Abstract
Using the Xenopus egg extract system, we investigated the involvement of DNA replication in activation of the DNA damage checkpoint. We show here that DNA damage slows replication in a checkpoint-independent manner and is accompanied by replication-dependent recruitment of ATR and Rad1 to chromatin. We also find that the replication proteins RPA and Polα accumulate on chromatin following DNA damage. Finally, damage-induced Chk1 phosphorylation and checkpoint arrest are abrogated when replication is inhibited. These data indicate that replication is required for activation of the DNA damage checkpoint and suggest a unifying model for ATR activation by diverse lesions during S phase.
Keywords: ATR, Rad1, DNA damage, Xenopus, checkpoint, replication
The cell cycle must be tightly regulated to ensure complete and accurate transmission of the genome from the parent to daughter cells. DNA damage checkpoints are conserved cellular mechanisms that slow or stall cell cycle progression, preventing entry of a cell into mitosis with an incompletely replicated or damaged genome. The phosphatidylinositol kinase-related kinases, ATM (ataxia-telangiectasia mutated) and ATR (ATM- and Rad3-related), as well as members of the Rad1 complex, Rad1, Rad9, and Hus1, have been shown to be essential components of the machinery that controls checkpoint activation in response to DNA damage (Zhou and Elledge 2000). These proteins appear to recognize the DNA damage signal and communicate its presence to the downstream kinases Chk1 and Chk2/Cds1, which in turn regulate a variety of downstream effectors (Zhou and Elledge 2000).
One primary difference between ATM and ATR is the types of damage to which each responds (Abraham 2001). Loss of ATM leads to increased radiosensitivity and a defect in the ionizing-radiation-induced DNA damage checkpoint, but the response of ATM-deficient cells to ultraviolet radiation (UV) and hydroxyurea appears to be intact (Zhou and Elledge 2000). In contrast, studies of ATR in mammalian cells suggest that ATR is required for the response to numerous chemical and physical DNA-damaging agents. Deletion of ATR in a conditional cell line leads to loss of the ionizing-radiation-induced G2 checkpoint (Cortez et al. 2001), and cells expressing a kinase-inactive ATR mutant are sensitive to ionizing radiation, UV, hydroxyurea, and methyl methanesulfonate (MMS; Cliby et al. 1998). In addition, depletion of Xenopus ATR (xATR) from Xenopus egg extracts leads to loss of the UV- and aphidicolin-induced checkpoints (Hekmat-Nejad et al. 2000; Zou et al. 2002).
Studies in yeast and mammalian cells suggest that components of the Rad1 complex are also required for the response of cells to multiple forms of DNA damage (Melo and Toczyski 2002). For example, Hus1-deficient mammalian cells are sensitive to lesions caused by UV and replication blocks, although the response of these cells to ionizing radiation appears intact (Weiss et al. 2000). Interestingly, each member of the Rad1 complex is distantly related to PCNA (proliferating cell nuclear antigen), a homotrimeric, ring-like complex that acts as a processivity factor for polymerase δ during replication and is loaded onto primed DNA by replication factor C (RFC). Structural modeling and biochemical studies suggest that Rad1, Hus1, and Rad9 form a heterotrimeric complex like PCNA (Melo and Toczyski 2002). The Rad1 complex is thought to be loaded onto damaged DNA by a complex containing Rad17 and several subunits of RFC (Melo et al. 2001; Zou et al. 2002). The apparent similarity between PCNA and the Rad1 complex may indicate that this complex also functions during DNA replication or that it recognizes a structure generated by DNA damage that is similar to that recognized by PCNA.
