern blots such as these with a wide variety of restriction enzymes led to the conclusion that there are 15 to 20 enJSRV proviruses in the sheep and goat genomes (12), but they do not provide any information on their distribution in the sheep or goat genome.
FIG. 1.
enJSRV sequences in the sheep genome. Sheep genomic DNA was digested with BamHI (B), EcoRI (E), HindIII (H), PstI (P), SacI (S), or XbaI (X) and subjected to agarose gel electrophoresis and Southern blot hybridization using a JSRV capsid region probe.
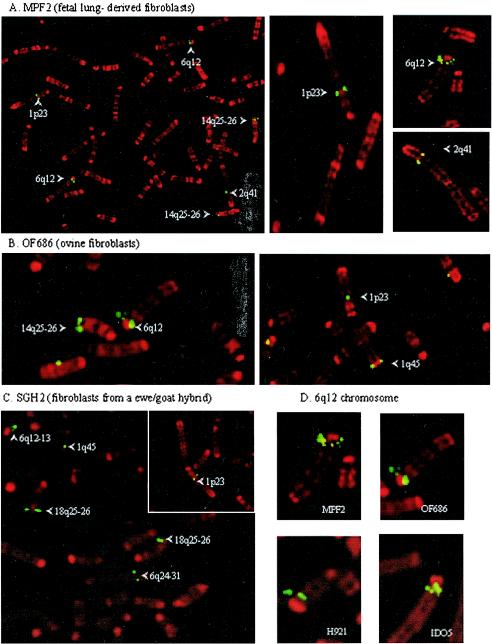
One way of examining the distribution of proviruses in the genome is by FISH. The enJSRV chromosomal distribution in goat (2n = 60) and sheep (2n = 54) chromosomes was analyzed by FISH on four ovine cell lines, two caprine cell lines, and one goat-sheep hybrid cell line. Chromosome identification was carried out using the R-banding karyotype. The positions were confirmed on multiple metaphase spreads for each cell line. For a given cell line, only the sites reproducibly found are reported. Using a biotinylated full-length JSRV genome as a probe, enJSRV copies were consistently localized on the two pairs of metacentric chromosomes 1 and 2 and on the acrocentric chromosomes 6, 8, 10, and 14 in sheep and on the acrocentric chromosomes 1, 2, 3, 6, 7, and 18 in goats (Fig. 2A and B; Table 1) Except for positions 1q45 and 2q41, enJSRV copies were integrated in different sites in goat and sheep genomes. All the species-specific copies were confirmed by the use of a goat-sheep hybrid cell line derived from a fertile goat-sheep hybrid (Fig. 2C; Table 1).
FIG. 2.
FISH analysis with a full-length JSRV probe on ovine and caprine metaphase spreads. Chromosomes were identified by R-banding, and integration sites are indicated by arrows. (A) Ovine MPF2 cells (fetal lung-derived fibroblasts). (B) Ovine OF686 cells. (C) Sheep-goat hybrid SGH2 cells. (D) Multicopy locus on position q12 of ovine chromosome 6 from four different lines.
TABLE 1.
