Mammalian reoviruses are nonenveloped viruses that contain a segmented double-stranded RNA genome. Most mammalian species, including humans, serve as hosts for reovirus infection, but reovirus-induced disease is restricted to the very young (reviewed in reference 61). Reovirus infections of newborn mice have been used as a tractable experimental system for studies of viral pathogenesis. The segmented genome of these viruses has allowed the genetic basis for complex viral phenotypes to be determined by analysis of reassortant viruses containing mixtures of gene segments derived from parental strains that exhibit biological polymorphisms of interest. One of the best characterized models of reovirus pathogenesis is infection of the murine central nervous system (CNS), in which serotype 1 (T1) and serotype 3 (T3) reoviruses display markedly different patterns of disease (Table 1).
TABLE 1.
Properties of reovirus pathogenesis attributable to σ1 protein
| Viral serotype | Functional property
|
||||
|---|---|---|---|---|---|
| Intestinal growth | Pathway of spread | CNS tropism | Tail receptor | Head receptor | |
| 1 | Yes | Hematogenous | Ependymal cells | Unknown carbohydrate | JAM1 |
| 3 | Variablea | Neural | Neurons | Sialic acid | JAM1 |
Some type 3 reovirus strains can infect the intestine and disseminate systemically. Others fail to infect intestinal tissue and do not spread to distant sites.
After oral inoculation of newborn mice, reovirus is taken up by intestinal M cells (72) and undergoes primary replication in lymphoid tissue of the Peyer's patches. The virus then invades the CNS, yet T1 and T3 strains use different routes of dissemination and manifest distinct pathological consequences. T1 reovirus spreads to the CNS hematogenously and infects ependymal cells (62, 69), resulting in hydrocephalus (68). In contrast, T3 reovirus spreads to the CNS neurally and infects neurons (38, 62, 69), causing lethal encephalitis (58, 68). Analysis of reassortant viruses obtained by coinfecting cells with prototype strains T1 Lang (T1L) and T3 Dearing (T3D) demonstrated that the pathways of viral spread in the host (62) and tropism for neural tissues (24, 69) segregate with the viral S1 gene, which encodes the viral attachment protein σ1 (33, 67). T1L × T3D reassortant viruses were also used to show that serotype-specific differences in virus binding to primary cultures of ependymal cells and neurons are determined by the S1 gene (24, 59). These studies suggest that σ1 dictates the CNS cell types that serve as targets for reovirus infection, presumably by its capacity to bind receptors expressed by specific CNS cells.
Reovirus pathogenesis is not restricted to the CNS, and σ1 is not the sole determinant of reovirus virulence. Reovirus infection causes pathology and physiologic dysfunction in a wide range of organs and tissues, including the hepatobiliary system, the myocardium, lungs, and endocrine tissues (reviewed in reference 65). Of these, myocarditis (54) has become a particularly well-established experimental model of reovirus-induced disease. Myocarditis caused by reovirus infection is unusual in comparison to other viral etiologies of myocarditis in that the pathogenesis is not immune mediated. Instead, reovirus cytopathicity is a direct cause of myocyte injury, which results from a complex interplay of the interferon (1, 42, 55) and apoptosis (22) pathways. Efficiency of viral RNA synthesis is a key factor in determining the extent of myocardial injury (51). Accordingly, viral gene segments encoding proteins involved in viral transcription and genome replication play important roles in determining strain-specific differences in the capacity of reovirus to induce myocarditis (52, 53, 55).
REOVIRUS ATTACHMENT PROTEIN σ1 BINDS TO CELL SURFACE CARBOHYDRATE AND JAM1
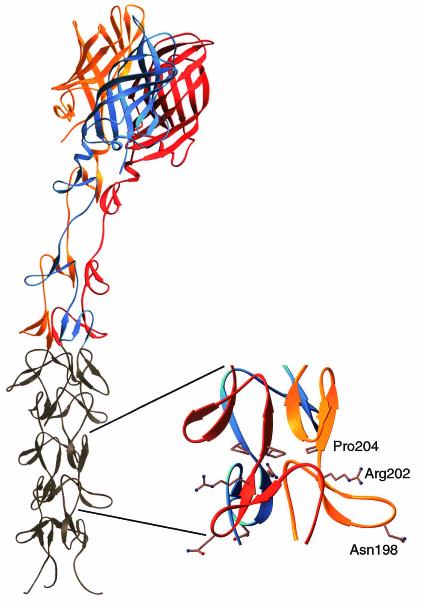
The σ1 protein is a fibrous trimer consisting of an elongated tail domain that inserts into the virion and a globular head domain that projects away from the virion surface (2, 27, 28). The recently determined crystal structure of the C-terminal half of T3D σ1 reveals that the tail is formed in part by a triple-β spiral and that the head is formed by a compact eight-stranded β-barrel (14) (Fig. 1). T3 σ1 contains receptor-binding domains in both the tail and head regions. A domain in the tail binds α-linked sialic acid (12, 13), whereas a domain in the head binds junctional adhesion molecule 1 (JAM1) (4). In T3D σ1, these domains are dissociable by treatment of σ1 with intestinal proteases, such as trypsin or chymotrypsin (11, 39), which likely accounts for the attenuated virulence of this strain after oral inoculation (9, 31, 49). The T1 σ1 tail also binds cell surface carbohydrate (12), but this molecule has not been identified.
