Abstract
Recent work points to the importance of changes in gene expression patterns in species-specific differences. Here, we investigate the evolution of the nematode lin-48 ovo gene. lin-48 is expressed in several cells in both Caenorhabditis elegans and Caenorhabditis briggsae, but acts in the excretory duct cell only in C. elegans. We find the differences result both from alterations in the cis-regulatory sequences and in proteins that mediate lin-48 expression. One factor that contributes to the species differences is the bZip protein CES-2. Our results indicate the accumulation of several regulatory changes affecting one gene can contribute to evolutionary change.
Keywords: Gene expression changes, Pax gene, EGL-38, CES-2
Multicellular organisms are composed of a variety of cell types that result from the expression of different genes in subsets of cells. This differential gene expression is mediated by cis-regulatory DNA sequences that can be bound by proteins such as transcription factors. Changes in gene-expression patterns play an important role in the molecular and morphological differences between species (Sucena and Stern 2000; Tautz 2000; Grandien and Sommer 2001). However, whether these changes result from alterations in the gene's cis-regulatory sequences or in the trans-acting proteins that bind to them, and whether a single major sequence change or multiple cumulative changes contribute to the species differences remain important questions, as they address the molecular mechanisms underlying evolutionary change. To understand the molecular changes responsible for the evolution of the expression pattern of a gene, we have investigated the function and expression of lin-48 in two Caenorhabditis nematode species. lin-48 encodes a C2H2 zinc-finger protein similar to the product of the Drosophila ovo gene (Johnson et al. 2001). In Caenorhabditis elegans, lin-48 has been shown to play an important role in hindgut development and its expression is limited to a subset of hindgut cells, the excretory duct cell, and several neuronal support cells. In preliminary studies, we found that reporter transgenes made from C. elegans lin-48 were expressed in a similar set of cells in Caenorhabditis briggsae, but the excretory duct cell expression was absent. The excretory duct cell is part of the nematode excretory system that is proposed to mediate osmotic and ionic regulation (Nelson et al. 1983; Nelson and Riddle 1984). We sought to understand the function of lin-48 in this cell, and to explore the molecular basis for the inter-species differences.
Results and Discussion
To investigate the function of lin-48 in development of the excretory duct cell, we visualized the cell in lin-48 mutants using transgenes containing the upstream regulatory sequences of C. elegans lin-48 (Ce-lin-48) directing the expression of the reporter gene gfp (green fluorescent protein; Fig. 1a,b). Although C. elegans lin-48 mutants develop a functional excretory duct cell, we found the cell morphology is different and the duct is more anterior than in wild type (Fig. 1c,d; Table 1). We observed the excretory duct placement in C. briggsae animals and found it is also more anterior than in wild-type C. elegans animals (Fig. 1e). Transgenes containing a Ce-lin-48 genomic clone rescue all lin-48 mutant phynotypes, whereas a comparable C. briggsae lin-48 (Cb-lin-48) genomic clone rescues lin-48 defects in hindgut and male tail development, but not duct morphogenesis (Fig. 1f,g; Table 1). Although we cannot rule out the possibility that additional regulatory elements in the C. briggsae gene lie outside of the tested genomic region, this experiment indicates that there are differences between the regulatory features of Ce-lin-48 and Cb-lin-48. Taken together, these results support the idea that Ce-lin-48 functions to alter duct cell morphogenesis and positioning, but this function is not found in C. briggsae.
Figure 1.
lin-48 expression and function in C. elegans and C. briggsae. (a) Nomarski DIC micrograph of wild-type L3 C. elegans animal showing the excretory system. (b) Epi-fluorescent micrograph of animal in a, showing expression of Ce-lin-48∷gfp. The location of the excretory cell and excretory duct cell are indicated by an asterisk and an arrow. The excretory duct is indicated by an arrowhead. The excretory duct forms more posterior in wild-type C. elegans (c) than in C. elegans lin-48 mutants (d) or C. briggsae (e). ces-2 mutant animals exhibit the same excretory duct cell morphological phenotype as lin-48(sa469) mutants. Bracket shows distance between the duct and the base of the posterior pharynx. (f) lin-48 mutant males develop abnormal ectopic spicule cells anterior to the normal spicule cells. (g) lin-48 mutant animal rescued by Cb-lin-48 genomic clone, resulting in development of normal spicule cells. Arrows indicate cells expressing spicule cuticle. Scale bars, 10μm. (Left) anterior; (top) dorsal.