enJSRV distribution in ovine and caprine chromosomes determined by fluorescent in situ hybridizationa
| Chromosome | Position of enJSRV sequence
|
||||||
|---|---|---|---|---|---|---|---|
| Ovine cells
|
Sheep- goat cells (SGH2) | Caprine cells
|
|||||
| MPF2 | IDO5 | 686 | H921 | CAT2 | TIGEF | ||
| 1 | q45 | q45 | q45 | q45 | q45 | q45 | q45 |
| p23 | p23 | p23 | p23 | p23 | |||
| 2 | q41 | q41 | q41 | q41 | q41 | q41 | q41 |
| q12.2 | |||||||
| 3 | q23 | q23 | q23 | ||||
| 4 | |||||||
| 5 | q15 | ||||||
| 6 | q12-13* | q12-13* | q12-13* | q12-13* | q12-13 | ||
| q24-31 | q24-31 | q24-31 | |||||
| 7 | q13 | q13 | |||||
| 8 | q22 | q22 | q22 | q22 | |||
| 9 | |||||||
| 10 | q22 | q22 | q22 | q22 | |||
| 11 | |||||||
| 12 | q15 | ||||||
| 13 | |||||||
| 14 | q25-26 | q25-26 | q25-26 | q25-26 | q25-26 | ||
| 15 | |||||||
| 16 | |||||||
| 17 | |||||||
| 18 | q25-26 | q25-26 | q25-26 | ||||
| 19 | |||||||
| 20 | |||||||
| 21 | |||||||
| 22 | |||||||
| 23 | |||||||
| 24 | |||||||
| 25 | |||||||
| 26 | |||||||
| X | q34 | ||||||
| Y | |||||||
Chromosome identification was carried out using R-banding karyotype. The positions were confirmed on multiple metaphase spreads for each cell line. For a given cell line, only the sites reproducibly found are reported. Asterisk indicates multi-copy enJSRV loci.
In the ovine cell lines, the signal was clearly stronger in position q12 of chromosome 6, suggesting a multicopy enJSRV locus. This was confirmed in ovine cell lines MPF2, OF686, H921, and IDO5 (Fig. 2D). Depending on the quality of the metaphase spreads and of the hybridization, three to four individual hybridization sites were observed. enJSRV was absent from the equivalent position in caprine chromosome 2. Additional sites were present in position 5q15 in the OF686 cell line and position 2q12.2 in the H921 cell line (Table 1). Despite the fact that these two cell lines were derived from nasal tumor and heart of a sheep with pulmonary adenocarcinoma, no exogenous betaretrovirus was detected by PCR and nested PCR (data not shown), suggesting that the additional copies were of endogenous origin.
Another way of examining the distribution of proviruses in the sheep genome is to make use of the panel of sheep-hamster cell lines constructed by fusing sheep cells with Chinese hamster ovary cells (4). Total genomic DNA was extracted from each of the cell lines and used as a template for PCR with primers from the gag region of JSRV. As indicated in Table 2 products of the expected size (229 bp) were amplified from all of the lines except R928-6 A3 (chromosomes 16 and 21), R891-16R4B (chromosome 16), and R891-16R5A (chromosome 17). Since many of the lines contain more than one sheep chromosome, it is not possible to determine which of the chromosomes in these lines carry proviruses. However, from these data it can be concluded that at least chromosomes 1, 2, 3, 4, 6, 7, 8, 9, 10, 13, 14, 25, 26, and X carry gag sequences of proviral loci.
TABLE 2.
Detection of enJSRV loci in sheep-hamster hybrid cell lines by PCRa
| Sheep-hamster hybrid cell line | Chromosome(s) | PCR result |
|---|---|---|
| QT131-2 | 1, 6 | + |
| QT131-13F | 1q, 9, 10, 22 | + |
| QT131-30 | 1q | + |
| R711-16P2A | 2 | + |
| R693-37A7R4 | 2p, 26 | + |
| Q576-15BX | 3q | + |
| R612-3H9 | 3, 7 | + |
| R928-19D1 | 4, 16 | + |
| R904-17J14R | 5, 9, 19 | + |
| R693-37AR12A | 16 | + |
| J104-30B (5)5 | 6, 24 | + |
| R612-3H39 | 7 | + |
| R928-19F | 8, 16 | + |
| R904-17D5 | 9 | + |
| RN96-6B17X6 | 10 | + |
| R914-10 | 4, 6, 10, 11, 16, 17, Y | + |
| R894-25C | 12, 13, 15 | + |
| R891-29R8A | 13, 19 | + |
| R891-27R1B | 13 | + |
| R891-5A1 | 14 | + |
| R928-6 A3 | 16, 21 | − |
| R891-16R4B | 16 | − |
| R891-16R5A | 17 | − |
| R891-29 | 3, 7, 13, 14, 18 | + |
| R894-33CR17 | 19, 23, 26 | + |
| R894-28CR20E | 10, 20 | + |
| R894-3B1 | 21, 24, X | + |
| RN596-6B17E | 25 | + |
| R612-3B12D | 26 | + |
| R612-3B10 | X | + |
The sheep-hamster cell lines developed by Burkin et al. and the sheep chromosomes contained in each of them are shown in the first two columns (4). The third column indicates whether a 229-bp gag PCR product was amplified from the DNA of the hybrid.