FIG. 1.
Crystal structure of reovirus attachment protein σ1. The crystal structure of T3D σ1 includes residues 245 to 455 (14). The three monomers of the σ1 trimer are shown in red, orange, and blue. Each monomer consists of a C-terminal head domain formed by a compact β-barrel and an N-terminal fibrous tail that contains three β-spiral repeats. Based on analysis of patterns in aligned σ1 sequences, the β-spiral likely begins at residue 167 of T3D σ1 and comprises eight repeats. The N-terminal five repeats, which are not included in the crystal structure, are shown in gray. The spiral has been extended using translated and rotated σ1 repeats to generate a model that depicts the approximate dimensions of the molecule. Amino acids Asn198, Arg202, and Pro204 have been implicated in the interaction of T3D σ1 with sialic acid (13). The approximate location of these residues in the model (shown in ball-and-stick representation on the right) suggests that they form a binding site for sialic acid. Residues 1 to 167 are not shown; these residues are predicted to form a triple α-helical coiled coil structure (6, 25, 27, 40). This figure was prepared by Thilo Stehle (Harvard University) (published with permission) with the program RIBBONS (10).
The capacity of T3 reovirus to bind sialic acid influences infection of cultured cells. Both T1 and T3 reoviruses can infect L929 cells, a murine fibroblast cell line commonly used to propagate reovirus. However, only T3 strains can infect murine erythroleukemia (MEL) cells (13, 50). This growth restriction is sialic acid dependent (13, 50), and serial passage of non-sialic-acid-binding T3 strains in these cells results in selection of viruses that have acquired the capacity to bind sialic acid (13). The MEL-adapted phenotype is conferred by single point mutations (13) in a region of the σ1 tail implicated in sialic acid binding (12). Although the sialic acid-binding region of T3D σ1 was not included in the crystal structure (14), molecular modeling suggests that this region is contained within the triple β-spiral (Fig. 1).
Substantial evidence has accumulated to suggest that the σ1 head binds to proteinaceous receptors on the cell surface (7, 26, 39, 60). A flow cytometry-based expression-cloning approach was used to identify such molecules by use of a non-sialic-acid-binding strain as an affinity ligand (4). A neural precursor cell (NT2) cDNA library was selectively enriched by fluorescence-activated cell sorting for cDNAs that confer binding of fluoresced virions to transfected cells. Four clones were identified that conferred virus binding to all transfected cells. Each encoded JAM1, a member of the immunoglobulin superfamily postulated to regulate formation of intercellular tight junctions (35, 37, 70). JAM1-specific monoclonal antibodies inhibit reovirus binding and infection, and expression of JAM1 in nonpermissive cells rescues reovirus growth (4). Most importantly, the σ1 protein binds directly to JAM1 with an apparent Kd of ∼6 × 10−8 M (4), providing confidence that JAM1 is a reovirus receptor. Surprisingly, JAM1 serves as a receptor for both prototype and field-isolate strains of all three reovirus serotypes (4; J. A. Campbell and T. S. Dermody, unpublished observations). Therefore, JAM1 does not appear to explain the serotype-dependent differences in reovirus tropism in the murine CNS. These observations suggest that proteinaceous receptors other than JAM1, or unique carbohydrate-based coreceptors, influence reovirus pathogenesis.
SIALIC ACID AS A DETERMINANT OF REOVIRUS ATTACHMENT AND DISEASE
To dissect the contribution of sialic acid to T3 reovirus attachment, infection, and disease, the S1 gene segments of a MEL-adapted strain, T3C44-MA, and its non-sialic-acid-binding parental strain, T3C44, were introduced into the genetic background of T1L by reassortment (3). The resultant viruses, termed T3SA+ and T3SA−, respectively, differ by a proline-to-leucine substitution at amino acid 204 in σ1 (3) which confers the capacity to bind sialic acid (13). HeLa cells are permissive for infection by both T3SA+ and T3SA−; however, viral yields from a single replication cycle are significantly higher for T3SA+ when HeLa cells are infected with equivalent multiplicities of infection of these strains (3). This enhanced growth is sialic acid dependent, as removal of cell surface sialic acid by neuraminidase or incubation of virions with the soluble sialic acid analog sialyllactose (SLL) decreases yields of T3SA+ to the same levels as those of T3SA− (3). Radioligand binding studies using T3SA+ and T3SA− indicate that the capacity to bind sialic acid enhances the association rate of virus for cells and increases the avidity of binding (3). Kinetic analyses using inhibitors of sialic acid and JAM1 binding demonstrate that sialic acid is engaged first in the adsorption process, as the inhibitory effect of SLL on infection by T3SA+ occurs at early but not late time points. However, a σ1-specific monoclonal antibody that blocks virus binding to JAM1 inhibits viral infectivity at both early and late times during adsorption. These data suggest that reovirus attaches to cells via an adhesion-strengthening mechanism by which initial low-affinity binding to sialic acid facilitates secondary higher-affinity binding to JAM1 (3).
