Abstract
Kaposi's sarcoma associated herpesvirus (KSHV)/human herpesvirus 8 (HHV-8) encodes an immediate early transcriptional activator, Rta, which mediates viral reactivation from latency and lytic viral replication. Here we report the purification and characterizations of HHV-8 Rta and its interaction with Rta-responsive DNA elements. The Rta response element (RtaRE) in the promoter of the KSHV/HHV-8 K8 open reading frame was mapped to a 47-bp sequence (RtaRE1) and a 60-bp sequence (RtaRE2) upstream of the TATA motif. A comparison of the K8 RtaREs with other viral RtaREs revealed a pattern of multiple A/T triplets spaced with a periodicity of 10 or 20 bp. Substitutions of the in-phase A/T trinucleotides of the RtaRE1 with G/C bases greatly diminished Rta responsiveness and Rta binding. By contrast, base substitutions in an out-of-phase A/T-trinucleotide sequence had no effect. Importantly, multimers of (A/T)3N7 and N5(A/T)5N6(A/T)4 motifs supported a strong Rta response in a copy number-dependent manner. No specific sequence motifs in the spacer regions could be discerned. Potent Rta response, however, was obtained with phased A/T trinucleotides with 7-bp spacers of arbitrary sequences with high G/C content. Lengthening of the phased A/T motifs or lowering of the G/C content of the spacers resulted in a reduction in Rta response. Finally, Escherichia coli-derived Rta is an oligomer of 440 kDa in molecular size and binds RtaRE as an oligomer. These results support a model of Rta transactivation wherein the subunits of the Rta oligomer make multiple contacts with a tandem array of phased A/T triplets in the configuration of (A/T)3(G/C)7 repeats.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), was first discovered in tumor tissues of Kaposi's sarcoma patients (4) and subsequently linked to several other human cancers, including an AIDS-associated malignancy called body cavity-based lymphoma or primary effusion lymphoma (2) and an atypical lymphoproliferative disorder known as multicentric Castleman's disease (23, 24). The nucleotide sequence of KSHV/HHV-8 (16, 18) shows it to be highly related to oncogenic, lymphotropic, gammaherpesviruses, such as Epstein-Barr virus (EBV) and the rhadinovirus subfamily of gammaherpesviruses, including simian herpesvirus saimiri and avian Marek's disease virus.
The regulatory circuit for KSHV/HHV-8 gene expression during the lytic cycle and reactivation from latency resembles that of EBV but with interesting differences. Based on protein sequence similarities and genome location with respect to their EBV counterparts, two putative KSHV/HHV-8 viral transcription activators, Rta and Zta (K-bZIP), encoded by open reading frame (ORF) 50 and ORF K8, respectively, have been identified (12, 13, 14, 25, 28). Like their EBV equivalents, the polypeptides for KSHV/HHV-8 Rta and Zta (K-bZIP) are derived from the splicing of a complex set of RNA transcripts (12, 25). While EBV Rta and Zta act synergistically and are both important for EBV reactivation in latently infected B cells, KSHV/HHV-8 Rta alone appears to be responsible for viral reactivation (7, 14, 15, 25).
The KSHV/HHV-8 Rta is a polypeptide of 691 amino acid residues in size. It shares protein sequence homology with the immediate early protein 2 (IE2) of the bovine herpesvirus 4 (27); with the putative transactivator, EDRF1(ORF 50), of the herpesvirus saimiri (6, 26, 28, 29); and with the EBV Rta (otherwise called BRLF1) (9, 15). The highest degree of sequence homology among these proteins lies in regions corresponding to amino acid residues 1 to 240 and 611 to 652 of KSHV/HHV-8 Rta. The sequence and functional similarities suggest structural and functional conservation for this family of proteins.
Rta is a sequence-specific DNA-binding protein that recognizes Rta-responsive DNA motifs located in the transcriptional regulatory regions of immediate early and early genes (3, 5, 10, 13, 21, 22, 30). This notwithstanding, the interaction between Rta and its responsive elements is incompletely understood. Unlike conventional sequence-specific transcription factors such as c-Jun, CREB/ATF-1, NF-κB, and the steroid hormone receptor, which bind well-defined cis-regulatory elements that are often no more than 8 to 10 bp in sequences, KSHV/HHV-8 Rta and the related bovine herpesvirus IE2 appear to interact with DNA elements that consist of longer and less-defined sequences (1, 5, 17, 19, 21, 22). Furthermore, how Rta interacts with cellular transcription factors, transcriptional coactivators, and components of the basal transcriptional machinery to augment mRNA transcription also remains unresolved.
Here, we describe the identification of a 47-bp sequence in the promoter of K8 ORF as a minimal Rta response element (RtaRE), which we termed RtaRE1. The 47-bp K8 RtaRE supports potent Rta response. Sequence comparison of the 47-bp K8 RtaRE with similar elements found in polyadenylated nuclear RNA (PAN), K12 ORF, and ORF 57 promoters revealed a consensus of triplet A/T motifs (underlined in Table 1) spaced every 7 or 17 nucleotides apart. Based on these data, another RtaRE (RtaRE2) with a similar sequence pattern was also identified in the region downstream of the 47-bp motif in the K8 promoter. Mutational analyses indicate that these phased A/T trinucleotides and, to a lesser extent, the G/C contents of the intervening spacer sequences are critical for potent Rta-mediated transactivation. We showed further that multimers of (A/T)3N7 and N5(A/T)5N6(A/T)4 motifs support strong Rta response in a copy number-dependent manner. Importantly, optimal Rta response was obtained with phased A/T trinucleotides with arbitrary 7-nucleotide spacer sequences of high G/C content. Biochemical analyses of Rta showed it to be a multisubunit, possibly hexameric DNA-binding protein. These results suggest that Rta oligomers make contacts with multiple A/T triplets repeated in phase every 10 or 20 bp in the context of G/C-rich sequences to bring about transcriptional activation.
TABLE 1.
Sequence comparison of RtaREsa
| Gene | RtaREs | Source or reference |
|---|---|---|
| ORF K8 RTA RE1 | TTTATTTTTAAC[AGTTTGGTGCAAAGTGGAGTTAACCTACAGATTCTACTTAAAATAGC]T | This study |
| ORF K8 RTA RE2 | CATTTTCTCACGAATCTGGTTGATTGTGACTATTTGTGAAACAATAATGATTAAAGGGGG | This study |
| PAN | CTTCCAA(AAATGGGTGGCTAACCTGTCCAAAA)TATG | 3, 21 |
| ORF K12 (kaposin) | GGAAATGGGTGGCTAACCCCTACACATAA | 3 |
| ORF 57 | AGCAAGTGTAACAATAATGTTCCCACGGCCCATTTTTCGTTTGTGGTACCA | 5 |
| Consensus | (A/T)3N7(A/T)3N7(A/T)3 or (A/T)3N17(A/T)3 | |
| Optimal RtaRE | [(A/T)3(G/C)7]3-6 |
A/T motifs are in bold. A/T triplets repeated every 10 or 17 nucleotides are underlined. G/C residues are preferred for the 7 or 17 nucleotides in the spacer regions between the A/T motifs. The 47-bp RtaRE sequence (top), characterized in depth here, is bracketed.
MATERIALS AND METHODS
Plasmid construction.
The coding sequence of Rta was PCR amplified from a BCBL1 cDNA library using primers 5′-AGATCTCCATGGCGCAAGATGACAAG and 5′-GAATTCTCAGTCTCGGAAGTAATTACG. The PCR product was inserted into PCR2.1 TA cloning vector (Invitrogen, Inc., San Diego, Calif.). A BglII-EcoRI fragment containing the cDNA of Rta was then purified and cloned into pcDNA3.1 vector via BamHI and EcoRI sites to obtain CMV-Rta.