Three endogenous proviruses have been cloned and sequenced (18). These have been designated enJS56A1, enJS59A1, and enJS5F16. Sheep genomic sequences adjacent to the proviruses were determined for each of these clones (GenBank accession numbers AY326472, AY326474, and AY326473, respectively). Primers were designed for these sequences and were used in PCR with DNA from each of the hybrid cell lines as a template. By determining which lines gave amplification products, it was possible to assign enJS56A1 to chromosome 6 and enJS59A1 to the X chromosome. The enJS5F16 flanking primers amplified a product from line R894-25C, which carries chromosomes 12, 13, and 15. It did not amplify a product from line R891-29R8A, which carries chromosomes 13 and 19, or line R891-27R1B, which carries chromosome 13. Thus, either chromosome 12 or chromosome 15 must carry enJS5F16 (data not shown). The assignment of enJS56A1 to chromosome 6 was further confirmed by FISH on metaphase chromosomes with a probe derived from the DNA flanking the provirus. This probe colocalized with virus probes that hybridize with the multicopy locus (data not shown).
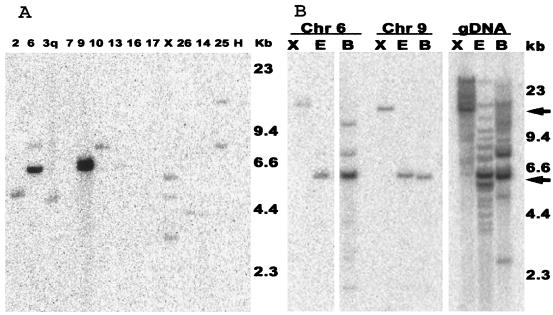
In order to further examine the loci on individual chromosomes, DNA preparations from sheep-hamster cell lines that contain a single sheep chromosome were digested with EcoRI and subjected to Southern blot hybridization with a capsid probe (Fig. 3A) All of the available JSRV and enJSRV sequences have an EcoRI site just downstream of the beginning of the pol open reading frame. The capsid probe should therefore detect junction fragments between the 5′ LTR and sheep DNA. Hybridization results indicate that cell lines had bands of various sizes, suggesting one or more sites of random enJSRV integration in the sheep chromosomes. No background hybridization was evident with the Chinese hamster ovary cell line (negative control) or with the lines containing chromosomes 16 and 17, which were negative for enJSRV by PCR (Table 2). The cell lines containing chromosomes 6 and 9 had bands, which migrated at approximately 6 kb, with intensities 5- to 10-fold higher than those of the other bands. This suggests that chromosomes 6 and 9 contain multiple copies of enJSRV that have the same size restriction fragment hybridizing to the capsid probe. A possible explanation for this is that the copies form part of a tandemly repeated array of enJSRV genomes.
FIG. 3.
Analysis by Southern blot hybridization on sheep-hamster hybrid cell lines. (A) DNA from cell lines containing single sheep chromosomes was digested with EcoRI, and Southern blots were probed with a JSRV capsid region probe. The sheep chromosome contained by the cell line is shown above the lane. (B) Sheep genomic DNA (gDNA) and DNA from hybrid cell lines containing single sheep chromosomes 6 or 9 were digested with XbaI (X), EcoRI (E), or BamHI (B), and Southern blots were probed with a JSRV capsid region probe.