In addition to attaching virus to the cell surface, the capacity of T3 reovirus to bind sialic acid influences tissue tropism and disease phenotypes. Both T3SA+ and T3SA− grow well in the intestine following peroral inoculation of newborn mice and disseminate to distant sites (5). However, T3SA+ produces higher titers in the brain and liver at early times after inoculation than those produced by T3SA−. Importantly, by day 12 following inoculation, though, titers of both viruses are equivalent in all organs tested (5). These findings correlate well with observations made using cultured cells and suggest that the enhancement of viral attachment due to sialic acid binding also occurs in vivo.
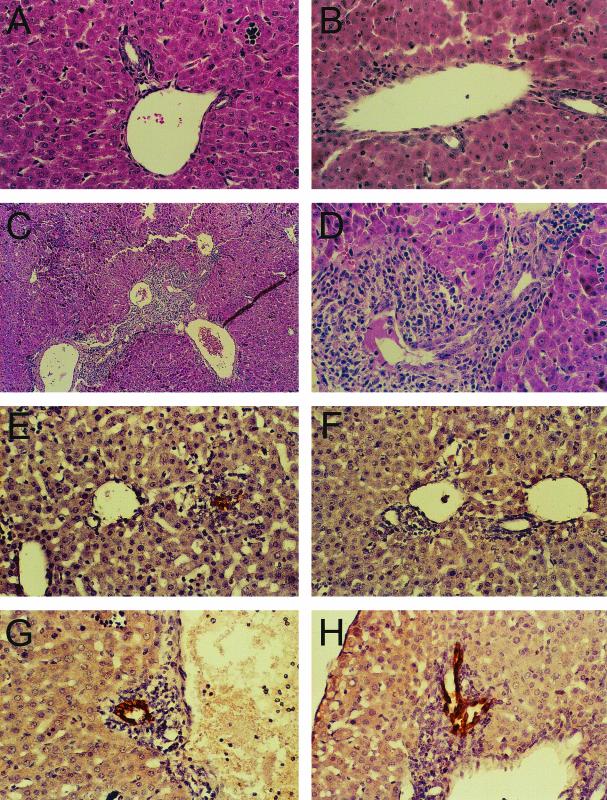
Despite the differences in the kinetics of spread exhibited by T3SA+ and T3SA− following infection of newborn mice, the disease phenotypes associated with these strains differ dramatically. Animals infected with T3SA+ but not T3SA− develop jaundice, steatorrhea, and oily fur (5). These findings are consistent with previous studies of reovirus infections of mice and correlate with injury to bile duct epithelium (45-47, 71). Histological analysis indicates that T3SA+ and T3SA− vary strikingly in the pattern of liver injury in infected mice (5) (Fig. 2). Liver sections from animals infected with T3SA+ demonstrate a robust inflammatory response concentrated in the portal areas, whereas liver sections from animals infected with T3SA− contain only mild inflammatory infiltrates (Fig. 2A to D). Concordantly, immunohistochemical staining reveals viral antigen in bile duct epithelial cells of animals infected with T3SA+ (Fig. 2E and F). Viral antigen is also present in liver tissue of animals infected with T3SA−, but the antigen is primarily localized to hepatocytes (Fig. 2G and H). These findings suggest that utilization of sialic acid as a coreceptor targets reovirus to bile duct epithelial cells. Interestingly, the disease produced by T3SA+ in mice is similar in some respects to biliary atresia in human infants, which has been linked to reovirus in approximately 50% of cases in one study (63). As such, infection of newborn mice by sialic acid-binding reoviruses may have utility as a model to dissect the pathogenesis of bile duct injury in humans.
FIG. 2.
(A to D) Liver histopathology in mice following infection with T3SA− and T3SA+. ND4 Swiss Webster mice (2 to 3 days old) were inoculated perorally with phosphate-buffered saline (A) or 2.5 × 103 PFU of either T3SA− (B) or T3SA+ (C and D). At 8 days postinoculation, liver tissue was harvested, embedded in paraffin, thin sectioned, and stained with hematoxylin and eosin. Magnification, ×100 (C) or ×400 (A, B, and D). (E to H) Immunohistochemical localization of reovirus antigen in bile duct epithelial cells. ND4 Swiss Webster mice (2 to 3 days old) were inoculated perorally with 2.5 × 103 PFU of either T3SA− (E and F) or T3SA+ (G and H). At 6 days postinoculation, liver tissue was harvested, embedded in paraffin, thin sectioned, and stained for reovirus antigen using rabbit anti-reovirus serum and horseradish peroxidase. Dark-brown staining indicates the presence of reovirus antigen. Magnification, ×400. Representative sections from two separate animals are shown. Modified from Barton et al. (5) with permission from the publisher.