Table 1.
lin-48 function in duct placement
|
|
Distance (μm)a
|
No. of animals
|
|---|---|---|
| C. elegans (N2) | 13.07 ± 1.55bc | 48 |
| C. elegans (CB4856) | 13.94 ± 1.70c | 48 |
| C. briggsae (AF16) | 17.73 ± 1.75d | 48 |
| C. briggsae (HK104) | 19.70 ± 2.05d | 48 |
| Ce-lin-48(sa469) | 20.12 ± 1.90d | 54 |
| Ce-ces-2(n732) | 17.16 ± 2.16d | 48 |
| Ce-ces-2(RNAi) | 16.92 ± 1.29d | 48 |
| Ce-lin-48(sa469);guEx(Ce-lin-48) | 13.93 ± 1.24c | 51 |
| Ce-lin-48(sa469);guEx(Cb-lin-48) | 20.03 ± 1.71d | 34 |
Excretory ducts of Caenorhabditis elegans lin-48 mutants, C. elegans ces-2 mutants, and Caenorhabditis briggsae locate more anterior compared with C. elegans wild-type. guEx(Ce-lin-48) and guEx(Cb-lin-48) are transgenes containing genomic clones of Ce-lin-48 and Cb-lin-48, respectively. These transgenes rescue the other functions of lin-48.
Distance is from excretory opening to the base of the terminal pharynx bulb. Values are means ± SD. There is no significant difference in the distance of the base of the terminal pharynx bulb to the P1.p nucleus between C. elegans and C. briggsae (12.74 ± 4.02 μm vs. 12.04 ± 3.09 μm, p = 0.262 by t-test).
All other strains (d) are significantly different from C. elegans wild type and lin-48 rescued animals (c) according to Bonferroni test (p < 0.05).
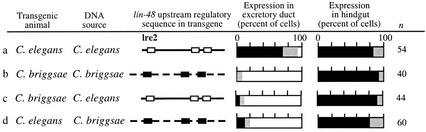
The upstream sequences of C. elegans lin-48 drive expression of GFP in the same pattern as gfp-tagged genomic clones capable of complementing lin-48 mutations (Johnson et al. 2001; data not shown). To identify the source of the differences in lin-48, we constructed gfp reporter transgenes for the C. briggsae lin-48 gene using upstream sequences. We tested the expression of Ce-lin-48∷gfp in C. briggsae, and vice versa, and found only the C. elegans gene in C. elegans animals is expressed in the excretory duct cell (Fig. 2a–d). All other aspects of expression were conserved in intra- and inter-species experiments. Because Ce-lin-48∷gfp is not expressed in the C. briggsae excretory duct cell (Fig. 2c), it suggests that there are differences in the potential regulatory proteins between the species. In addition, these experiments show that there are differences in the lin-48 upstream region as the Cb-lin-48∷gfp is not expressed in C. elegans (Fig. 2d).
Figure 2.
lin-48∷gfp transgenes from C. elegans and C. briggsae tested in each species indicate only the C. elegans gene in C. elegans animals is expressed in the excretory duct cell. All other expression patterns are conserved. The 2.4 kb of Ce-lin-48 upstream regulatory sequences is indicated with a solid line and white blocks. The 3.0 kb of Cb-lin-48 upstream regulatory sequences is indicated by a broken line and black blocks. Blocks correspond to conserved DNA sequences in the area of the previously defined regulatory element lre2 and two additional regions more proximal to the transcriptional start site.