In order to test this, DNA samples from the cell lines containing chromosomes 6 and 9 were then cleaved with EcoRI, BamHI, or XbaI and subjected to Southern blot hybridization. None of the available provirus sequences contains an XbaI site; therefore, the number of XbaI bands that hybridize with the probe should indicate the number of integrations. For chromosome 6 and 9, there are single XbaI bands that migrate in the high-molecular-weight region of the gel where the size resolution is poor (Fig. 3B). Therefore, all of the proviruses appear to reside on large XbaI fragments. All of the available provirus sequences contain a single BamHI site near the end of pol. Thus, left-end junction fragments and full-length copies of the putative tandem repeat sequence should hybridize with the gag probe. For chromosome 6, there is a strong band of hybridization around 6 kb and two weaker bands of hybridization at higher molecular weights (Fig. 3B). This would be consistent with a tandem array of 6-kb units and another locus. For chromosome 9, all three digests gave a single strong band of hybridization. This is again consistent with a single tandem array of 6-kb repeats. The junction fragments either are not resolved from the 6-kb band in the EcoRI and BamHI digests or they are two dilute to be visualized in these blots. These results suggest that chromosomes 6 and 9 each have several enJSRVs integrated in a tandem fashion. The 6-kb bands seen for chromosomes 6 and 9 correspond well with prominent bands seen in EcoRI and BamHI digests of sheep genomic DNA (Fig. 3B).
DISCUSSION
The presence of an endogenous retrovirus family related to JSRV has been known since the first JSRV nucleic acid probes were used on Southern blots of sheep genomic DNA. The distribution of the proviruses in the sheep genome appears to be quite stable, since the pattern of hybridization shows little variation among domestic sheep breeds and even among wild sheep (12). This suggests that there has not been much movement of these viruses for many thousands of years. This is quite different from the type C mouse endogenous viruses, which vary considerably from strain to strain (5). The Southern blot pattern, however, does not provide information on the location of proviruses on the sheep genome or allow the identification of individual loci, which may be of considerable interest in assessing their possible effects on the host physiology or their potential for interaction with exogenous JSRV. For example, there is a high level of expression from one or more enJSRV loci in the reproductive tract of the ewe (17). However, little is known about which loci are expressed, and the significance of this expression can only be speculated at this time.
The chromosomal sites of enJSRV were physically mapped by FISH in sheep and goat chromosomes using a biotinylated full-length JSRV probe. Using seven ovine and caprine cell lines, we consistently mapped enJSRV to seven chromosomal positions in sheep and eight in goat. Only two chromosomal positions (1q45 and 2q41) are common in the sheep and goat genomes, suggesting that some of the enJSRV copies entered into the small ruminants genomes after the speciation. The 7.5-kb probe we used reliably detected single proviral copies. A multicopy locus was identified on ovine chromosome 6, in position q12-13, with clear multiple sites of hybridization probably corresponding to at least three or four enJSRV sequences.
Additional insight into the distribution of viruses among the chromosomes was gained by determining whether a gag PCR product could be amplified from a panel of sheep-hamster cell lines, each of which carries one or a few sheep chromosomes. The results indicate that at least half of the sheep chromosomes carry at least one copy of the gag portion of the proviral genome. This is consistent with what is predicted by the Poisson distribution for random integration of 15 to 20 endogenous proviruses on the 28 chromosomes of the sheep genome.
All of the chromosomes identified as having proviral loci by FISH were also identified by PCR on the sheep-hamster hybrids. It is not surprising that more proviral loci were identified by PCR, since it should be a more sensitive technique due to the exponential amplification of the signal. In addition, the PCR primers were designed to detect the relatively conserved gag sequences of the virus. The FISH hybridization probe was a full-length clone of the exogenous JSRV genome and may not be sensitive enough to reliably detect truncated and/or relatively divergent enJSRV genomes.