It is not understood how ATR and the components of the Rad1 complex respond to multiple types of DNA damage. One possibility is that activation of ATR and other checkpoint proteins is coupled to a cellular process, such as DNA replication, the disruption of which generates a signal for checkpoint activation. Here, we tested this hypothesis using the Xenopus egg extract system. We show that UV and MMS, both of which lead to activation of an ATR-dependent checkpoint, cause a decrease in the rate of DNA replication. We also find that the recruitment of ATR and Rad1 to UV- and MMS-damaged chromatin requires initiation of DNA replication. The induction of DNA damage by these agents is also accompanied by the accumulation on chromatin of two replication proteins, replication protein A (RPA) and DNA polymerase α (Polα). Finally, we show that the damage-inducible phosphorylation of Chk1 and inhibition of mitotic entry requires initiation of DNA replication. These results indicate that initiation of DNA replication must occur in order for damage caused by UV or MMS to activate the checkpoint in Xenopus egg extracts. They also suggest that disruption of DNA replication by UV or MMS may be necessary for generation and/or recognition of the signal that activates ATR.
Results and Discussion
UV damage slows replication in a checkpoint-independent manner
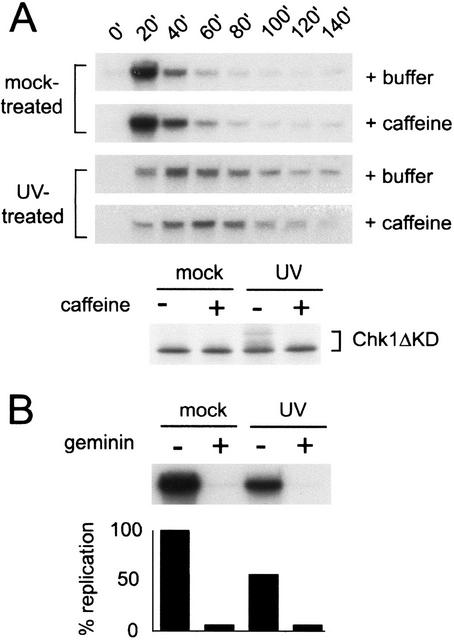
To test the possibility that DNA damage disrupts replication in Xenopus egg extracts, we investigated the effect of UV damage on replication by measuring the incorporation of radioactive nucleotides into chromatin. To do so, we removed aliquots from the extract at 20-min intervals and labeled the chromatin for 15 min in the presence of [α-32P]dCTP. If UV damage slows DNA replication, a decrease in the rate of [α-32P]dCTP incorporation should occur. Consistent with this hypothesis, we found that addition of UV-treated chromatin to Xenopus interphase egg extracts significantly decreased the rate of nucleotide incorporation relative to that observed for mock-treated chromatin (Fig. 1A). UV damage did not affect the timing of nuclear assembly (data not shown) or the loading of xORC2 onto chromatin (Fig. 2). Therefore, these data suggest that UV damage slows the rate of DNA replication.
Figure 1.
Replication is slowed in response to UV treatment. (A) Sperm nuclei (2000/μL) were mock-treated or UV-treated and added to Xenopus interphase extract in the presence (+caffeine) or absence (+buffer) of caffeine (4 mM), and the extract was divided into two samples. To assay replication, aliquots were removed from one sample at the given times, incubated with [α-32P]dCTP for 15 min, terminated, separated on a 0.8% agarose gel, and analyzed by autoradiography. To assay phosphorylation of xChk1, an in vitro translated, [35S]methionine-labeled fragment of xChk1 (Chk1ΔKD) was added to the second sample (5% reaction volume). Nuclei were isolated from this sample at 100 min, then proteins were separated by SDS-PAGE and analyzed by autoradiography. (B) CSF extract (10 μL) was preincubated with recombinant geminin or an equal volume of buffer. The extract was then supplemented with [α-32P]dCTP and mock- or UV-treated sperm nuclei, incubated at room temperature for 100 min, and processed as described in A.