A DNA fragment containing the 220-bp sequence upstream of the K8 coding region was generated by PCR by using genomic DNA of TPA-induced BCBL1 cells as a source of the DNA template together with the primer pair 5′-CTCGAGAGTGTTCGCAAGGGCGTCTG and 5′-AGATCTTTGGCAGGGTTACACGTTTA. The PCR product was first cloned into a TA cloning vector, PCR2.1, released by XhoI and BglII digestion, and inserted into a promoterless reporter plasmid, pA3Pluc, to produce RtaRE220-Luc. The RtaRE deletions were also derived by PCR by using the RtaRE220-Luc as a template. The nucleotide sequences for upstream primers are as follows: primer I, CTCGAGGTTTGGTGCAAAGTG; primer II, CTCGAGGCTCATTTTCTCACG; primer III, CTCGAGTTTGTGAAACAATAATGA; primer 1, CTCGAGGAGTTAACCTACAGAT; and primer 2, CTCGAGCTACTTAAAATAGCTC. The same downstream primer, namely, AAGCTTGGCAGGGTTACACGT, was used in all deletion constructs. Again, PCR products were inserted into pCR2.1 TA cloning vector, isolated as XhoI and HindIII fragments, and then cloned into SalI- and HindIII-digested PA3Luc vector.
Plus- and minus-strand oligonucleotides corresponding to regions a, b, c, and d of RtaRE220 were chemically synthesized. The plus strand of each region contains a 5′ BamHI protruding end, and the minus strand contains a 3′ BglII protruding end. After the plus- and minus-strand oligonucleotides had been annealed, the double-stranded DNAs were phosphorylated at the 5′ end with T4 polynucleotide kinase (New England Biolabs). They were then ligated in tandem, digested with BamHI and BglII restriction endonucleases exhaustively, and inserted into the BamHI site of the pUC19 vector. Plasmid clones that harbor oligonucleotide inserts were identified by PCR with the universal pUC primer pair. The copy number of each oligonucleotide insert was determined by polyacrylamide gel electrophoresis (PAGE) of the PCR products against appropriate DNA markers. SalI/XmaI fragments containing multimers of RtaRE regions a to d were inserted into an enhancerless reporter plasmid, pΔE-RSV-luc, via the same sites.
To alter the non-A/T bases in the K8 ORF promoter region randomly, a 90-mer oligonucleotide, 5′-CTAGGGTACCATCCCGGGTTTATTTTTAANNNTTTNNNNAAANNGNNNTTAANNNNNNNATTNNNCNNAAAATAGTCGACAGATCTGTCA-3′ (N represents a mixture of all four bases equally represented in the reaction), flanked by KpnI and SmaI restriction endonuclease sequences in the 5′ end and SalI and BglII sequences in the 3′ end, was generated by chemical synthesis. Double-stranded DNA was obtained by primer extension by using the 90-mer as a template and a primer of the sequence 5′-TGACAGATCTGTCGACTA-3′. The double-stranded DNA was then digested by XmaI/SalI and inserted into similarly digested pΔE-RSV-luc plasmid. Thirty clones were picked up and analyzed for response to Rta-mediated transactivation. Results from five representative clones are shown. Mutagenesis of the A/T motifs in the 47-bp RtaRE was preformed similarly. The sequences of the wild-type and mutant (M1 to M5) oligonucleotides used are 5′-CTAGGGTACCATCCCGGGTTTATTTTTAACAGTTT(CCC for M1)GGTGCAAA(GGG for M2)GTGGAGTTAA(CGG for M3)CCTACAGATT(GCC for M4) CTACTTAAAA(GGG for M5) TAGTCGACAGATCTGTCA-3′, with the mutated bases underlined and the respective substitutions (C for T and G for A in M1 to M5) in parentheses. Complementary oligonucleotide pairs for Con1, Con2, and RtaRE2 sequences were made to contain 5′-phosphorylated SmaI/SalI, BglII/BamHI, and SmaI/SalI protruding ends, respectively. Con1 sequences 5′-CCGGGAAAGCGACCTTAAGCGACCGATTGCGACCGAAAGCGACCG-3′ (plus strand) and5′-TCGACGGTCGCTTTCGGTCGCAATCGGTCGTTAACGGTCGCTTTC-3′ (minus strand) and RtaRE2 sequences 5′-CCGGGCATTTTCTCACGAATCTGGTTGATTGTGACTATTTGTGAAACAATAATGATTAAAGGGG-3′(plus strand) and 5′-TCGACCCCTTTAATCATTATTGTTTCACAAATAGTCACAATCAACCAGATTCGTGAGAAAATGC-3′ (minus strand) were annealed by incubation in a water bath at 70°C and allowed to cool to 40°C over a period of 45 min. The double-stranded Con1 DNA was then inserted directly into pΔE-RSV-luc via SmaI/SalI sites. Reporter plasmids containing tandem repeats of the Con2 sequence, namely, 5′-GATCTAAAAGTCGACAAAAG-3′ (plus strand) and 5′-GATCCTTTTGTCGACTTTTA-3′(minus strand), were generated similarly, as described earlier for the constructs of regions a to d. The sequences of all constructs were confirmed by DNA sequencing.
Cell lines.
Human embryonic kidney 293 (HEK293) cells were grown in Dulbecco's modified Eagle medium supplemented with 100 IU of penicillin/ml, 0.1 mg of streptomycin (STR)/ml, and 10% heat-inactivated fetal bovine serum and cultured at 37°C in a humidified incubator with 5% CO2.
Transfection and luciferase assays.
Lipofectamine (GIBCO BRL) was used in DNA transfection of HEK293 cells according to the manufacturer's protocol. Briefly, cells were plated at a density of 1.0 × 105 cells/well in 12-well plates 1 day before transfection. For each well, 0.5 μg each of CMV-Rta or Rta-null pcDNA3.1 and 0.5 μg of the reporter plasmid were used. To control for variability in transfection, 0.1 μg of pRL-TK containing an HSV thymidine kinase (TK) promoter-driven Renilla luciferase reporter cassette (Promega) was included as an internal control. Cells were harvested 48 h after DNA transfection, and the firefly luciferase activity was assayed and normalized against Renilla luciferase activity. For luciferase assays, cell lysates were prepared by dissolving the DNA-transfected cells from each well in 200 μl of the reporter lysis buffer (Promega). Twenty microliters of the lysate was placed in each well of a 96-well plate. After injection of 100 μl of a luciferase substrate buffer, the luciferase activity was measured immediately in an MLX microtiter plate luminometer (DYNEX Technologies).
Immunofluorescence analysis.
Forty-eight hours after DNA transfection, cells on coverslips were fixed with 4% paraformaldehyde for 10 min on ice, followed by incubation with 0.1% Triton X-100 solution for 10 min on ice. The coverslips were washed with phosphate-buffered saline (PBS) three times, immersed in 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature, and incubated overnight at 4°C with a rabbit polyclonal antibody against Rta (1:500) in the same buffer. After five washes with PBS, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (Sigma; 1:10,000) in PBS containing 3% BSA was applied for 1 h at room temperature. Cells were washed again with PBS five times, then mounted with Fluoromount-G (Southern Biotechnology Associates, Inc., Birmingham, Ala.) solution containing 0.5 μg of 4′-6-diamidine-2-phenylindole (DAPI)/ml for fluorescence microscopy. Photographs were taken with appropriate filters and images overlaid in a computer to create two-color images.
Expression and purification of KSHV/HHV-8 Rta.