An unexpected aspect of the enJSRV distribution in the sheep genome is the presence of two loci, which apparently contain several copies of a proviral genome. Two lines of evidence suggest that they may exist in a tandem repeat configuration. First, prior to the use of high-resolution R-banding for karyotyping the sheep chromosomes as described above, two to four loci were consistently detected by FISH on sheep metaphase chromosomes with probes of 2 and 4 kb, representing the 5′ and 3′ portions of the genome, respectively (data not shown). These probes did not reliably detect single proviral copies but readily detected these loci, suggesting multiple copies of provirus. Second, Southern blot analysis of DNA from sheep-hamster cell lines that carry either chromosome 6 or chromosome 9 suggests that multiple copies of enJSRV are present and that they may be organized in a head-to-tail tandem array. This type of organization in retroviral proviruses is unusual but not unprecedented. For example, a tandem HERV-K proviral locus has recently been described in humans (20), and a tandem duplication of avian sarcoma virus has been found in transformed quail cells (13). The significance of the arrays must await their further characterization and the determination of whether or not they are transcribed and expressed at some time during the life of the sheep.
Acknowledgments
We thank Dave Geier and David Patterson of the Eleanor Roosevelt Institute for Cancer Research for providing the panel of sheep-hamster cell lines. We thank Andrew Paterson for technical contributions to the project. We also thank Massimo Palmarini for providing clones of the endogenous proviruses, enJS56A1, enJS59A1, and enJS5F16.
This work was supported by grant 1R01 CA 59116 from the National Cancer Institute, National Institutes of Health, by the Rhône, Drôme, Loire, and Ardèche departmental committees of the Ligue Contre le Cancer, and by a grant from the Rhône Alpes Region. C.L. was a recipient of an ARC postdoctoral fellowship.
REFERENCES
- 1.Bahri-Darwich, I., D. Vaiman, I. Olsaker, A. Oustry, and E. P. Cribiu. 1994. Assignment of bovine synteny groups U27 and U8 to R-banded chromosome 12 and 27, respectively. Hereditas 120:261-265. [DOI] [PubMed]
- 2.Bai, J., J. V. Bishop, J. O. Carlson, and J. C. DeMartini. 1999. Sequence comparison of JSRV with endogenous proviruses: envelope genotypes and a novel ORF with similarity to a G-protein-coupled receptor. Virology 258:333-343. [DOI] [PubMed] [Google Scholar]
- 3.Bai, J., R. Y. Zhu, K. Stedman, C. Cousens, J. Carlson, J. M. Sharp, and J. C. DeMartini. 1996. Unique long terminal repeat U3 sequences distinguish exogenous jaagsiekte sheep retroviruses associated with ovine pulmonary carcinoma from endogenous loci in the sheep genome. J. Virol. 70:3159-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkin, D. J., T. E. Broad, M. R. Lambeth, H. R. Burkin, and C. Jones. 1998. New gene assignments using a complete, characterized sheep-hamster somatic cell hybrid panel. Anim. Genet. 29:48-54. [DOI] [PubMed] [Google Scholar]
- 5.Coffin, J. M. 1995. Retrovirus variation and evolution in the DNA provirus, p. 221-243. In G. M. Cooper, R. G. Temin, and B. Sugden (ed.), Howard Temin's Scientific Legacy. ASM Press, Washington D.C.