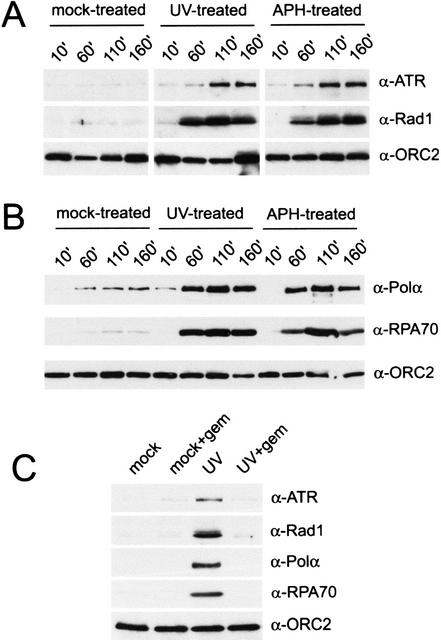
Figure 2.
Geminin blocks UV-induced accumulation of checkpoint and replication proteins on chromatin. (A) Binding of xATR, xRad1, and xORC2 to chromatin in interphase extract was assayed after mock, UV, or aphidicolin (APH) treatment. Chromatin (2000/μL) was isolated at the given time points, and chromatin-bound proteins were analyzed by Western blotting with antibodies to xATR, xRad1, or xORC2. (B) Chromatin binding of the p180 subunit of xPolα, xRPA70, and xORC2 was assayed after mock, UV, and aphidicolin treatment by Western blotting. Samples were treated and prepared as described in A. (C) Interphase extract was pretreated for 10 min with geminin (+gem) before addition of mock- or UV-treated chromatin (2000/μL). Chromatin was isolated after 100 min, and chromatin-bound proteins were analyzed as described above.
To determine if the delay observed in our experiments is a checkpoint-dependent event, we examined the effect of caffeine, an inhibitor of ATM and ATR function, on the rate of S-phase progression. When added to extracts containing UV-damaged chromatin, we found that caffeine did not restore the rate of nucleotide incorporation to that observed in mock-treated extracts (Fig. 1A). To confirm that caffeine was inhibiting xATR, we examined its effect on the ATR-dependent phosphorylation of xChk1 in the same extract (Fig. 1A). We monitored the mobility of a kinase-deleted, [35S]-labeled xChk1 fragment (xChk1ΔKD), which has been shown to reliably reflect xChk1 phosphorylation and checkpoint activation (Michael et al. 2000). Phosphorylation of this xChk1 fragment decreases its mobility by SDS-PAGE. We found that phosphorylation of xChk1 in response to UV was inhibited by caffeine in these extracts. Taken together, these data suggest that the observed slowing of S phase does not involve a caffeine-sensitive pathway, indicating that neither xATR nor xATM is involved.
Incorporation of radioactive nucleotides into DNA could also result from a UV-induced arrest prior to initiation of replication and subsequent repair of UV-damaged DNA. To address this possibility, recombinant Xenopus geminin was used to inhibit DNA replication in a cytostatic factor-arrested (CSF) extract. Geminin inhibits pre-replication complex (pre-RC) formation by blocking MCM loading onto chromatin (McGarry and Kirschner 1998). When added to extracts prior to the addition of chromatin, geminin completely abolished [α-32P]dCTP incorporation into both untreated and UV-treated chromatin (Fig. 1B). These data suggest that the incorporation of radioactivity observed after UV treatment is caused by DNA replication rather than pre-replication repair.
Requirement for replication in loading of checkpoint proteins after UV damage
Previous studies in Xenopus egg extracts suggest that both UV damage and aphidicolin treatment lead to xATR-dependent phosphorylation of xChk1 (Guo et al. 2000; Hekmat-Nejad et al. 2000). We have also found that the recruitment of xATR to chromatin after aphidicolin treatment is dependent on DNA replication (Hekmat-Nejad et al. 2000), consistent with the observation that the aphidicolin-induced phosphorylation of xChk1 is replication-dependent (Michael et al. 2000). To determine if the checkpoint response to UV damage is also dependent on replication, we compared the effects of UV or aphidicolin treatment on the chromatin-binding properties of xATR and Xenopus Rad1 (xRad1). Although both of these proteins are thought to be required for the response to multiple forms of DNA damage, recent studies suggest that they are recruited to sites of damage independently (Melo et al. 2001; You et al. 2002; Zou et al. 2002).