The complete coding sequence of Rta was inserted into an expression vector, pTrc2His2-TOPO (Invitrogen), whose product is tagged with a c-Myc epitope and a hexahistidine extension at the COOH terminus, to produce pTrc2His2-TOPO-Rta. Competent Escherichia coli BL21 cells harboring the pTrc2His2-TOPO-Rta plasmid were grown in 10 ml of Luria-Bertani (LB) broth containing 100 μg of ampicillin/ml overnight at 37°C. The overnight culture was used to inoculate 1 liter of LB broth containing 100 μg of ampicillin/ml, and the culture was incubated for 3 to 4 h until it reached an A600 of 0.8 to 1.0. At this point, Rta expression was induced for 6 to 7 h with 1 mM isopropylthio-β-d-galactoside (IPTG) at 37°C with shaking. The Rta-expressing cells were then harvested by centrifugation and resuspended in 10 ml of lysis buffer (1 M NaCl, 10 mM imidazole, 25 μM phenylmethylsulfonyl fluoride [PMSF] in PBS buffer [pH 7.4]). After sonication with a microtip at 70% duty cycle for four bursts of 60 s each, the cell suspension was centrifuged in a Sorvall SS-34 rotor at 12,000 rpm for 30 min at 4°C to pellet unbroken cells and debris. The supernatant was then incubated with 1 ml of Ni2+-nitrilotriacetic acid (NTA) resin (Qiagen) overnight at 4°C. The protein-bound gel matrix was packed into a column (1.5 by 10 cm) and washed with 40 volumes of the same buffer containing 20 mM imidazole. Rta protein was then eluted with a 10-ml gradient of 40 to 160 mM imidazole. Proteins in each fraction were analyzed by sodium dodecyl sulfate (SDS)-12% PAGE followed by immunoblotting with an anti-c-Myc antibody (Invitrogen). Fractions containing Rta were dialyzed against buffer D (20 mM HEPES [pH 7.9], 150 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM dithiothreitol [DTT], 20% glycerol) and stored frozen at −70°C.
Gel electrophoretic mobility shift assay (EMSA).
A 116-bp EcoRI/HindIII fragment containing the 47-bp RtaRE from pUC19-RtaRE (see Fig. 4 and 5) and a 69-bp XmaI/SalI fragment containing the mutant or wild-type RtaRE derived from the corresponding ΔE-RSV-luc constructs (see Fig. 7) were labeled with [α-32P]dATP and [α-32P]dCTP, respectively, by Klenow enzyme, resolved in a 6% polyacrylamide gel, and isolated. Protein-DNA binding reactions were carried out as described previously with minor modifications (20, 31). Typically, each binding reaction was in 20 μl, in a binding buffer containing 10 mM HEPES [pH 7.9], 40 mM KCl, 5 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol, 0.5 mg of BSA/ml, and 0.1 mM EDTA. Each reaction mixture contained 2 μl of α-32P-labeled DNA (∼5 ng) and 5 μl (∼0.1 μg) of Rta protein in the presence of other DNA competitors in indicated amounts and was incubated at room temperature for 1 h. A rabbit antibody generated against a peptide containing amino acid residues 527 to 539 (KKRKALTVPEADT) of Rta (a generous gift of Gary Hayward) was used in the supershift experiment. Reaction mixtures were electrophoresed at 35 mA in a 5% nondenaturing polyacrylamide gel (30:1 acrylamide to bisacrylamide) in Tris-glycine-EDTA buffer at 4°C for 1.5 h. The gel was then dried on a piece of Whatman filter paper and autoradiographed.
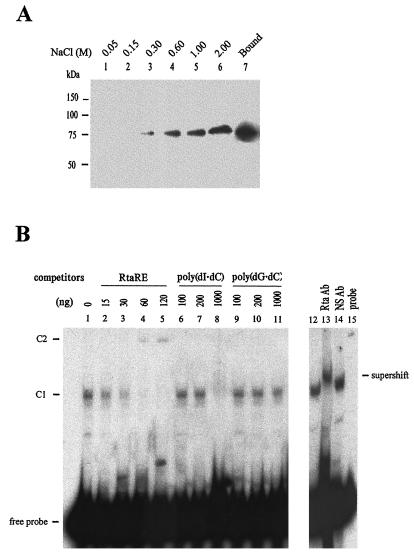
FIG. 4.
Rta binds RtaRE specifically. (A) Rta is a DNA-binding protein. Fifty microliters of partially purified Myc- and hexahistidine-tagged Rta was incubated with 0.5 ml of double-stranded DNA cellulose and washed three times with a buffer containing 100 mM NaCl, 20% (vol/vol) glycerol, 5 mM MgCl2, 0.1 mM EDTA, and 20 mM HEPES. The DNA cellulose-bound proteins (lane 7) were eluted with 0.5 ml of the same buffer solution containing increasing concentrations of NaCl, as indicated (lanes 1 to 6). The eluted fractions were immunoblotted with an antibody against c-Myc. (B) Rta forms a protein/DNA complex with RtaRE. EMSAs were carried out as described in Materials and Methods by using partially purified Rta (Fig. 3B) and a 32P-labeled DNA fragment (1xRtaRE) containing a single copy of the 47-bp minimal RtaRE. DNA competitors that included unlabeled RtaRE DNA fragment (lanes 2 to 5), poly(dI-dC) (lanes 6 to 8), and poly(dG-dC) (lanes 9 to 11) were added in the amounts indicated to binding reactions. C1 denotes the Rta/RtaRE complex. C2 denotes a slower-migrating protein/DNA complex in reactions where excess unlabeled RtaRE was used as a competitor (lanes 4 and 5). Lanes 13 and 14 contained an antibody specific for Rta (Rta Ab) and a control antibody against the E. coli maltose binding protein (NS Ab), respectively.
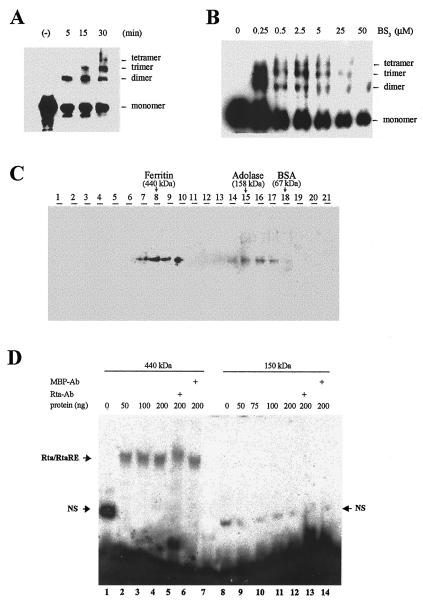
FIG. 5.
Rta is an oligomeric DNA-binding protein. (A and B) Chemical cross-linking of Rta. Purified E. coli-derived Rta was chemically cross-linked with BS3 (see Materials and Methods for details) at room temperature with increasing time (A) or amount of BS3 (B), resolved in 4 to 20% polyacrylamide gradient gels, and analyzed by immunoblotting with the c-Myc antibody. Cross-linked Rta species with molecular sizes corresponding to different oligomers are as indicated. (C) Sucrose density gradient sedimentation analysis of Rta. Partially purified Rta protein, together with a protein standard containing ferritin (440 kDa), adolase (158 kDa), and BSA (67 kDa), was applied over a sucrose gradient and centrifuged in a Beckman SW40Ti rotor at 32,000 rpm at 4°C for 20 h. The peak positions (lanes 8, 15, and 18) of the protein markers in the gradient are indicated. (D) The oligomeric form of Rta is responsible for RtaRE binding. The gradient fractions corresponding to the 440- and 150-kDa species were pooled separately, concentrated by Microcon YM-30 filtration, and assayed for RtaRE binding (lanes 1 to 6 and 8 to 14, respectively) as described in the legend for Fig. 4B. Increasing amounts of 440- and 150-kDa Rta were used in lanes 2 to 5 and 9 to 12, respectively. Lanes 1 and 8 contain free probe in the binding buffer. Lane 7 contains the free probe only. Rta antibody was included in reactions in lanes 5 and 13. An antibody against the E. coli maltose binding protein (MBP-Ab) was included in lanes 6 and 14 as a nonspecific antibody control. The Rta/RtaRE complex is as indicated. A fast-migrating nonspecific band (NS) is as noted.