REOVIRUS ATTACHMENT AND DISASSEMBLY ARE REQUIRED FOR INDUCTION OF APOPTOSIS
In addition to conferring viral attachment, engagement of reovirus receptors also induces postbinding signaling events that may influence disease pathogenesis. Reovirus induces apoptosis in cultured cells (15, 21, 48, 64) and in vivo (22, 43). Insight into mechanisms by which reovirus elicits apoptosis was first developed from studies of viral prototype strains that vary in the capacity to induce this cellular response. T3D induces apoptosis to a greater extent than T1L in L cells (64), Madin-Darby canine kidney (MDCK) cells (48), and HeLa cells (19). Differences in the capacity of these strains to induce apoptosis are determined primarily by the viral S1 gene (19, 48, 64); however, the M2 gene makes a secondary contribution to the magnitude of the apoptotic response (48, 64).
Linkage of the S1 gene to the efficiency of reovirus-induced apoptosis suggests that the capacity of attachment protein σ1 to bind different receptors regulates proapoptotic signaling. Sialic acid-binding strain T3SA+ induces apoptosis to a much greater extent than non-sialic-acid-binding strain T3SA− in both HeLa cells and L cells (19). Removal of cell surface sialic acid with neuraminidase or blockade of virus binding to sialic acid by SLL abolishes the capacity of T3SA+ to induce apoptosis (19). These findings indicate that the capacity of T3 reovirus to bind sialic acid significantly enhances its capacity to induce an apoptotic response. However, JAM1 also plays a critical role in apoptosis. Although T3SA+ can bind and enter cells via a JAM1-independent pathway mediated by binding sialic acid, T3SA+ is incapable of inducing apoptosis in the absence of JAM1 binding (4). Thus, reovirus binding to both sialic acid and JAM1 is required to induce maximal levels of apoptosis. Since T1 and T3 reoviruses bind different carbohydrate coreceptors, it is possible that serotype-specific differences in disease are attributable to differences in the induction of apoptosis in vivo.
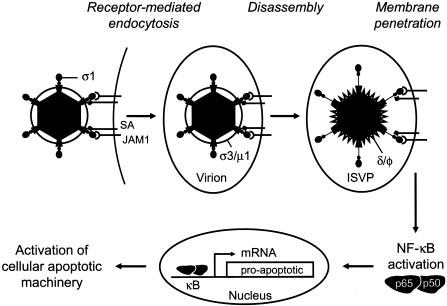
Although ligation of sialic acid and JAM1 is necessary for apoptosis induced by reovirus, viral attachment to the cell surface alone is not sufficient. Inhibitors of acid-dependent viral disassembly block apoptosis by reovirus (20), indicating a requirement for postattachment entry steps. However, viral transcription is dispensable, as inhibitors of viral RNA synthesis do not diminish the capacity of reovirus to induce apoptosis (20, 64). Linkage of the M2 gene with the efficiency of apoptosis induction by reovirus provides additional support for the idea that viral entry steps are required for proapoptotic signaling (48, 64). The M2 gene encodes the μ1 protein, which mediates penetration of the virus into the cytosol following viral disassembly in endosomes (30, 34, 36, 41). These findings suggest that receptor binding and disassembly must occur within the same cellular compartment to elicit an apoptotic response. It is also possible that virus-receptor interactions couple with μ1-mediated membrane penetration to elicit apoptosis (Fig. 3).
FIG. 3.
Model of reovirus-induced apoptosis. Reovirus infection is initiated by attachment of the virus to carbohydrate coreceptors and JAM1. For T3 reoviruses, the carbohydrate bound is sialic acid (SA). After attachment to cellular receptors, reovirus enters cells by receptor-mediated endocytosis. Within an endocytic compartment, the viral outer capsid is removed to generate infectious subvirion particles (ISVPs). During virion-to-ISVP conversion, σ3 is degraded and lost from virions, σ1 undergoes a conformational change, and μ1 is cleaved to form particle-associated fragments δ and φ. Removal of σ3 exposes hydrophobic domains in μ1 that facilitate interactions of ISVPs with endosomal membranes, leading to delivery of core particles into the cytoplasm and concomitant activation of the viral transcriptase. Viral attachment and disassembly must occur within the same cellular compartment to activate NF-κB. Activation of NF-κB also might be achieved by μ1-mediated membrane penetration acting in synergy with viral receptor engagement. Once activated, NF-κB translocates to the nucleus, where it induces the expression of proapoptotic genes.