As shown in Figure 2A, UV damage stimulates the recruitment of xATR to chromatin with kinetics similar to that observed after aphidicolin treatment. xORC2, a member of the origin recognition complex that loads onto chromatin prior to initiation of DNA replication, was unaffected by DNA damage and was used as a control for the amount of DNA loaded in each lane (Fig. 2A). To investigate the chromatin-binding properties of Rad1, we cloned the Xenopus Rad1 cDNA, which encodes a 281-amino-acid protein with 85% sequence identity to human Rad1. Antisera raised against recombinant xRad1 protein indicate that xRad1 is also recruited to UV- and aphidicolin-treated chromatin with kinetics similar to those observed for xATR (Fig. 2A). Interestingly, unlike xORC2, neither xATR nor xRad1 accumulates on UV-damaged chromatin immediately after the addition of chromatin. However, the kinetics of xATR and xRad1 recruitment to UV- and aphidicolin-treated chromatin are remarkably similar. Moreover, each protein accumulates to a similar level in the UV- and aphidicolin-treated extracts.
Two proteins involved in DNA replication, the single-stranded DNA-binding protein RPA and DNA Polα, are known to accumulate on chromatin upon treatment with aphidicolin (Michael et al. 2000). This observation and others have led to the hypothesis that aphidicolin treatment functionally uncouples the helicase from the stalled polymerase, causing the accumulation of unreplicated, single-stranded DNA, which serves as a template for the binding of RPA and Polα (Michael et al. 2000; Walter and Newport 2000). If similar processes occur when the replication fork encounters a UV lesion, RPA and Polα would be expected to accumulate under these conditions as well. Indeed, we found that UV damage caused the accumulation of xPolα and xRPA70 on chromatin with kinetics similar to that observed for aphidicolin (Fig. 2B).
Activation of the replication checkpoint has been shown to require initiation of DNA replication (Hekmat-Nejad et al. 2000; Michael et al. 2000). If replication-associated events are required to create the signal to which xATR, xRad1, xRPA, and xPolα bind after DNA damage, inhibition of replication initiation should prevent the association of these proteins with damaged chromatin. To test this hypothesis, we again used geminin to inhibit DNA replication. Recruitment to UV-damaged chromatin of the checkpoint proteins xATR and xRad1, as well as the replication factors xRPA and xPolα, was abolished when replication was inhibited by geminin (Fig. 2C). In contrast, xORC2 binding was unaffected by geminin addition. Taken together, these data indicate that replication-associated events are necessary for recognition of UV damage by xATR, xRad1, xRPA, and xPolα.
UV-induced checkpoint activation requires DNA replication
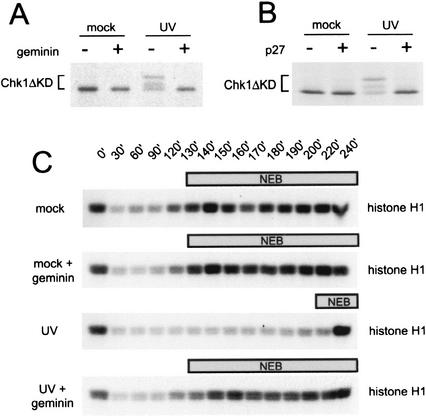
Although the recruitment of xATR and xRad1 to UV-damaged chromatin correlates with checkpoint activation, it is possible that checkpoint activation can occur in the absence of a detectable level of chromatin-bound checkpoint proteins. The ATR-dependent phosphorylation of Chk1 has been shown to be necessary for the activation of Chk1 kinase activity (Kumagai and Dunphy 2000) and subsequent downstream inactivation of Cdc2 (Zhou and Elledge 2000). To determine if the UV-induced activation of xChk1 also requires initiation of replication, we examined the phosphorylation state of the xChk1 protein after UV damage in geminin-treated extracts. We found that phosphorylation of xChk1 was induced by UV treatment, but geminin addition inhibited this phosphorylation (Fig. 3A). To confirm that the inhibition of UV-induced xChk1 phosphorylation after geminin treatment is due to its effect on DNA replication, we also used the Cdk inhibitor p27kip to inhibit DNA replication in this assay. In the Xenopus system, replication initiation requires the assembly of a mature pre-RC on chromatin, followed by the activation of Cdk2 kinase (Jackson et al. 1995). We found that recombinant p27kip also blocked UV-induced phosphorylation of xChk1 (Fig. 3B), indicating that the effect of geminin is specific to replication. This suggests that initiation of replication is also necessary to activate downstream components of the checkpoint signaling pathway in response to UV damage.