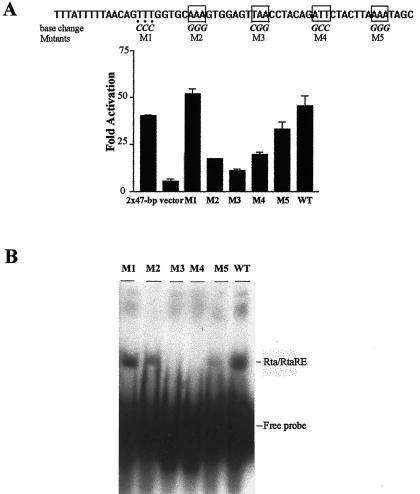
FIG. 7.
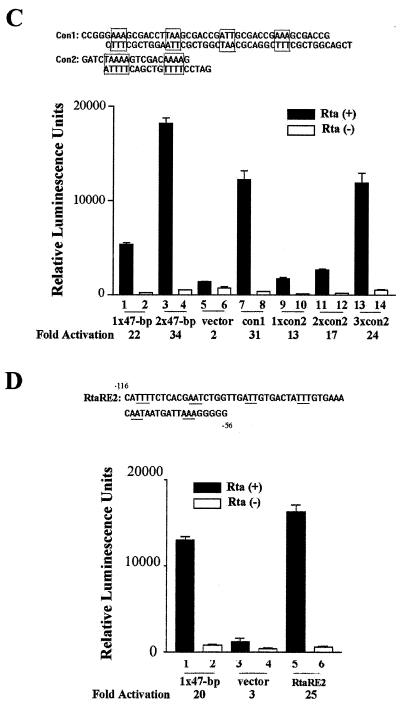
Phased A/T triplets flanked by G/C-rich spacers are critical for potent Rta transactivation. (A) Multiple phased A/T triplets are critical for Rta response. Six 90-mer oligonucleotides (wild-type and M1 to M5) that contain the sequence of the 47-bp K8 RtaRE (bracketed) and 12 additional upstream nucleotides were generated by chemical synthesis, as described in the legend for Fig. 6. Each of the oligonucleotides M1 to M5 contains three specific base substitutions, with the A and T bases replaced with G and C residues, respectively (indicated below the sequence). After primer extension, the double-stranded DNA was then inserted into pΔE-RSV-luc and confirmed by DNA sequence analyses. The constructs were then cotransfected with CMV-Rta into 293 cells, as described in the legend for Fig. 1. Activation of each construct was calculated and is shown. A pΔE-RSV-luc-derived reporter containing two copies of the 47-bp RtaRE (2×47-bp) (same as 2×d in Fig. 2) and pΔE-RSV-luc (vector) were included as controls. Phased A/T motifs critical for Rta response are in boxes, and the nonessential T trinucleotide is marked with dots beneath it. (B) Rta-binding activities of mutant RtaREs. EMSAs were as described in the legend for Fig. 4B, except that the various wild-type and mutant RtaREs were 69 bp in length. Wild-type and mutant RtaREs were labeled to similar specific activities, as in described in Materials and Methods. Approximately the same amount of DNA probe was used in each reaction. Possibly because of the decreased size of the DNA probe, the Rta/DNA complexes migrated faster than the complex in Fig. 4B. (C) Phased A/T triplets with G/C-rich heptanucleotide spacers support potent Rta transactivation. Complementary oligonucleotide pairs for Con1 and Con2 sequences were made to contain 5′-phosphorylated SmaI/SalI and BglII/BamHI protruding ends, respectively. A/T motifs are boxed. Con1 sequence was inserted directly into pΔE-RSV-luc via SmaI/SalI sites. Luciferase reporter constructs containing the indicated numbers of Con2 sequence were produced as described in Materials and Methods and characterized. The Rta response of each construct as determined by firefly luciferase activity was measured as described above and marked as appropriate (lanes 7 to 14). All firefly luciferase reporter assays were normalized against TK-Renilla luciferase reporter activities, as described in the legend for Fig. 1. The 1×47-bp, 2×47-bp, and pΔE-RSV-luc empty vector controls (lanes 1 and 2, 3 and 4, and 5 and 6, respectively) are as described in the legend for panel A. (D) RtaRE2 supports strong Rta transactivation in ΔE-RSV-luc. A complementary oligonucleotide pair of RtaRE2 with 5′-phosphorylated SmaI/SalI sites was inserted directly into pΔE-RSV-luc and analyzed for Rta response. A reporter containing the 47-bp RtaRE1 sequence was included for comparison. Luciferase assays were as described in the legend for panel C.
Sucrose gradient centrifugation.
Rta protein (100 μl, ∼2 μg), together with 30 μl of a protein standard containing ferritin (440 kDa), adolase (158 kDa), and BSA (67 kDa) at a concentration of 4 mg/ml each, was gently layered on top of a 10-ml sucrose gradient generated in a 12-ml ultracentrifuge tube by the freezing and thawing of a 15% sucrose solution containing 300 mM ammonium acetate, 1 mM DTT, and 1× protease inhibitor cocktail (Roche Molecular Biochemicals). Ultracentrifugation was carried out in a Beckman SW40Ti rotor at 32,000 rpm at 4°C for 20 h. Five hundred-microliter fractions were collected from the bottom of the centrifuge tubes after puncturing with a needle. Protein fractions were lyophilized, resuspended in 50 μl of SDS-PAGE loading buffer, and resolved in a 4 to 20% SDS-polyacrylamide gradient gel. Coomassie blue staining and immunoblotting were used to identify protein markers and Rta, respectively.
Chemical cross-linking.
Bis-sulfosuccinimidyl suberate (BS3), a water-soluble, homobifunctional chemical cross-linker, was used in protein cross-linking studies. Purified wild-type Rta (approximately 0.2 μg in 10 μl) was incubated at room temperature with 1 μl of freshly prepared BS3 at different concentrations (0.25, 0.5, 2.5, 5, 25, and 50 mM) for 30 min or at 0.5 mM for different times (5 to 30 min). The reaction was quenched by adding 1 μl of 1 M Tris buffer (pH 8.0) followed by incubation at room temperature for 15 min. The samples were then mixed with SDS-PAGE loading buffer containing 100 mM β-mercaptoethanol, heated at 100°C for 5 min, resolved in 4 to 20% SDS-PAGE gels, and analyzed by immunoblotting with the Myc epitope antibody.
RESULTS
Characterizations of a KSHV/HHV-8 RtaRE.
