Abstract
The ubiquitin-like protein, Nedd8, covalently modifies members of the Cullin family. Cullins are the major components of a series of ubiquitin ligases that control the degradation of a broad range of proteins. We found that Nedd8 modifies Cul1 in Drosophila. In Drosophila Nedd8 and Cul1 mutants, protein levels of the signal transduction effectors, Cubitus interruptus (Ci) and Armadillo (Arm), and the cell cycle regulator, Cyclin E (CycE), are highly accumulated, suggesting that the Cul1-based SCF complex requires Nedd8 modification for the degradation processes of Ci, Arm, and CycE in vivo. We further show that two distinct degradation mechanisms modulating Ci stability in the developing eye disc are separated by the morphogenetic furrow (MF) in which retinal differentiation is initiated. In cells anterior to the MF, Ci proteolytic processing promoted by PKA requires the activity of the Nedd8-modified Cul1-based SCFSlimb complex. In posterior cells, Ci degradation is controlled by a mechanism that requires the activity of Cul3, another member of the Cullin family. This posterior Ci degradation mechanism, which partially requires Nedd8 modification, is activated by Hedgehog (Hh) signaling and is PKA-independent.
Keywords: Nedd8, Cullins, SCF, Ci, protein degradation, eye development
Ubiquitin-mediated protein degradation mechanisms control the stability of various proteins that are essential for cellular function. Nedd8 is a ubiquitin-like small protein modifier. The Nedd8 conjugation process, called neddylation, is similar to ubiquitination. Neddylation utilizes the E1 activating-enzyme complex composed of two subunits, APP-BP1 and UBA3, and the E2 conjugating-enzyme, UBC12 (Yeh et al. 2000). The only known substrates of neddylation are Cullin family proteins, Cul1, Cul2, Cul3, Cul4A, Cul4B, and Cul5, which have been shown to be modified by Nedd8 in mammalian cells (Osaka et al. 1998; Hori et al. 1999). Cullins directly interact with Roc1, a Ring finger protein, and the Cullin-Roc1 complex comprises the core module of a series of ubiquitin E3 ligases, which confer substrate specificity and therefore regulate the degradation process (Kamura et al. 1999b; Ohta et al. 1999; Seol et al. 1999; Skowyra et al. 1999). Among Cullins, many studies focused on Cul1, an essential component of the SCF complex which functions as ubiquitin E3 ligase. The SCF complex consists of core subunits: Cul1/Cdc53, Skp1, Roc1/Hrt/Rbx1, and a substrate-recognition F-box protein. Cul1 functions as a scaffold protein within the SCF complex; the N-terminal domain of Cul1 interacts with the adaptor protein Skp1 that links with the F-box protein, and the C-terminal domain interacts with Roc1 and the ubiquitin E2 enzyme.
In vitro, neddylation of Cul1 is required for ubiquitination of IκBα and p27Kip1 (Morimoto et al. 2000; Podust et al. 2000; Read et al. 2000). In addition, neddylation enhances E2-ubiquitin recruitment to SCF (Kawakami et al. 2001). In fission yeast, Nedd8 is essential for the SCF-mediated degradation of Rum-1, a cyclin-dependent kinase inhibitor (Osaka et al. 2000). In Arabidopsis thaliana, the Nedd8 pathway is required for SCF-mediated Auxin response (Pozo et al. 1998; Schwechheimer et al. 2001). In mice deficient for UBA3, a subunit of the E1 enzyme in neddylation, embryonic development is aberrant, with accumulation of two putative SCF substrates, β-catenin and cyclin E (CycE; Tateishi et al. 2001).
In the SCF complex, F-box proteins convey substrate specificity by direct interaction with substrates for degradation. Many F-box proteins have been characterized in metazoans, and increasing numbers of specific targets for F-box proteins are being found (Deshaies 1999). Among them, the Drosophila F-box protein Slimb and its mammalian homolog β-TrCP are well characterized for their target specificity (for review, see Maniatis 1999). The specific targets for Slimb/β-TrCP are pIκBα in the Dorsal/NFκB pathway, Arm/β-catenin in the Wg/Wnt pathway, and Ci/Gli in the Hedgehog (Hh) pathway (Jiang and Struhl 1998; Yaron et al. 1998; Spencer et al. 1999; Winston et al. 1999).