Figure 3.
Loss of the DNA damage checkpoint response after inhibition of DNA replication. (A) Phosphorylation of xChk1 (Chk1ΔKD) in Xenopus interphase extract after geminin treatment. Mock-treated or UV-damaged sperm chromatin (2000/μL) was added to mock- or geminin-pretreated extract containing Chk1ΔKD, and samples were analyzed as described in Figure 1A. (B) Phosphorylation of xChk1 (Chk1ΔKD) in Xenopus interphase extract after p27 treatment. Mock-treated or UV-damaged sperm chromatin (2000/μL) was added to mock- or p27-pretreated extract containing Chk1ΔKD, and samples were analyzed as described in Figure 1A. (C) Phosphorylation of histone H1 in mock- or geminin-treated cytostatic-factor-arrested (CSF) extract. CSF extract pretreated with buffer or recombinant geminin was supplemented with mock- or UV-treated sperm nuclei and 1 mM CaCl2. At the indicated time points, samples were removed to assay histone H1 kinase activity. The time at which nuclear envelope breakdown (NEB) was >50% is indicated by the bar above each panel.
To further characterize the effect of replication inhibition on activation of the UV damage checkpoint, we examined the ability of geminin to abrogate the UV-induced delay in the cell cycle using CSF-arrested extracts. We monitored progress through the in vitro cell cycle by examining the activity of Cdc2–cyclin B and nuclear envelope breakdown (NEB), both of which occur upon entry into mitosis. Treatment of chromatin with UV induced a significant delay in activation of Cdc2, which was monitored by phosphorylation of histone H1 (Fig. 3C). Importantly, geminin treatment abrogated the UV-induced delay. The rise in histone H1 kinase activity tightly correlated with the timing of NEB (Fig. 3C). This shows that replication is required for the mitotic delay induced by UV damage.
Replication-dependent activation of the MMS-induced checkpoint
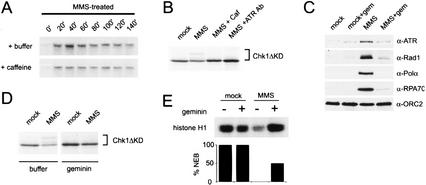
To determine if the requirement for replication in checkpoint activation is limited to damage caused by UV, we also examined the need for replication after treatment with methyl methanesulfonate (MMS). MMS is an alkylating agent known to inhibit replication as well as activate the DNA damage checkpoint (Kato and Strauss 1974; Paulovich and Hartwell 1995). After treatment with MMS, we observed a caffeine-independent slowing of replication similar to that seen for UV damage (Fig. 4A). The mock-treated control for this experiment is shown in Figure 1A. xChk1 phosphorylation was also observed (Fig. 4B). To confirm that xATR has a role in the MMS checkpoint, we examined the phosphorylation of xChk1 in the presence of either caffeine or an ATR-specific neutralizing antibody. MMS-induced xChk1 phosphorylation was abrogated by both of these reagents, suggesting that xATR does regulate the MMS-induced activation of xChk1 (Fig. 4B).
Figure 4.