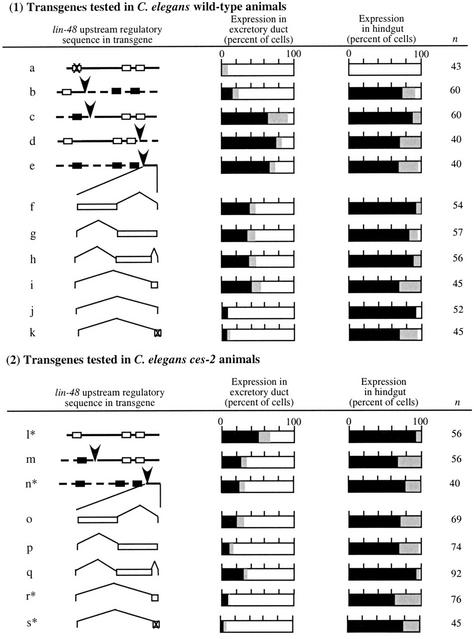
A mutation analysis of the Ce-lin-48 upstream regulatory region identified an element essential for excretory duct cell expression that is conserved between C. elegans and C. briggsae (Fig. 3a). This site (termed lre2) binds the Pax protein EGL-38 and is required for expression in both the excretory duct cell and hindgut cells (Johnson et al. 2001). However, this site cannot be sufficient for duct cell expression, because it is present in both C. elegans and C. briggsae genes. To identify additional sequences important for duct cell expression and responsible for the differences in the lin-48 cis-regulatory region, we made chimeric clones by swapping regions between Ce-lin-48 and Cb-lin-48, and tested these clones for expression in C. elegans animals. The results indicated that the more proximal 1.5 kb of Ce-lin-48 upstream sequences also contains cis-regulatory sequences necessary for excretory duct cell expression that are absent from the C. briggsae gene (Fig. 3b,c). Further analysis of this region showed that it includes at least four independent sites (one distal and three proximal) that are each sufficient to increase expression levels. This conclusion results from the following observations. First, either a more distal or more proximal portion of this region is sufficient for the expression (Fig. 3d,e). Second, deletions in the clone containing the most proximal 525-bp C. elegans sequence and the remaining sequence from C. briggsae (Fig. 3f–j) revealed at least three elements that confer partial duct cell expression activity. Each tested subdivision of this region retained some activity (Fig. 3f–i). Although several distinct sites are present in the proximal region of lin-48, the sequence does not contain obvious EGL-38 recognition sites. This result suggests that a different factor or factors is also essential for excretory duct cell expression.
Figure 3.
(1) Experiments to identify cis-regulatory sequences important for excretory duct cell expression in C. elegans. (a) A mutation in the Ce-lin-48 promoter (indicated by X) shows the conserved lre2 sequence is important for expression in both excretory duct and hindgut cells (data from Johnson et al. 2001). (b–j) Chimeric promoter sequences resulting from DNA swap between C. elegans and C. briggsae and deletion analysis of the chimeric promoter identify at least four additional regulatory regions that contribute to Ce-lin-48 expression in the duct cell. (k) Mutations in putative CES-2-binding sites in the Ce-lin-48 promoter reduced the excretory duct cell expression. (2) Experiments to show that ces-2 affects lin-48 excretory duct cell expression. (l–s) ces-2(RNAi) reduced lin-48 expression in transgenic animals. Reduction of lin-48 expression by transgenes marked with asterisk was also confirmed in ces-2(n732) mutant background. The percentages of cells expressing GFP (black bar), expressing very low, but detectable levels of GFP (gray bar) or not expressing GFP (white bar), are indicated for each construct. (n) Number of animals scored for expression. Source of DNA in transgene indicated as in Figure 2. For chimeric clones, arrowhead indicates the point of the DNA swap. GFP expression in hindgut was used as an internal control for expression level and presence of the transgene in all experiments.
To identify additional factors important for Ce-lin-48 expression in excretory duct cell, we used MatInspector software (Quandt et al. 1995) to search for potential transcription factor-binding sites in the ∼60-bp C. elegans most proximal sequences contained in the constructs of Figure 3i. This identified two sites with the potential to bind the C. elegans bZip protein CES-2 (Fig. 4). ces-2 was originally characterized for its role in mediating a developmental decision between survival and apoptosis (Metzstein et al. 1996). To test whether ces-2 plays a role in Ce-lin-48 excretory duct cell expression, we observed the excretory duct morphology in ces-2(n732) and ces-2(RNAi) animals and found it is similar to lin-48 mutants (Table 1). The ces-1 gene has been shown to act downstream of ces-2 in regulating the cell death decision (Ellis and Horvitz 1991; Metzstein and Horvitz 1999), but we found no effect on excretory duct morphogenesis in ces-1 mutants (data not shown). Thus, the role of ces-2 in duct morphogenesis is distinct from its role in apoptosis.
Figure 4.