A critical component of the signaling cascade that leads to apoptosis of reovirus-infected cells is the transcription factor NF-κB, which is known to play important roles in regulating cellular stress responses, including apoptosis (44). Reovirus activates NF-κB in several cell lines, including L cells, MDCK cells, and HeLa cells (21). NF-κB complexes activated by reovirus are comprised of NF-κB subunits p50 and p65 (RelA) (21). Apoptosis induced by reovirus is significantly reduced in cells treated with a proteasome inhibitor and in cells expressing a transdominant inhibitor of NF-κB (21). In addition, reovirus-induced apoptosis is blocked in cells deficient in the expression of the p50 or p65 NF-κB subunits. These results demonstrate that NF-κB plays a proapoptotic role during reovirus infection (Fig. 3). Reovirus also activates c-Jun N-terminal kinase and extracellular signal-related kinase (17), but the involvement of these signaling molecules in NF-κB activation and apoptosis induction is not understood.
In addition to NF-κB, several other cellular molecules have been implicated in reovirus-induced apoptosis. The calcium-dependent protease calpain is activated during reovirus infection, and calpain inhibitors block apoptosis induced by reovirus (23). It is not known how the calpain and NF-κB pathways couple to cause apoptosis during reovirus infection. Cellular gene expression is required for apoptosis induced by reovirus (21). In some cell types, reovirus infection leads to expression of death receptors DR4 and DR5 and their proapoptotic ligand TRAIL (15, 16), suggesting that apoptosis is induced by autocrine or paracrine mechanisms. However, mitochondrial injury during reovirus infection has been documented (32), providing evidence that intrinsic pathways are also involved in apoptosis induced by reovirus infection.
CONCLUSIONS
A precise understanding of the serotype-dependent differences in tropism exhibited by reovirus in the murine CNS remains elusive. T1 and T3 reoviruses vary in the types of cell surface carbohydrate used as coreceptors (12), but both serotypes bind JAM1 (4). These observations make it unlikely that JAM1 is the sole determinant of reovirus tropism in the murine CNS. It is possible that JAM1 serves as a serotype-independent reovirus receptor at some sites within the host and that other as yet unidentified receptors confer serotype-dependent tropism in the CNS. Definitive assessment of the role of JAM1 in reovirus pathogenesis awaits the results of studies using JAM1-null mice.
The nature of the carbohydrate bound by reovirus plays an important role in viral attachment (3) and apoptosis induction (19), and new evidence indicates that reovirus strains that vary in sialic acid utilization also vary in the capacity to produce biliary tract disease (5). However, both T3SA+ and T3SA− infect neurons, and these viruses display equivalent 50% lethal dose values following peroral inoculation of mice (5). These data suggest that sialic acid binding is not the primary determinant of neural tropism exhibited by T3 reovirus. It is possible that the unidentified carbohydrate bound by T1 reovirus directs infection to ependymal cells in the CNS. If so, engagement of JAM1 on neurons by non-sialic-acid-binding T3 reovirus would be expected, with a lethal outcome as the result. Studies using viruses containing chimeric σ1 proteins in which the T1 and T3 carbohydrate-binding domains are reciprocally exchanged (12) will help to clarify the role of carbohydrate coreceptors in reovirus disease.
Differences in receptor utilization might also influence pathogenesis by virtue of activating different types of signaling pathways. Since T1 and T3 differ in the capacity to induce apoptosis (21, 48, 64), a property linked to receptor binding (4, 19), it is possible that postattachment signaling plays a role in the production of disease. Support for this idea comes from studies of reovirus-induced myocarditis in which treatment of mice with inhibitors of calpain to inhibit proapoptotic signaling ameliorates tissue injury (22). Additional studies are required to confirm an association of apoptosis with reovirus-induced disease. However, it appears that the role of reovirus receptors in disease pathogenesis is more complex than simply mediating the virus docking event.
A final remarkable observation is the localization of JAM1 to tight junctions. In addition to reovirus, several other viruses bind receptors expressed at regions of cell-cell contact (56). Like JAM1, the coxsackievirus and adenovirus receptor CAR (8) is expressed at tight junctions (18). Nectins, which serve as receptors for herpes simplex virus (29, 66), are expressed at adherens junctions (57, 73). Interestingly, each of these viruses is capable of infecting both epithelial surfaces and neurons in some types of host organisms. Junctional regions are sites of enhanced membrane recycling, endocytic uptake, and intracellular signaling (74). Therefore, it is possible that viruses have selected junction-associated proteins as receptors to usurp the physiologic functions of these molecules. Such viruses would be expected to display common themes in attachment and internalization, which may extend to conserved modes of pathogenesis and disease production.
Acknowledgments
We thank members of our laboratory for many useful discussions and Jim Chappell, Sean O'Donnell, and Tim Peters for reviews of the manuscript.