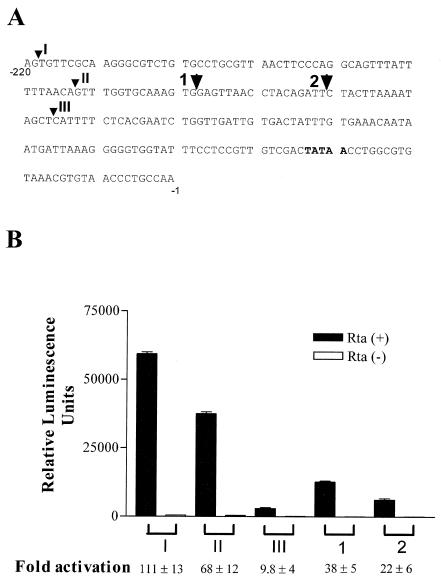
KSHV/HHV-8 Rta (referred to as Rta herein) potently activates the transcription of KSHV/HHV-8 K8 ORF and several other KSHV/HHV-8 early genes (5, 10, 13, 14, 21, 22, 30). To study the interaction between Rta and its response elements, we generated a 220-bp DNA fragment containing the sequence that lies immediately upstream of the K8 coding region by PCR and fused it to a promoterless firefly luciferase reporter plasmid, pA3Pluc-FL, to produce RtaRE220-Luc. To determine if the 220-bp sequence can mediate transactivation by Rta, HEK293 cells were cotransfected with RtaRE220-Luc and an Rta expression plasmid, CMV-Rta, in which the Rta cDNA was placed under the control of the human cytomegalovirus immediate early enhancer/promoter. As expected, under these conditions, CMV-Rta induced a greater than 100-fold increase in gene expression compared to the Rta-null, pcDNA3.1 vector control (Fig. 1A and B), indicating that the 220-bp region contains DNA elements that mediate a potent Rta response. To delineate further the cis element responsible for Rta transactivation, four 5′ deletion mutants (deletion mutants II, III, 1, and 2) in the 220-bp sequence were constructed, and the ability of each mutant to mediate transactivation by Rta was determined after cotransfection with CMV-Rta. Deletion mutant II, lacking 57 bp in the 5′ region of the 220-bp fragment, showed approximately 70 to 80% of the activity of the full-length fragment. A deletion of 104 bp (deletion mutant III) greatly reduced the absolute level of Rta-mediated transcription and the relative level of transactivation (factor of activation) that compares the promoter activity in the presence or absence of Rta (Fig. 1, compare II and III). Smaller deletions (constructs 1 and 2) in the sequence spanning nucleotides −163 and −117 also diminished the overall promoter activity but diminished the activation levels to a lesser extent. Approximately 6- to 10-fold transactivation could still be observed with deletion construct III, although on the whole, the promoter was largely inactive. These results suggest that multiple cooperating Rta-responsive regions reside in the 220-bp sequence.
FIG. 1.
Identification of RtaREs in K8 promoter. (A) DNA sequence of 220-bp upstream of the K8 ORF. A DNA fragment containing a 220-bp sequence immediately upstream of the K8 coding region and four nested 5′ deletions (II, III, 1, and 2) were generated by PCR with appropriate primers (see Materials and Methods) and then fused to a promoterless firefly luciferase reporter plasmid, pA3Pluc-FL, to produce RtaRE220-Luc and four other luciferase reporter plasmids. Arrows mark the 5′ end of each construct. (B) Localization of RtaREs. HEK293 cells were cotransfected with luciferase reporter plasmid and an Rta expression plasmid, CMV-Rta, as described in Materials and Methods. At least three independent sets of luciferase assays, each with triplicate DNA transfection, were performed (a total of nine transfections) for each plasmid construct. All firefly luciferase reporter assays were normalized against HSV TK promoter-driven Renilla luciferase reporter activities. Standard deviations of the reporter activities and the extent of transactivation by Rta are also shown.
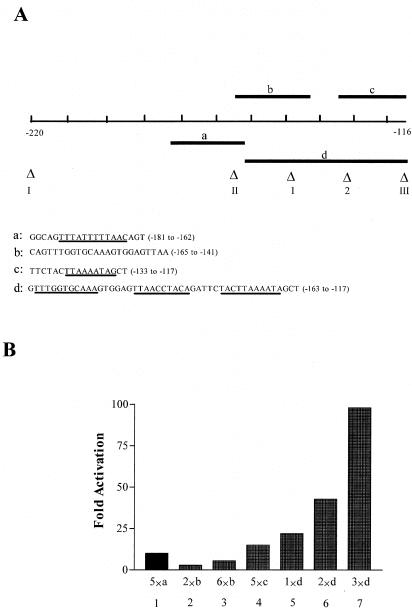
In an effort to locate the minimal RtaRE, we derived synthetic oligonucleotides that cover different parts (Fig. 2A, a to d) of the −181 to −117 regions. These oligonucleotides were ligated as tandem multimers and inserted into a reporter plasmid, pΔE-RSV-luc, which contains an enhancer-less promoter, derived from the long terminal repeat of the Rous sarcoma virus, upstream of the firefly luciferase reporter gene. This promoter extends 142 nucleotides upstream of the Rous sarcoma virus mRNA start site, retains elements that are potential NF-Y and C/EBP binding sites, and has a relatively low basal level activity. As show in Fig. 2, only the 47-bp sequence in region d supported a potent Rta response. Further, the levels of Rta transactivation increased significantly with an increase in the copy number of the 47-bp sequence (Fig. 2B, lanes 5 to 7). Tandem copies of sequences that contain the TTTATTTTTAA (region a) or the TTAAAATA (region c) motifs showed reduced, albeit detectable, response to Rta (Fig. 2B, lanes 1 and 4), while multimers of the oligonucleotide containing the GTTAA sequence (region b) showed lower Rta response (Fig. 2B, lanes 2 and 3). These results suggest that the 47-bp sequence corresponding to region d contains a minimal RtaRE, and subregions of this element support only partial or reduced Rta response. Finally, multimers of the 47-bp sequence promote potent Rta-mediated transactivation.
FIG. 2.
Mapping of minimal RtaRE. (A) The sequences of the oligonucleotides (a to d) covering different parts of the −181 to −116 region are as indicated, with the A/T-rich regions underlined. (B) These oligonucleotides were synthesized with 5′ BamHI and 3′ BglII protruding ends. They were ligated as tandem multimers and inserted upstream of the TATA box of an enhancerless promoter in a reporter plasmid, pΔE-RSV-luc (see Materials and Methods). DNA transfections were carried out with or without CMV-Rta, and luciferase assays were as described in the legend for Fig. 1. The copy numbers, ranging from 1 (1×) to 6 (6×), of the individual sequences tested (a to d) are indicated below each column.
Partial purification and characterizations of Rta derived from E. coli.
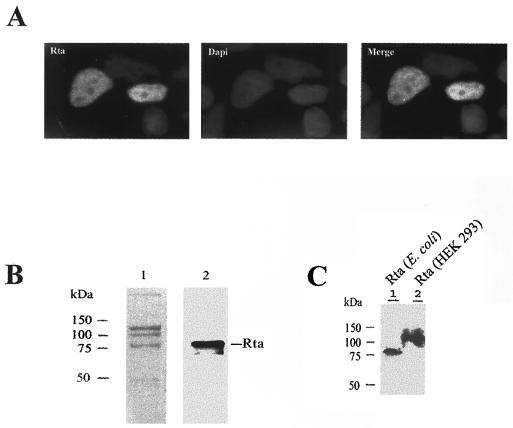
Several gammaherpesviruses encode Rta-like proteins. Earlier studies have indicated that Rta is a nuclear protein that functions as a sequence-specific DNA-binding transcription factor (5, 14, 19, 21, 22). The molecular details of how Rta interacts with its target DNA element remain incompletely understood. Consistent with a role of Rta in transcriptional activation, an immunofluorescence assay revealed that Rta localized to the nucleus in a diffuse staining pattern (Fig. 3A). To derive and purify Rta protein in sufficient quantity for biochemical studies, we expressed the full-length Rta with a c-Myc epitope tag and a hexahistidine extension in E. coli. The recombinant Rta protein was then partially purified by Ni2+-NTA Sepharose affinity chromatography and detected by Coomassie blue staining and immunoblotting (Fig. 3B, lanes 1 and 2, respectively). The E. coli-derived Rta is approximately 74 kDa in size, in agreement with its predicted size based on amino acid sequence (Fig. 3C, lane 1). The Rta expressed in BCBL-1 cells, however, has been reported to have an apparent size that is much greater (14). To compare the Rta derived from different expression systems, we expressed it in 293 cells after DNA transfection. As shown in Fig. 3C, the Rta expressed in 293 cells after DNA transfection has an apparent molecular size of 120 kDa, significantly larger than its E. coli-derived counterpart (Fig. 3C, lane 2), suggesting that it is posttranslationally modified as previously described (14).