Potential CES-2-binding sites in Ce-lin-48 promoter. (1) DNA sequence of ∼60 bp Ce-lin-48 proximal region. CES-2-binding sites are shown by arrows under the sequences. (2) Alignment between the CES-2-binding sites in the Ce-lin-48 proximal region, ces-1 upstream element (Metzstein and Horvitz 1999), and the CES-2/PAR family consensus (Metzstein et al. 1996). Y = T or C; R = A or G.
To test the function of ces-2 in mediating lin-48 duct cell expression, we tested lin-48∷gfp expression in ces-2(n732) and ces-2(RNAi) mutant animals, and found lin-48∷gfp expression is notably reduced in these animals (Fig. 3l–s). Finally, mutations disrupting the potential CES-2-binding sites in transgenes containing the 60-bp C. elegans proximal sequences eliminated the excretory duct cell expression (Fig. 3k). Taken together, these results indicate CES-2 mediates expression of lin-48 in the C. elegans excretory duct cell, and alteration of CES-2 regulatory sequences contribute to the differences between C. elegans and C. briggsae. Because mutations in ces-2 do not completely eliminate excretory duct expression (Fig. 3l–q), it suggests that other factors also contribute to the differences in lin-48 expression. This observation is consistent with the identification of multiple regulatory sites in the chimeric lin-48∷gfp reporter transgenes.
Using inter-specific gene expression assays, we have found that changes in cis-regulatory sequences and in trans-acting factors contribute to differences in Caenorhabditis lin-48 expression and function in the excretory duct cell. Two types of lin-48 cis-regulatory sequences are important for excretory duct cell expression: several in the proximal region that differ between C. elegans and C. briggsae, and the more distal lre2 that is conserved. Although both are necessary, the lre2 sequence from either C. elegans or C. briggsae can function with the C. elegans proximal sequences to mediate expression. Our results provide experimental evidence that the modular architecture of cis-regulatory sequences allow for gene evolution (Arnone and Davidson 1997). Specifically, evolutionary changes have affected the proximal sequences important only for duct cell expression, whereas lre2 (important for expression in two cell types) is unchanged. Our results suggest that the bZip transcription factor CES-2 is one factor that acts through the proximal sequences that differ between C. elegans and C. briggsae. Further work is necessary to determine whether ces-2 function also differs between C. elegans and C. briggsae, or whether it is change of other cis-regulatory factors that prevents the expression of Ce-lin-48∷gfp in the C. briggsae excretory duct.
Analysis of chimeric enhancers from Drosophila species suggests that cis-regulatory sequences are subject to complex stabilizing selection (Ludwig et al. 2000). These experiments showed that conservation of a gene expression pattern between species results from multiple compensatory nucleotide changes. Our experiments address the question of what molecular changes occur in the evolution of a gene expression pattern. A regulatory change could result from a sequence change in a single cis-regulatory element, or several sequence changes affecting multiple cis-regulatory elements. Evolution of gene expression by a single change might be associated with a dramatic phenotypic alteration fixed by strong selection, whereas the accumulation of multiple smaller changes might underlie a less stringent selection process or genetic drift. These small changes would act to stabilize an initial altered expression pattern. Our results with Caenorhabditis lin-48 genes are consistent with the latter mechanism. We speculate that the selective forces that influence cis-regulatory sequences and promote stabilization of gene expression can also act to stabilize gene expression differences between species.
Materials and methods
Morphology of the excretory duct cell
We measured the distance from the excretory pore to the base of the posterior pharynx bulb in L3 animals to quantify the morphological change of the excretory duct. Control measurements between the posterior pharynx bulb and the P1.p nucleus indicate that the bulb distance from other epidermal landmarks is the same in the two species (Table 1).