This research was supported by Public Health Service awards AI38296 and AI50080 and the Elizabeth B. Lamb Center for Pediatric Research.
REFERENCES
- 1.Azzam-Smoak, K., D. L. Noah, M. J. Stewart, M. A. Blum, and B. Sherry. 2002. Interferon regulatory factor-1, interferon-beta, and reovirus-induced myocarditis. Virology 298:20-29. [DOI] [PubMed] [Google Scholar]
- 2.Banerjea, A. C., K. A. Brechling, C. A. Ray, H. Erikson, D. J. Pickup, and W. K. Joklik. 1988. High-level synthesis of biologically active reovirus protein sigma 1 in a mammalian expression vector system. Virology 167:601-612. [PubMed] [Google Scholar]
- 3.Barton, E. S., J. L. Connolly, J. C. Forrest, J. D. Chappell, and T. S. Dermody. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 276:2200-2211. [DOI] [PubMed] [Google Scholar]
- 4.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 5.Barton, E. S., B. E. Youree, D. H. Ebert, J. C. Forrest, J. L. Connolly, T. Valyi-Nagy, K. Washington, J. D. Wetzel, and T. S. Dermody. 2003. Utilization of sialic acid as a coreceptor is required for reovirus-induced biliary disease. J. Clin. Investig. 111:1823-1833. [DOI] [PMC free article] [PubMed]
- 6.Bassel-Duby, R., A. Jayasuriya, D. Chatterjee, N. Sonenberg, J. V. Maizel, Jr., and B. N. Fields. 1985. Sequence of reovirus haemagglutinin predicts a coiled-coil structure. Nature 315:421-423. [DOI] [PubMed] [Google Scholar]
- 7.Bassel-Duby, R., D. R. Spriggs, K. L. Tyler, and B. N. Fields. 1986. Identification of attenuating mutations on the reovirus type 3 S1 double-stranded RNA segment with a rapid sequencing technique. J. Virol. 60:64-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 9.Bodkin, D. K., and B. N. Fields. 1989. Growth and survival of reovirus in intestinal tissue: role of the L2 and S1 genes. J. Virol. 63:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson, M. 1987. Ribbon models of macromolecules. J. Mol. Graph. 5:103-106. [Google Scholar]
- 11.Chappell, J. D., E. S. Barton, T. H. Smith, G. S. Baer, D. T. Duong, M. L. Nibert, and T. S. Dermody. 1998. Cleavage susceptibility of reovirus attachment protein σ1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the σ1 neck. J. Virol. 72:8205-8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chappell, J. D., J. L. Duong, B. W. Wright, and T. S. Dermody. 2000. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 74:8472-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chappell, J. D., V. L. Gunn, J. D. Wetzel, G. S. Baer, and T. S. Dermody. 1997. Mutations in type 3 reovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein σ1. J. Virol. 71:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chappell, J. D., A. Prota, T. S. Dermody, and T. Stehle. 2002. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke, P., S. M. Meintzer, S. Gibson, C. Widmann, T. P. Garrington, G. L. Johnson, and K. L. Tyler. 2000. Reovirus-induced apoptosis is mediated by TRAIL. J. Virol. 74:8135-8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke, P., S. M. Meintzer, A. C. Spalding, G. L. Johnson, and K. L. Tyler. 2001. Caspase 8-dependent sensitization of cancer cells to TRAIL-induced apoptosis following reovirus-infection. Oncogene 20:6910-6919. [DOI] [PubMed] [Google Scholar]
- 17.Clarke, P., S. M. Meintzer, C. Widmann, G. L. Johnson, and K. L. Tyler. 2001. Reovirus infection activates JNK and the JNK-dependent transcription factor c-Jun. J. Virol. 75:11275-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, C. J., J. T. Shieh, R. J. Pickles, T. Okegawa, J. T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly, J. L., E. S. Barton, and T. S. Dermody. 2001. Reovirus binding to cell surface sialic acid potentiates virus-induced apoptosis. J. Virol. 75:4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connolly, J. L., and T. S. Dermody. 2002. Virion disassembly is required for apoptosis induced by reovirus. J. Virol. 76:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connolly, J. L., S. E. Rodgers, P. Clarke, D. W. Ballard, L. D. Kerr, K. L. Tyler, and T. S. Dermody. 2000. Reovirus-induced apoptosis requires activation of transcription factor NF-κB. J. Virol. 74:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeBiasi, R., C. Edelstein, B. Sherry, and K. Tyler. 2001. Calpain inhibition protects against virus-induced apoptotic myocardial injury. J. Virol. 75:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBiasi, R. L., M. K. T. Squier, B. Pike, M. Wynes, T. S. Dermody, J. J. Cohen, and K. L. Tyler. 1999. Reovirus-induced apoptosis is preceded by increased cellular calpain activity and is blocked by calpain inhibitors. J. Virol. 73:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dichter, M. A., and H. L. Weiner. 1984. Infection of neuronal cell cultures with reovirus mimics in vitro patterns of neurotropism. Ann. Neurol. 16:603-610. [DOI] [PubMed] [Google Scholar]