Activation of the MMS checkpoint is replication-dependent. (A) DNA replication is slowed after MMS damage. Chromatin was pretreated with MMS, and replication was assayed as described in Figure 1A. See Figure 1A for untreated control (+/−) caffeine. (B) Phosphorylation of xChk1ΔKD induced by MMS was assayed after treatment of the extract with the checkpoint kinase inhibitor caffeine (4 mM) or an xATR-neutralizing antibody (10% reaction volume) as described in Figure 1A. (C) Chromatin was isolated from mock- or geminin-treated extract containing mock- or MMS-treated chromatin after 100 min, and chromatin-bound proteins were analyzed as described in Figure 2. (D) MMS-induced phosphorylation of xChk1ΔKD was assayed after geminin treatment. Geminin-treated interphase extract was supplemented with xChk1ΔKD and sperm chromatin (3000/μL), and samples were harvested at 100 min. Sperm nuclei were treated with MMS prior to addition to the extract. (E) Histone H1 kinase activity and percentage of nuclear envelope breakdown (%NEB) were assayed in CSF extract at 160 min. Extract was pretreated with buffer or geminin, and sperm chromatin was mock-treated or treated with MMS prior to addition to extract.
We also found that xATR and xRad1 bound chromatin after MMS treatment (Fig. 4C). Like UV and aphidicolin, MMS also induced the binding of xRPA and xPolα to chromatin (Fig. 4C). When initiation of DNA replication was blocked by geminin treatment, binding of the checkpoint proteins xATR and xRad1 as well as xRPA70 and xPolα was blocked, whereas the xORC2 loading control was unaffected (Fig. 4C). Downstream events associated with the MMS-induced checkpoint were also blocked by inhibition of DNA replication. MMS-induced xChk1 phosphorylation was prevented by geminin or p27 treatment (Fig. 4D; data not shown), and geminin abrogated the MMS-induced cell cycle delay as determined by histone H1 kinase activity and nuclear envelope breakdown (Fig. 4E). Taken together, these data suggest that MMS lesions, like UV lesions, require replication to initiate recruitment of checkpoint proteins to chromatin and to activate downstream effectors.
Our data show that replication is required for activation of the DNA damage checkpoint in Xenopus egg extracts. Blocking the initiation of replication with either geminin or p27 prevents the damage-induced phosphorylation of xChk1 and the recruitment of xRad1, xATR, xPolα, and xRPA to chromatin. More importantly, it leads to loss of the checkpoint and progression of the extract into mitosis. This suggests that some event that occurs upon activation of Cdk2 and/or initiation of DNA replication is required to generate the signal that leads to checkpoint activation. This is consistent with several studies in yeast model systems that show a role for replication proteins in activation of the checkpoint (D'Urso et al. 1995; Navas et al. 1995).
Activation of the replication checkpoint by aphidicolin in Xenopus egg extracts is thought to result from stalling of a DNA polymerase and the subsequent generation of a signal(s) that recruits checkpoint proteins to chromatin (Michael et al. 2000). Our data suggest UV and MMS may have effects on DNA replication similar to those of aphidicolin. Both UV and MMS slow replication in a caffeine-insensitive manner, indicating that the delay does not depend on xATR or xATM. Consistent with this observation, studies in yeast have shown that MMS slows the rate of fork progression in a manner that is independent of Mec1, the homolog of ATR in Saccharomyces cerevisiae (Tercero and Diffley 2001). It seems likely that the slowing caused by UV and MMS results from a block to replication fork progression, similar to the block caused by aphidicolin. This suggests that aphidicolin, UV, and MMS share the ability to slow DNA polymerases, raising the possibility that they also share a common mechanism of checkpoint activation. The recruitment to chromatin of xATR, xRad1, xPolα, and xRPA in response to both aphidicolin and DNA-damaging agents further supports this possibility.
The accumulation of xRPA and xPolα on damaged chromatin may also provide some insight into the events leading up to activation of the checkpoint after DNA damage. xRPA accumulation suggests that single-stranded DNA is generated by these types of damage, and this is supported by experiments in human tissue culture cells treated with UV and MMS (Kato and Strauss 1974; Ventura and Meneghini 1984). xPolα accumulation suggests that further modification of the single-stranded DNA also occurs. Although we have not shown that either xRPA or xPolα is required for checkpoint activation after DNA damage, other recent studies have shown that depletion of xRPA or xPolα prevents the aphidicolin-induced association of xATR and xHus1 with chromatin and subsequent activation of the replication checkpoint (You et al. 2002). Thus, single-stranded DNA is not sufficient for activation of the replication checkpoint, but it appears to be necessary and may serve as a template for the association of additional proteins and generation of a checkpoint-activating structure. Whether or not this is the case after UV or MMS damage remains to be seen, but it seems plausible that there are similar requirements for the DNA damage checkpoint.