FIG. 3.
Characterizations of Rta. (A) Rta is a nuclear protein. Immunofluorescence was carried out as detailed in Materials and Methods by using rabbit anti-Rta antibody (Rta). Cells were also stained with DAPI (Dapi). (B) Coomassie blue staining (lane 1) and immunoblot (lane 2) of partially purified E. coli-derived Rta. (C) Immunoblot analysis of E. coli-derived Rta (lane 1) and Rta expressed in 293 cells (lane 2).
Rta/RtaRE interaction.
To determine if Rta is a DNA-binding protein, we incubated the partially purified Rta with double-stranded DNA cellulose and eluted the bound protein with buffer solutions containing increasing concentrations of NaCl (Fig. 4A). Consistent with the notion that Rta is a DNA-binding protein, the E. coli-derived Rta bound tightly to double-stranded DNA cellulose and eluted preferentially with buffer solutions that contained NaCl at 0.6 M or greater concentrations (Fig. 4A, lanes 4 to 6). The interaction between Rta and RtaRE was examined by EMSA using the partially purified Rta protein and a p32-labeled DNA fragment (1xRtaRE) containing a single copy of the 47-bp minimal RtaRE. As shown in Fig. 4B, Rta bound to RtaRE to produce a specific protein/DNA complex, C1 (Fig. 4B, lane 1). An unlabeled RtaRE-containing DNA fragment competed specifically for Rta binding (Fig. 4B, lanes 1 to 5). By contrast, nonspecific competitors such as poly(dG-dC) did not compete with RtaRE for Rta binding even when added in 200-fold molar excess (1,000 ng) of the probe (lanes 9 to 11). Interestingly, poly(dI-dC) competed with RtaRE when used in excess (lane 8). Likewise, poly(dA-dT) also competed for Rta binding (data not shown). Inclusion of a specific antibody generated against amino acid residues 527 to 539 (KKRKALTVPEADT) of Rta in the binding reaction resulted in a supershift (lane 13), while addition of a nonspecific control antibody had no effect (lane 14), indicating that Rta is present in the complex. Unexpectedly, a slower-migrating protein/DNA complex, C2, was often detected in Rta EMSA when excess unlabeled RtaRE was used as a competitor (lanes 4 and 5).
Rta is an oligomeric DNA-binding protein.
The appearance of the C2 complex in EMSA prompted us to consider the possibility that native Rta may be oligomeric and comprise multiple RtaRE-binding sites such that extra copies of RtaRE DNA may be incorporated into holo-Rta to produce the C2 complex. To determine if Rta is oligomeric, both chemical cross-linking and sucrose gradient centrifugation were carried out. Purified Rta protein was incubated at room temperature with increasing concentrations of a chemical cross-linker, BS3, for 30 min or with 0.5 μM BS3 for increasing duration. The cross-linked Rta was then resolved in 4 to 20% polyacrylamide gradient gels (Invitrogen) and analyzed by immunoblotting with the c-Myc antibody. As shown, protein species with molecular sizes that correspond to Rta dimer, trimer, and tetramer appeared in increasing abundance that correlated with the duration of incubation (Fig. 5A) and the concentration of BS3 used (Fig. 5B). Minor bands suggestive of larger Rta species were also seen, but they could not be adequately resolved in the gel, and their abundances were low, possibly because of the inefficiency of the cross-linker to covalently join all subunits and the difficulty in transferring high-molecular-size protein species to a nitrocellulose membrane. Consistent with the notion that Rta is an oligomer, sucrose gradient centrifugation further revealed the presence of a major Rta species with a Stokes radius slightly smaller than that of ferritin (440 kDa) and a minor species slightly smaller than adolase (158 kDa) (Fig. 5C). Judging from the predicted molecular size of 74 kDa for Rta, it appears that the major Rta species may be a hexamer and that the minor species may be a dimer. To demonstrate that the oligomeric form of Rta is the protein species responsible for RtaRE binding, the gradient fractions containing the 440- and 150-kDa species were separately pooled and concentrated by filtration through a Microcon centrifugal filter unit (molecular weight cutoff, 10,000; Microcon YM-10, Millipore Corp.). The sucrose-containing buffer in which these protein fractions were kept was also exchanged with an appropriate storage buffer (buffer D) following filtration and concentration. The protein preparations were then assayed for RtaRE-binding activity by EMSA. As shown in Fig. 5D, the 440-kDa species, but not the 150-kDa species, bound the RtaRE probe to produce a protein/DNA complex that is indistinguishable from the unfractionated Rta preparation (Fig. 4B). Rta antibody supershift further indicates that this complex contains Rta. These results show that each native Rta molecule most likely exists as a 440-kDa hexamer and possibly contains multiple DNA binding sites as a result.
A/T nucleotides are critical for mediating Rta response.
The oligomeric structure of Rta suggests that one possible way by which it interacts with DNA is via binding to multiple short nucleotide sequence motifs spaced over arbitrary intervening sequences. This would explain why a consensus Rta-binding motif is not immediately apparent. Assuming that Rta is a hexamer or a trimer of dimers, a complete RtaRE may consist of three or six Rta-binding motifs. In view of these results, we compared the DNA sequences of the 47-bp K8 RtaRE with RtaREs of PAN, K12, and ORF 57 promoters previously reported in the literature (3, 5, 21). As shown in Table 1, multiple stretches of A/T motifs that range from 3 to 11 residues long can be identified in the RtaREs of all four promoters. Furthermore, a sequence periodicity of (A/T)3N7(A/T)3N7(A/T)3N7—i.e., triplet A/T motifs (underlined in Table 1) spaced 7 or 17 nucleotides apart—can be discerned, with the RtaREs of K8 and PAN showing the greatest conformity to this pattern and the RtaRE of K8 containing the highest number of repeats.