- 25.Duncan, R., D. Horne, L. W. Cashdollar, W. K. Joklik, and P. W. K. Lee. 1990. Identification of conserved domains in the cell attachment proteins of the three serotypes of reovirus. Virology 174:399-409. [DOI] [PubMed] [Google Scholar]
- 26.Duncan, R., D. Horne, J. E. Strong, G. Leone, R. T. Pon, M. C. Yeung, and P. W. K. Lee. 1991. Conformational and functional analysis of the C-terminal globular head of the reovirus cell attachment protein. Virology 182:810-819. [DOI] [PubMed] [Google Scholar]
- 27.Fraser, R. D. B., D. B. Furlong, B. L. Trus, M. L. Nibert, B. N. Fields, and A. C. Steven. 1990. Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J. Virol. 64:2990-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 30.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keroack, M., and B. N. Fields. 1986. Viral shedding and transmission between hosts determined by reovirus L2 gene. Science 232:1635-1638. [DOI] [PubMed] [Google Scholar]
- 32.Kominsky, D. J., R. J. Bickel, and K. L. Tyler. 2002. Reovirus-induced apoptosis requires mitochondrial release of Smac/DIABLO and involves reduction of cellular inhibitor of apoptosis protein levels. J. Virol. 76:11414-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, P. W., E. C. Hayes, and W. K. Joklik. 1981. Protein σ1 is the reovirus cell attachment protein. Virology 108:156-163. [DOI] [PubMed] [Google Scholar]
- 34.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, Y., A. Nusrat, F. J. Schnell, T. A. Reaves, S. Walsh, M. Ponchet, and C. A. Parkos. 2000. Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 113:1-11. [DOI] [PubMed] [Google Scholar]
- 36.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Padura, I., S. Lostaglio, M. Schneemann, L. Williams, M. Romano, P. Fruscella, C. Panzeri, A. Stoppacciaro, L. Ruco, A. Villa, D. Simmons, and E. Dejana. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison, L. A., R. L. Sidman, and B. N. Fields. 1991. Direct spread of reovirus from the intestinal lumen to the central nervous system through vagal autonomic nerve fibers. Proc. Natl. Acad. Sci. USA 88:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nibert, M. L., J. D. Chappell, and T. S. Dermody. 1995. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved σ1 protein. J. Virol. 69:5057-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nibert, M. L., T. S. Dermody, and B. N. Fields. 1990. Structure of the reovirus cell-attachment protein: a model for the domain organization of σ1. J. Virol. 64:2976-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noah, D. L., M. A. Blum, and B. Sherry. 1999. Interferon regulatory factor 3 is required for viral induction of beta interferon in primary cardiac myocyte cultures. J. Virol. 73:10208-10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oberhaus, S. M., R. L. Smith, G. H. Clayton, T. S. Dermody, and K. L. Tyler. 1997. Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J. Virol. 71:2100-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pahl, H. L. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 45.Papadimitriou, J. M. 1968. The biliary tract in acute murine reovirus 3 infection. Am. J. Pathol. 52:595-601. [PMC free article] [PubMed] [Google Scholar]
- 46.Parashar, K., M. J. Tarlow, and M. A. McCrae. 1992. Experimental reovirus type 3-induced murine biliary tract disease. J. Pediatr. Surg. 27:843-847. [DOI] [PubMed] [Google Scholar]
- 47.Phillips, P. A., D. Keast, J. M. Papadimitriou, M. N. Walters, and N. F. Stanley. 1969. Chronic obstructive jaundice induced by reovirus type 3 in weanling mice. Pathology 1:193-203. [DOI] [PubMed] [Google Scholar]
- 48.Rodgers, S. E., E. S. Barton, S. M. Oberhaus, B. Pike, C. A. Gibson, K. L. Tyler, and T. S. Dermody. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 71:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubin, D. H., and B. N. Fields. 1980. Molecular basis of reovirus virulence: role of the M2 gene. J. Exp. Med. 152:853-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubin, D. H., J. D. Wetzel, W. V. Williams, J. A. Cohen, C. Dworkin, and T. S. Dermody. 1992. Binding of type 3 reovirus by a domain of the σ1 protein important for hemagglutination leads to infection of murine erythroleukemia cells. J. Clin. Investig. 90:2536-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherry, B., C. J. Baty, and M. A. Blum. 1996. Reovirus-induced acute myocarditis in mice correlates with viral RNA synthesis rather than generation of infectious virus in cardiac myocytes. J. Virol. 