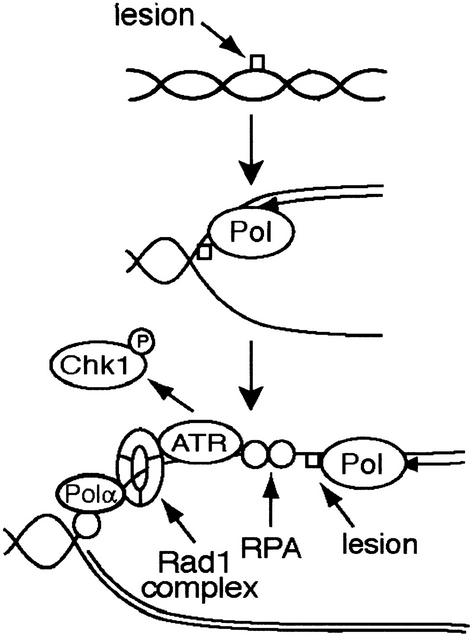
Our data together with other recent studies lead us to hypothesize that replication may convert polymerase-blocking lesions into a common structure that is recognized by checkpoint proteins. Single-stranded DNA may accumulate when replication is blocked by DNA damage or with aphidicolin, possibly through functional uncoupling of the helicase from the polymerase or through the action of an exonuclease involved in DNA repair or the resolution of stalled replication intermediates (Walter and Newport 2000). The association of xRPA, xPolα, and possibly other proteins with single-stranded DNA may then facilitate the recruitment of xATR and the xRad1 complex (Fig. 5). Inhibition of replication prevents the formation of this structure, and consequently blocks xATR- and xRad1-dependent chromatin binding and activation of the checkpoint. Thus, the ability of xATR to respond to a diverse array of lesions may result from the ability of many agents to disrupt replication and generate a common intermediate. Further investigation will be required to test this model and to reveal the identity of the structure and proteins involved.
Figure 5.
A model for activation of the DNA damage checkpoint in Xenopus egg extracts. UV and MMS lesions do not activate the checkpoint prior to initiation of DNA replication. During DNA replication, we suggest that encounter of a polymerase with a lesion leads to slowing of replication. Accumulation of single-stranded DNA is followed by association of ATR and the Rad1 complex with a protein and/or nucleic acid structure that is dependent on the presence of RPA, Polα, and possibly additional downstream events. Activation of ATR and Chk1 phosphorylation follow.
Materials and methods
Cloning and production of recombinant xRad1
A 450-bp fragment of xRad1 was cloned from Xenopus oocyte cDNA using degenerate PCR, followed by 5′ and 3′ RACE. Three independent full-length clones were amplified to generate a consensus sequence. This sequence is available from GenBank (accession no. AY126669). xRad1 was expressed in insect cells with a His6 epitope tag and purified using Ni-Agarose (QIAGEN).
Production of antibodies, recombinant geminin
Rabbit polyclonal xRad1 and xATR antibodies were raised at a commercial facility (Josman, Napa, CA). Antibodies used to neutralize xATR were raised against a C-terminal epitope fused to GST and affinity-purified, and antibodies to xRad1 were raised against recombinant full-length protein. Recombinant His6-geminin was expressed in bacteria and purified using Ni-Agarose and anion-exchange chromatography. Antibodies to xORC2 and xRPA (p70) have been described previously (Jackson et al. 1995; Furstenthal et al. 2001).