- 6.Cribiu, E. P., D. Di Berardino, G. P. Di Meo, A. Eggen, D. S. Gallagher, I. Gustavsson, H. Hayes, L. Iannuzzi, C. P. Popescu, J. Rubes, S. Schmutz, G. Stranzinger, A. Vaiman, and J. Womack. 2001. International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet. Cell Genet. 92:283-299. [DOI] [PubMed] [Google Scholar]
- 7.Cribiu, E. P., M. Matejka, B. Dennis, and X. Malher. 1988. Etude chromosomique d'un hybride chèvre x mouton fertile. Genet. Sel. Evol. 20:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Da Silva Teixeira, M. F., V. Lambert, L. Mselli-Lakahl, A. Chettab, Y. Chebloune, and J. F. Mornex. 1997. Immortalization of caprine fibroblasts permissive for replication of small ruminant lentiviruses. Am. J. Vet. Res. 58:579-584. [PubMed] [Google Scholar]
- 9.Eggen, A., A. Oustry, D. Vaiman, L. Ferretti, R. Fries, and E. P. Cribiu. 1994. Bovine synteny group U7, previously assigned to G-banded chromosome 25 in the ISCNDA nomenclature, assigns to R-banded chromosome 29. Hereditas 121:295-300. [DOI] [PubMed] [Google Scholar]
- 10.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 11.Guillemot, E., F. Gary, H. M. Berland, V. Durand, R. Darré, and E. P. Cribiu. 1991. Cytogenetic investigation in Saanen and Alpine artificial insemination bucks. Identification of robertsonian translocation. Genet. Sel. Evol. 23:449-454. [Google Scholar]
- 12.Hecht, S. J., K. E. Stedman, J. O. Carlson, and J. C. DeMartini. 1996. Distribution of endogenous type B and type D sheep retrovirus sequences in ungulates and other mammals Proc. Natl. Acad. Sci. USA 93:3297-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, T. W., J. M. Taylor, C. Aldrich, J. B. Townsend, G. Seal, and W. S. Mason. 1981. Tandem duplication of the proviral DNA in an avian sarcoma virus-transformed quail clone. J. Virol. 38:219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemieux, N., B. Dutrillaux, and E. Viegas-Pequignot. 1992. A simple method for simultaneous R- or G-banding and fluorescence in situ hybridization of small single-copy genes. Cytogenet. Cell Genet. 59:311-312. [DOI] [PubMed] [Google Scholar]
- 15.Palmarini, M., C. Cousens, R. G. Dalziel, J. Bai, K. Stedman, J. C. DeMartini, and J. M. Sharp. 1996. The exogenous form of Jaagsiekte retrovirus is specifically associated with a contagious lung cancer of sheep. J. Virol. 70:1618-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmarini, M., S. Datta, R. Omid, C. Murgia, and H. Fan. 2000. The long terminal repeat of Jaagsiekte sheep retrovirus is preferentially active in differentiated epithelial cells of the lungs. J. Virol. 74:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmarini, M., C. A. Gray, K. Carpenter, H. Fan, F. W. Bazer, and T. E. Spencer. 2001. Expression of endogenous betaretroviruses in the ovine uterus: effects of neonatal age, estrous cycle, pregnancy, and progesterone. J. Virol. 75:11319-11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmarini, M., C. Hallwirth, D. York, C. Murgia, T. de Oliveira, T. Spencer, and H. Fan. 2000. Molecular cloning and functional analysis of three type D endogenous retroviruses of sheep reveal a different cell tropism from that of the highly related exogenous jaagsiekte sheep retrovirus. J. Virol. 74:8065-8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmarini, M., J. M. Sharp, M. de las Heras, and H. Fan. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep J. Virol. 73:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reus, K., J. Mayer, M. Sauter, D. Scherer, N. Muller-Lantzsch, and E. Meese. 2001. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics 72:314-320. [DOI] [PubMed] [Google Scholar]
- 21.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 22.Spencer, T. E., A. G. Stagg, M. M. Joyce, G. Jenster, C. G. Wood, F. W. Bazer, A. A. Wiley, and F. F. Bartol. 1999. Discovery and characterization of endometrial epithelial messenger ribonucleic acids using the ovine uterine gland knockout model. Endocrinology 140:4070-4080. [DOI] [PubMed] [Google Scholar]
- 23.York, D. F., R. Vigne, D. W. Verwoerd, and G. Querat. 1992. Nucleotide sequence of the jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J. Virol. 66:4930-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]