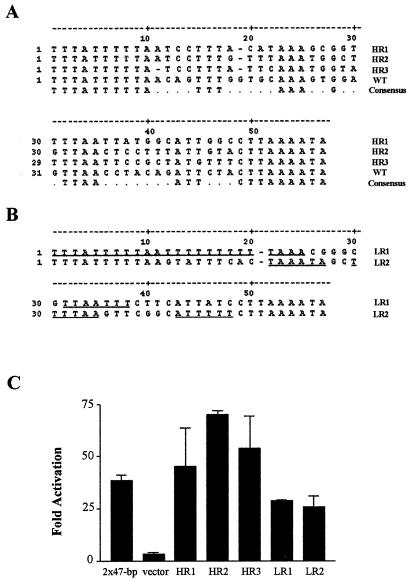
To test if the A/T motifs in the K8 ORF promoter mediate RtaRE, we generated a 90-mer oligonucleotide that contained the sequence of the 47-bp RtaRE and 12 additional upstream nucleotides flanked by KpnI and SmaI restriction endonuclease sequences in the 5′ end and SalI and BglII sequences in the 3′ end. During chemical synthesis of the 90-mer, the A/T bases of the K8 ORF promoter sequence were kept unchanged while most of the G/C bases except for two (Fig. 6A, positions 28 and 49) were replaced with 25% each of A, T, G, and C bases. The 90-mer was then made double stranded by using a primer that annealed to the 3′ sequence corresponding to the SalI and BglII sites. The duplex DNA was then inserted into pΔE-RSV-luc via SmaI and SalI sites. Multiple plasmid clones were picked, sequenced, and cotransfected with CMV-Rta to determine their abilities to support Rta-mediated transactivation. Three highly Rta-responsive (HR) clones and two clones of lower Rta responsiveness (LR) (Fig. 6A and B)—with levels of Rta transactivation varying from 20- to 100-fold—were sequenced and compared (Fig. 6C). The DNA sequences of all five clones conformed to the original 90-mer design, with the A/T-rich sequences largely unchanged expect for one base at position 19, which was fortuitously omitted during the synthesis. This single-base deletion occurred within an in-phase A trinucleotide and an out-of-phase T trinucleotide (see below) and did not affect the activity of the element, possibly because it did not alter the phasing of the critical A/T motifs (see below). The DNA sequences outside of the A/T motifs in the HR clones deviated significantly from that of the original K8 RtaRE, indicating that they are not absolutely critical for Rta response. The most obvious difference between the HR and LR clones resides in the presence of exceedingly long runs of A/T in the LR clones (Fig. 6B, underlined), suggesting that while A/T bases are important, they need to be kept at a specific length for optimal Rta response.
FIG. 6.
A/T nucleotides are critical for Rta response. (A and B) DNA sequences of chemically synthesized RtaREs. A 90-mer oligonucleotide that contains the sequence of the 47-bp RtaRE and 12 additional upstream nucleotides flanked by KpnI and SmaI sites in the 5′ end and SalI and BglII sites in the 3′ end was generated by chemical synthesis with the A/T bases kept unchanged and most of the G/C bases except for two (positions 28 and 49) replaced with 25% each of A, T, G, and C bases. After primer extension, double-stranded nucleotides were inserted into pΔE-RSV-luc. Multiple clones were sequenced and screened for Rta responsiveness after DNA transfection and reporter assays. The sequences of five representative clones, of which three (HR1 to HR3) (A) were highly responsive to Rta and two (LR1 and LR2) (B) showed lower response to Rta, are compared with the original sequence in the K8 ORF promoter (WT). The consensus sequence contains the unaltered bases of the original oligonucleotide. Long runs of A/T bases in LR clones are underlined. (C) Rta responses of HR and LR sequences. HEK293 cells were cotransfected with an Rta expression plasmid, CMV-Rta, and individual luciferase reporter plasmids containing the HR and LR sequences, as described in the legend for Fig. 1. Reporter activities of each construct with or without Rta were measured, and activation was computed. Standard deviations of activation are also shown.
The RtaRE consists of phased A/T triplets, quadruplets, or quintets spaced with a periodicity of 10 or 20 bp.
To demonstrate further the importance of the A/T motifs in the (A/T)3N7 repeats, we replaced both in-phase (M2 to M5) and out-of-phase A/T motifs (M1) found in the K8 RtaRE with G/C residues and at the same time kept the remainder of the sequence unchanged. Each mutant element was then inserted into the pΔE-RSV-luc reporter and tested for its response to Rta. In keeping with the notion that phased A/T trinucleotides are critical for a strong Rta transactivation, G/C substitutions of the in-phase A/T motifs greatly reduced Rta responsiveness (Fig. 7A, M2 to M5), while the out-of-phase TTT trinucleotide could be altered without significant loss of Rta response (Fig. 7A, M1). As expected, the reporter activity of each mutant RtaRE sequence in response to Rta (Fig. 7A) also correlated with its strength of Rta binding (Fig. 7B).
The importance of the phased A/T trinucleotides is shown further by using a consensus sequence, Con1 (AAAGCGACCTTAAGCGACCGATTGCGACCGAAAGCGACCG). Con1 contains four phased A/T nucleotides (underlined) whose sequences are identical to those in the 47-bp K8 RtaRE. However, unlike the 47-bp RtaRE, the A/T motifs in Con1 are placed adjacent to arbitrary heptanucleotide sequences of high G/C content. As shown in Fig. 7C, when inserted into pΔE-RSV-luc, Con1 supported potent Rta-mediated transactivation, whose level significantly exceeded that of a solo 47-bp K8 RtaRE (compare lanes 1 and 7).
With the idea that RtaRE consists of in-phase A/T motifs in mind, we constructed luciferase reporters that contain tandem copies of the N5T(A)4N6(A)4 motif sequence GGATCTAAAAGTCGACAAAA (termed Con2) and tested their abilities to mediate Rta response. As indicated in Fig. 7C, weak but significant Rta responses (especially in transactivation) were seen with single and double Con2 constructs (1xCon2 and 2xCon2, lanes 9 and 10 and lanes 11 and 12), while a potent Rta response, with activity comparable to that of Con1 (lane 7) and exceeding that of the 47-bp K8 RtaRE (lane 1), was seen with triple Con2 (3xCon2, lanes 13 and 14). A close inspection of the DNA sequence in deletion construct III (Fig. 1) also revealed a stretch of sequence (−116 to −56) that conforms to the [(A/T)3(G/C)7]3-6 pattern. This element (termed RtaRE2), when alone in the context of the K8 promoter, had a relatively modest response to Rta (Fig. 1, deletion III). However, when inserted into the pΔE-RSV-luc vector, it supported strong Rta transactivation (Fig. 7D). In aggregate, these data indicate that the K8 promoter contains two RtaREs. These two elements act synergistically to promote Rta-mediated mRNA transcription. When alone, each of these two elements responds moderately to Rta. However, when inserted upstream of a relatively robust promoter, such as that derived from the Rous sarcoma virus long terminal repeat, potent Rta response can be achieved. Finally, these data support the notion that Rta preferentially interacts with multiple, phased A/T triplets and, to a lesser extent, A/T quadruplets and quintets with G/C-rich intervening spacer sequences to promote strong transactivation.
DISCUSSION
Immediate early proteins that exhibit sequence and functional similarity to KSHV/HHV-8 Rta are common among herpesviruses; the R transcriptional activator encoded by the EBV BRLF1 gene and similar proteins encoded by bovine herpesvirus (BHV-4 IE2) and simian herpesvirus saimiri are examples (15, 27, 28). To date, the exact mechanism of action of this family of viral transcriptional activator remains incompletely understood. Rta appears to be able to activate transcription through two distinct mechanisms, namely, by direct DNA binding and via association with a DNA-binding transcription factor RBP-Jκ protein, which interacts with RBP-Jκ sites in several Rta-targeted promoters (11). In this study, we have analyzed the promoter of the K8 ORF and identified a region of 47 bp that responds to Rta-mediated transactivation. This 47-bp sequence, by itself or in the form of multimers, promotes potent transcriptional activation by Rta in a copy number-dependent manner. In agreement with published results (22), EMSAs using the 47-bp element as a probe indicate that Rta is a sequence-specific DNA-binding protein. The E. coli-derived Rta is oligomeric, possibly consisting of six subunits, and binds DNA as an oligomer. A DNA sequence comparison between the 47-bp RtaRE and similar elements found in PAN, K12 ORF, and ORF 57 promoters revealed the presence of multiple A/T-trinucleotide motifs spaced 7 or 17 nucleotides apart. Mutational analyses indicate that the phased A/T motifs are critical for Rta response, while the spacer regions and the out-of-phase A/T motifs can be base substituted without adverse effects on the activity of the element. Most importantly, tandem copies of (A/T)3N7 motif (A/T)3GCGACCG sequences and N5T(A)4N6(A)4 motif GGATCTAAAAGTCGACAAAA sequences promote strong Rta-mediated transactivation. Based on this sequence pattern, another RtaRE in the K8 promoter (termed RtaRE2) downstream of the 47-bp element was also identified. RtaRE2 also supported a strong Rta response when inserted upstream of the RSV promoter (Fig. 7D). Taken together, these results suggest that Rta oligomers (hexamers?) bind multiple, phased A/T triplets—i.e., [(A/T)3(G/C)7]3-6—in RtaRE to bring about potent transcriptional activation. This KSHV/HHV-8 Rta recognition sequence differs significantly from the previously reported consensus recognition site (GNCCN9GGNG) of the R transactivator of EBV (8). We think that RtaRE1 most likely extends upstream of the 47-bp repeat and includes the long stretch of A/T sequence as indicated in Table 1, judging from the reduction in promoter activity when the latter region was excluded (Fig. 1, construct II). RtaREs act synergistically when multimerized, as indicated by the combined action of RtaRE1 and RtaRE2 in the K8 promoter (Fig. 1) and that of multimers of the 47-bp motif (Fig. 2). Finally, RtaREs support a strong Rta response when incorporated in the context of a strong promoter, such as that of the Rous sarcoma virus.