The Hh pathway controls growth and pattern formation in many developmental processes in both vertebrates and invertebrates (for review, see Ingham and McMahon 2001). The Hh signal is transmitted through a receptor complex consisting of Patched (Ptc) and Smoothened (Smo). In the absence of Hh, Ptc inhibits Smo activity, and the effector Cubitus interruptus (Ci) is phosphorylated by PKA, leading to the proteolysis of Ci, which is converted into Ci75 with the C terminus truncated. Ci75 functions as a transcriptional repressor in the Hh signaling pathway. Upon binding to Ptc, Hh relieves Smo from its repression state. Activated Smo mediates signaling to prohibit proteolytic processing of Ci. The intact full-length Ci (CiFL) functions as a transcriptional activator for expression of target genes of the Hh pathway.
In Drosophila, Hh signaling functions in patterning the A/P compartments in developing tissues such as embryonic segments and wing and leg imaginal discs. In development of the eye imaginal disc, Hh signaling is a major driving force of the retinal differentiation wave, the morphogenetic furrow (MF), which is caused by transient constriction in cell apical surface (Heberlein et al. 1993; Ma et al. 1993). The MF progresses anteriorly from the posterior margin of the eye disc during the third instar larval and early pupal stages (Ready et al. 1976). Anterior to the advancing MF, cells are proliferating, whereas posterior cells differentiate sequentially into photoreceptor, cone, or pigment cells which produce and secrete Hh proteins. Transduction of Hh signaling in the MF is revealed by the accumulation of CiFL, which activates expression of target genes such as dpp and atonal in the MF (Heberlein et al. 1993; Dominguez and Hafen 1997; Greenwood and Struhl 1999). The induced MF cells soon differentiate and produce Hh proteins for further induction of more anterior cells, thus making the MF move forward.
The effect of neddylation on a broad spectrum of E3 ligases remains largely unknown. To investigate the role of neddylation in protein degradation control during developmental processes, we identified and analyzed Nedd8 and Cul1 mutants in Drosophila. Our results suggest that neddylation is required for Cul1-mediated protein downregulation of the signaling pathway effectors Ci and Armadillo (Arm) and the cell cycle regulator CycE. Using the developing eye disc as a model system to study the regulation of CiFL stability, we found that there is mechanistic difference in controlling CiFL stability between anterior and posterior cells separated by the MF. Whereas the Cul1-based SCFSlimb complex controls CiFL stability in anterior cells, a Cul3-dependent protein degradation mechanism controls CiFL stability in posterior cells. We further investigated the differences between these two protein degradation mechanisms.
Results
Effects of Nedd8 mutations on growth and protein stability
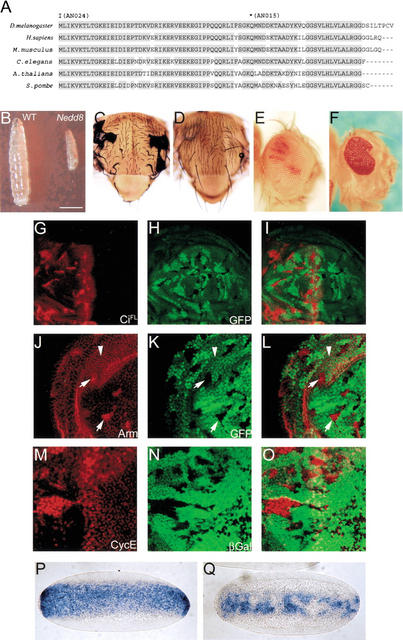
Nedd8 is highly conserved from yeast to mammals (Fig. 1A). We identified several Nedd8 alleles in Drosophila, including two null alleles Nedd8AN015 and Nedd8AN024 that were used in the present study (see Materials and Methods). The Nedd8 null mutants were growth-arrested in the first-instar larval stage and died within several days without further growth (Fig. 1B). We generated mutant clones to analyze Nedd8 loss-of-function phenotypes, and observed in the adult flies very few Nedd8AN015 mutant cells (Fig. 1D,F), whereas in control experiments, large Nedd8+ clones were frequently recovered (Fig. 1C,E). Nedd8 mutant clones of small size, however, were present in the developing discs, suggesting that Nedd8 mutant cells were defective in proliferation and survival.