70:6709-6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherry, B., and M. A. Blum. 1994. Multiple viral core proteins are determinants of reovirus-induced acute myocarditis. J. Virol. 68:8461-8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherry, B., and B. N. Fields. 1989. The reovirus M1 gene, encoding a viral core protein, is associated with the myocarditic phenotype of a reovirus variant. J. Virol. 63:4850-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherry, B., F. J. Schoen, E. Wenske, and B. N. Fields. 1989. Derivation and characterization of an efficiently myocarditic reovirus variant. J. Virol. 63:4840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherry, B., J. Torres, and M. A. Blum. 1998. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J. Virol. 72:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spear, P. G. 2002. Viral interactions with receptors in cell junctions and effects on junctional stability. Dev. Cell 3:462-464. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi, K., H. Nakanishi, M. Miyahara, K. Mandai, K. Satoh, A. Satoh, H. Nishioka, J. Aoki, A. Nomoto, A. Mizoguchi, and Y. Takai. 1999. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J. Cell Biol. 145:539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tardieu, M., M. L. Powers, and H. L. Weiner. 1983. Age-dependent susceptibility to reovirus type 3 encephalitis: role of viral and host factors. Ann. Neurol. 13:602-607. [DOI] [PubMed] [Google Scholar]
- 59.Tardieu, M., and H. L. Weiner. 1982. Viral receptors on isolated murine and human ependymal cells. Science 215:419-421. [DOI] [PubMed] [Google Scholar]
- 60.Turner, D. L., R. Duncan, and P. W. Lee. 1992. Site-directed mutagenesis of the C-terminal portion of reovirus protein σ1: evidence for a conformation-dependent receptor binding domain. Virology 186:219-227. [DOI] [PubMed] [Google Scholar]
- 61.Tyler, K. L. 2001. Mammalian reoviruses, p. 1729-1945. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 62.Tyler, K. L., D. A. McPhee, and B. N. Fields. 1986. Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science 233:770-774. [DOI] [PubMed] [Google Scholar]
- 63.Tyler, K. L., R. J. Sokol, S. M. Oberhaus, M. Le, F. M. Karrer, M. R. Narkewicz, R. W. Tyson, J. R. Murphy, R. Low, and W. R. Brown. 1998. Detection of reovirus RNA in hepatobiliary tissues from patients with extrahepatic biliary atresia and choledochal cysts. Hepatology 27:1475-1482. [DOI] [PubMed] [Google Scholar]
- 64.Tyler, K. L., M. K. Squier, S. E. Rodgers, S. E. Schneider, S. M. Oberhaus, T. A. Grdina, J. J. Cohen, and T. S. Dermody. 1995. Differences in the capacity of reovirus strains to induce apoptosis are determined by the viral attachment protein σ1. J. Virol. 69:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Virgin, H. W., K. L. Tyler, and T. S. Dermody. 1997. Reovirus, p. 669-699. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven, Philadelphia, Pa.
- 66.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 67.Weiner, H. L., K. A. Ault, and B. N. Fields. 1980. Interaction of reovirus with cell surface receptors. I. Murine and human lymphocytes have a receptor for the hemagglutinin of reovirus type 3. J. Immunol. 124:2143-2148. [PubMed] [Google Scholar]
- 68.Weiner, H. L., D. Drayna, D. R. Averill, Jr., and B. N. Fields. 1977. Molecular basis of reovirus virulence: role of the S1 gene. Proc. Natl. Acad. Sci. USA 74:5744-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiner, H. L., M. L. Powers, and B. N. Fields. 1980. Absolute linkage of virulence and central nervous system tropism of reoviruses to viral hemagglutinin. J. Infect. Dis. 141:609-616. [DOI] [PubMed] [Google Scholar]
- 70.Williams, L. A., I. Martin-Padura, E. Dejana, N. Hogg, and D. L. Simmons. 1999. Identification and characterisation of human junctional adhesion molecule (JAM). Mol. Immunol. 36:1175-1188. [DOI] [PubMed] [Google Scholar]
- 71.Wilson, G. A. R., L. A. Morrison, and B. N. Fields. 1994. Association of the reovirus S1 gene with serotype 3-induced biliary atresia in mice. J. Virol. 68:6458-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf, J. L., D. H. Rubin, R. Finberg, R. S. Kaufman, A. H. Sharpe, J. S. Trier, and B. N. Fields. 1981. Intestinal M cells: a pathway of entry of reovirus into the host. Science 212:471-472. [DOI] [PubMed] [Google Scholar]
- 73.Yoon, M., and P. G. Spear. 2002. Disruption of adherens junctions liberates nectin-1 to serve as receptor for herpes simplex virus and pseudorabies virus entry. J. Virol. 76:7203-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zahraoui, A., D. Louvard, and T. Galli. 2000. Tight junction, a platform for trafficking and signaling protein complexes. J. Cell Biol. 151:F31-F36. [DOI] [PMC free article] [PubMed]