The phasing of the A/T nucleotides in RtaRE provides an explanation for the moderate Rta response of A/T sequences linked together arbitrarily (Fig. 2). It further explains a recent methylation interference analysis of Rta and the RtaRE of PAN promoter, which revealed little contact of G/C nucleotides with Rta (22). Consistent with our conclusion, extensive analyses of the PAN RtaRE have indicated the center TAA motif (see Table 1) to be critical for both Rta binding and transactivation (22). The longer A/T motifs at the 5′ and 3′ borders of PAN promoter have also been shown to contribute to optimal Rta activation, although they are more tolerant of base substitutions (22). This is similar to what we describe here for the M5 mutation in K8 promoter (Fig. 7A). It should be pointed out that the mutations we introduced into the K8 RtaRE (Fig. 7A) altered three A/T bases simultaneously and may have a greater and more unambiguous effect than the single and double base substitutions in the PAN promoter reported by Song et al. (22).
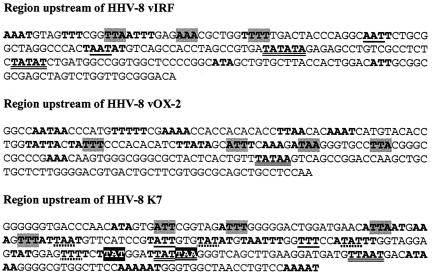
The nucleotide sequences of PAN and K12 RtaREs share extensive homology (22) (also see Table 1). The sequence of the K8 RtaRE, however, deviates significantly from them, especially in the spacers in between the phased A/T motifs (Fig. 6A). Our results indicate that optimal Rta response is obtained with phased A/T trinucleotides with heptanucleotide spacers of high G/C content. Increase in the length of the A/T motifs (Fig. 7C, Con1 versus Con2) or in the A/T content in the spacer regions diminishes (Fig. 6, HR versus LR), while increase in the G/C content of the spacers augments, Rta responsiveness (Fig. 7C). The need for three copies of Con2 (six A/T motifs) to support a potent Rta response may reflect a lower affinity of the A/T motifs in Con2 for Rta due to the increased length of A/T nucleotides; thus, additional binding motifs are needed for optimal interaction with Rta. Consistent with this notion, phased A/T hexanucleotides and heptanucleotides showed little Rta response (data not shown). Support for this conclusion can also be found in the G/C-rich nature of the spacers adjacent to the center TAA trinucleotide of the PAN RtaRE (see Table 1). A/T substitutions in these spacer regions have been shown to attenuate both Rta binding and transactivation (22). As might be expected, multiple phased A/T triplets can be readily discerned in the upstream regions of three immediate early genes of KSHV/HHV-8, namely, the vIRF, vOX-2, and K7 genes (Fig. 8). Whether these elements indeed mediate Rta transactivation remains to be determined.
FIG. 8.
DNA sequences upstream of the translational start sites of KSHV/HHV-8 vIRF, vOX-2, and K7 immediate early genes contain multiple phased A/T triplets. Continuous strings of A/T bases are denoted in bold. A/T triplets that are in the same phase are shaded, underlined with solid or dotted lines, or indicated in white over a black background. The potential TATA elements are double underlined.
The exact subunit stoichiometry in each Rta oligomer is not clear at present. Chemical cross-linking and fluid dynamic analyses of the holo-Rta suggest that it may be a hexamer. The oligomeric nature of Rta suggests that it has the potential to contact multiple DNA motifs and is consistent with its requirement for multiple (three or more) phased A/T trinucleotides for potent transactivation. Finally, the critical importance of the A/T trinucleotides for Rta response and the sensitivity of Rta/RtaRE interaction to competition by poly(dI-dC) and poly(dA-dT) (Fig. 4B) are reminiscent of the interaction between TATA box and TATA-binding protein (TBP). Because the minor groove structure of the dI-dC base pair is identical to that of the dA-dT base pair and TBP binds TATA sequence in the minor groove, TBP binding to the TATA motif is strongly competed by poly(dI-dC) (20). Whether each subunit of the Rta oligomer contacts the minor groove of each of the phased A/T motifs remains to be determined.
Acknowledgments
We thank Gary Hayward for the Rta antibody, Bala Chandran for the BCBL-1 cDNA library, Aviva Symes for the pA3Pluc plasmid, and Xin Xiang for assistance with fluorescence microscopy.
This work was supported by an in-house research grant RO73FH from the USUHS.
REFERENCES
- 1.Bermudez-Cruz, R., L. Zhang, and V. L. van Santen. 1998. Characterization of a bovine herpesvirus 4 (BHV-4) 1.1-kb RNA and its transactivation by BHV-4 immediate-early 2 gene product. Arch. Virol. 143:2391-2412. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 3.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 5.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 6.Goodwin, D. J., M. S. Walters, P. G. Smith, M. Thurau, H. Fickenscher, and A. Whitehouse. 2001. Herpesvirus saimiri open reading frame 50 (Rta) protein reactivates the lytic replication cycle in a persistently infected A549 cell line. J. Virol. 75:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruffat, H., and A. Sergeant. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 22:1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 16.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen, Y., G. A. Kassavetis, G. O. Bryant, and A. J. Berk. 1998. Polymerase (Pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated Pol II transcription. Mol. Cell. Biol. 18:1692-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, and L. Degos. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 24.Soulier, J., L. Grollet, E. Oksenhendler, J. M. Miclea, P. Cacoub, A. Baruchel, P. Brice, J. P. Clauvel, M. F. d'Agay, and M. Raphael. 1995. Molecular analysis of clonality in Castleman's disease. Blood 86:1131-1138. [PubMed] [Google Scholar]
- 25.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurau, M., A. Whitehouse, S. Wittmann, D. Meredith, and H. Fickenscher. 2000. Distinct transcriptional and functional properties of the R transactivator gene orf50 of the transforming herpesvirus saimiri strain C488. Virology 268:167-177. [DOI] [PubMed] [Google Scholar]
- 27.van Santen, V. L. 1993. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J. Virol. 67:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitehouse, A., M. Cooper, K. T. Hall, and D. M. Meredith. 1998. The open reading frame (ORF) 50a gene product regulates ORF 57 gene expression in herpesvirus saimiri. J. Virol. 72:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, L., J. Chiu, and J. C. Lin. 1998. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 17:735-742. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, L. J., and C. Z. Giam. 1991. Interaction of the human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc. Natl. Acad. Sci. USA 88:11445-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]