Chromatin-binding assay
At the given time points, 100 μL of Xenopus egg extract containing 2000 sperm nuclei/μL was layered on a 0.75 M sucrose-ELB cushion (50 mM KCl, 2.5 mM MgCl2, 10 mM HEPES at pH 7.7) and spun at 11,700g for 90 sec to isolate nuclei. Nuclei were lysed in 500 μL of chromatin extraction buffer (CEB, 50 mM KCl, 50 mM HEPES at pH 7.7, 5 mM MgCl2, 5 mM EGTA, 0.1% NP-40, 0.5 mM spermidine, 0.15 mM spermine, 2 mM β-mercaptoethanol). After 15 min on ice, the nuclear lysate was spun as described above. Isolated chromatin was resuspended in sample buffer.
Xenopus checkpoint and replication assays
Xenopus interphase extracts were prepared as described (Walter and Newport 1999). Cytostatic-factor-arrested (CSF) extracts and Xenopus sperm chromatin were prepared as outlined (Murray 1991). UV- or MMS-treated chromatin was damaged with 1000 J/m2 of UV light or 1% MMS prior to addition to extract. Aphidicolin was used at 100 μg/mL. Geminin (250 nM) and p27 (300 nM) were preincubated with extract at room temperature for 10 min. To assay xChk1ΔKD phosphorylation, nuclei were isolated from interphase extract, and mobility was monitored as described (Kumagai et al. 1998; Michael et al. 2000). Histone H1 kinase and nuclear envelope breakdown assays were performed as outlined (Murray 1991). Replication assays were carried out as described (Walter and Newport 1999).
Acknowledgments
We thank Peter Jackson for p27 as well as xORC2 and xRPA70 antibodies, Shou Waga for xPolα antibodies, and Matt Michael for the xChk1ΔKD and xGeminin constructs. We thank Peter Jackson and James Ferrell for critical reading of the manuscript, and members of the Cimprich and Ferrell labs for their advice and support. This work was funded by a fellowship from the CA BCRP (8GB-0091) to P.J.L. and grants from the ACS (CCG-97492) and NIH (GM62193) to K.A.C. K.A.C. is a Burroughs Wellcome Fund New Investigator and a Beckman Young Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cimprich@stanford.edu; FAX (650) 725-2952.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1013502.
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: Partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- D'Urso G, Grallert B, Nurse P. DNA polymerase α, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108:3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- Furstenthal L, Kaiser BK, Swanson C, Jackson PK. Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J Cell Biol. 2001;152:1267–1278. doi: 10.1083/jcb.152.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for ATR in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes & Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Nejad M, You Z, Yee M-C, Newport J, Cimprich KA. Xenopus ATR is a replication-dependent chromatin binding protein required for the DNA replication checkpoint. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Jackson PK, Chevalier S, Philippe M, Kirschner MW. Early events in DNA replication require cyclin E and are blocked by p21CIP1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Strauss B. Accumulation of an intermediate in DNA synthesis by HEp.2 cells treated with methyl methanesulfonate. Proc Natl Acad Sci. 1974;71:1969–1973. doi: 10.1073/pnas.71.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:836–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Guo ZJ, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Ott R, Fanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell-cycle extracts. Meth Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ. DNA polymerase ɛ links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S-phase in Saccharomyces cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Ventura AM, Meneghini R. Inhibition and recovery of the rate of DNA synthesis in V79 Chinese hamster cells following ultraviolet light irradiation. Variation in the rate of movement of the replication fork. Mutat Res. 1984;131:81–88. doi: 10.1016/0167-8817(84)90014-2. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. The use of Xenopus laevis interphase extracts to study genomic DNA replication. In: Cotterill S, editor. Eukaryotic DNA replication. Oxford: Oxford University Press; 1999. pp. 201–222. [Google Scholar]
- ————— Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45; RPA; and DNA polymerase α. Mol Cell. 2000;3:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes & Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- You Z, Kong L, Newport J. The role of single-stranded DNA and Polα in establishing the ATR, Hus1 DNA replication checkpoint. J Biol Chem. 2002;15:15. doi: 10.1074/jbc.M204120200. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes & Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]