Construction of DNA clones
A C. briggsae probe for the lin-48 gene was generated using PCR with C. briggsae genomic DNA and degenerate primers corresponding to the C. elegans gene, and used to screen a C. briggsae genomic fosmid library (Incyte Genomics, Inc). A 6.0-kb SalI fragment (pXW4) that hybridized to the probe was subcloned from fosmid 33J1. The clone was confirmed to be Cb-lin-48 by rescuing male tail defects of C. elegans lin-48 (sa469);him-5(e1490) mutant animals and by sequencing. pXW6 is a plasmid containing Cb-lin-48, 3.0-kb upstream sequence cloned into vector pPD95.73 provided by Andy Fire (Department of Embryology, Carnegie Institution of Washington, Baltimore, MD). pAJ49 is 2.4-kb upstream sequence of Ce-lin-48 cloned into vector pPD95.67 and shown previously to express in all lin-48-expressing cells (Johnson et al. 2001). Swapping clones were derived from these two plasmids by incorporating unique restriction enzyme sites at the exchange points by use of the Quickchange mutagenesis protocol (Strategene). Each clone with the introduced restriction enzyme site was shown to retain activity prior to the swap (data not shown). Deletions (Fig. 3f–j) were generated by use of the ExSite mutagenesis method (Strategene). pXW66 and pXW67 are genomic clones that include the same extent of upstream sequences as pXW6 and pAJ49. These clones were used for rescue analysis in Table 1.
DNA transformation and RNA interference
Transgenic C. elegans strains were generated as described (Johnson et al. 2001). For the rescue experiment, pXW4, pXW66, or pXW67 was coinjected with 15 ng/μL pDP#MM016 [unc-119(+)] plasmid into the mitotic germ line of hermaphrodites of C. elegans lin-48(sa469);unc-119(e2498);him-5(e1490) animals. For all experiments that test expression of lin-48∷gfp reporter constructs in C. elegans, 70–90 ng/μL reporter clone was coinjected with unc-119(+) plasmid into unc-119(e2498);him-5(e1490) animals. For C. briggsae lin-48∷gfp, reporter clones were coinjected with pRF4 [C. elegans rol-6 (d)] at a concentration of 120 ng/μL into C. briggsae (AF16). At least two independent lines were tested for each construct.
RNA interference was performed as described (Fire et al. 1998). dsRNA of ces-2 was obtained from the cDNA clone, yk91e8, kindly provided by Yuji Kohara (Gene Network Library, National Institute of Genetics, Mishima, Japan).
Acknowledgments
We thank W. Deng for assistance with statistical analysis, M.A. Browning and A.D. Johnson for assistance in the early stages of this project, and A. Simcox and Chamberlin laboratory members for comments on the manuscript. We also thank Yuji Kohara for ces-2 cDNA clone and Therasa Stiernagle from Caenorhabditis Genetics Center for worm strains; the CGC is supported by the NIH National Center for Research Resources. This work is supported by grants from NSF (IBN-0092603) and NIH (GM42336).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL chamberlin.27@osu.edu; FAX (614) 292-4466
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.996302.
References
- Arnone MI, Davidson EH. The hardwiring of development: Organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Horvitz HR. Two C. elegans genes control the programmed deaths of specific cells in the pharynx. Development. 1991;112:591–603. doi: 10.1242/dev.112.2.591. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montegomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Grandien K, Sommer RJ. Functional comparison of the nemtode Hox gene lin-39 in C. elegans and P. Pacificus reveals evolutionary conservation of protein function despite divergence of primary sequences. Genes & Dev. 2001;15:2161–2172. doi: 10.1101/gad.200601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Fitzsimmons D, Hagman J, Chamberlin HM. EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development. 2001;128:2857–2865. doi: 10.1242/dev.128.15.2857. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer elemant. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Horvitz HR. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol Cell. 1999;4:309–319. doi: 10.1016/s1097-2765(00)80333-0. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Hengartner MO, Tsung N, Ellis RE, Horvitz HR. Transcriptional regulator of programmed cell death encoded by Caenorhabditis elegans gene ces-2. Nature. 1996;382:545–547. doi: 10.1038/382545a0. [DOI] [PubMed] [Google Scholar]
- Nelson FK, Riddle DL. Functional study of the Caenorhabditis elegans secretory-excretory system. J Exp Zool. 1984;231:45–56. doi: 10.1002/jez.1402310107. [DOI] [PubMed] [Google Scholar]
- Nelson FK, Albert PS, Riddle DL. Fine structure of the Caenorhabditis elegans secretory-excretory system. J Ultrastruct Res. 1983;82:156–171. doi: 10.1016/s0022-5320(83)90050-3. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector - New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E, Stern D. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc Natl Acad Sci. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Evolution of transcriptional regulation. Curr Opin Genet Dev. 2000;10:575–579. doi: 10.1016/s0959-437x(00)00130-1. [DOI] [PubMed] [Google Scholar]