Figure 1.
Conservation of Nedd8 in evolution and Drosophila Nedd8 mutant phenotypes. (A) Sequence comparison of Nedd8. Drosophila Nedd8 shares 88%–98% identity to other Nedd8 from yeast to mammals. Also indicated are the point mutations in Nedd8AN024 and Nedd8AN015 alleles. Conceptually, Nedd8AN015 encodes a C-terminus-truncated protein by a nonsense mutation at aa 49. Thus, Gly76, the essential residue required for conjugation to the substrate protein (Wada et al. 1998), was missing in this mutant. In Nedd8AN024, the translation start codon was replaced by a missense mutation. (B) The 4-d-old larvae of wild-type (left) and Nedd8AN024 (right). Note growth of the Nedd8AN024 larva was arrested in the first instar stage. Bar, 0.5mm. (C–F) The growth and survival defect of Nedd8 mutant cells. (C,D) The adult nota of w f hs-FLP; + FRT40A/M2(Z)f+30b FRT40A (C) and w f hs-FLP; Nedd8AN015 FRT40A/M2(Z)f+30b FRT40A (D). (C) Almost all cells in the adult notum are marked with forked (note curly forked bristles) compared to D, in which no Nedd8AN015 mutant clones can be observed. (E,F) The adult eyes of w ey-FLP; + FRT40A/cl2L3 p[w+30C] FRT40A (E) and w ey-FLP; Nedd8AN015 FRT40A/cl2L3 p[w+30C] FRT40A (F). The wild-type clones containing white ommatidia occupy almost the whole eye in E, compared to F in which Nedd8AN015 clones (marked by white ommatidia) are absent, leaving a defective eye that contains only Nedd8AN015/+ cells in red ommatidia. (G–Q) Accumulation of CiFL, Arm, and CycE, and disruption of twist mRNA expression in Nedd8 mutant cells. The wing (G–L) or eye discs (M–O) carrying Nedd8 mutant clones marked by the lack of GFP (H,K) or β-galactosidase expression (N). Accumulation of CiFL (red in G,I) appears only in mutant clones located in the anterior compartment of the wing disc (for imaginal discs in this and following figures, anterior is to the left). Accumulation of Arm (red in J,L) appears in the cytosol of Nedd8 mutant cells (arrows), in contrast to the cell surface-associated Arm in wild-type cells (arrowheads). (M, O) Accumulation of CycE is also detected in the Nedd8 mutant clones in the eye disc. (P,Q) In situ hybridization of twist mRNA in wild-type embryos (P) and Nedd8 mutant embryos laid by Nedd8AN015/Nedd8203 females (Q).
To study the relationship between Nedd8 and the F-box protein Slimb-mediated protein degradation, we examined the protein stability for substrates of Slimb in Nedd8 mutant cells. As shown in Figure 1G–I and J–L, respectively, Nedd8 mutant cells in developing wing discs accumulated high levels of full-length Ci (CiFL) and Arm proteins, phenotypes identical to those observed in the slimb mutants (Jiang and Struhl 1998). In Drosophila embryonic development, the signaling pathway mediated by the NFκB homolog Dorsal is required for patterning the dorsoventral identity. Accumulation of pIκBα/Cactus inhibits Dorsal activation, leading to repression of the downstream target gene, twist, an effect that has been observed in slimb mutants (Spencer et al. 1999). We examined twist expression in embryos laid by Nedd8AN015/Nedd8203 females in which Nedd8203 is a hypomorphic allele (see Materials and Methods). In such embryos, the twist expression domain was reduced along the dorsoventral axis and often found missing in many cells (Fig. 1Q), revealing a requirement for Nedd8 in Dorsal signaling.
We further tested whether Nedd8 affects the protein level of CycE that is regulated by the F-box protein, Archipelago (Ago; Moberg et al. 2001) . As shown in Figure 1M–O, CycE accumulated in Nedd8 mutant cells in the eye disc. Our results suggest that Nedd8 might affect the stability of a broad range of proteins through F-box proteins in flies.
Consequence of CiFL accumulation in Nedd8 mutant cells in the developing eye disc and its response to Hh signaling
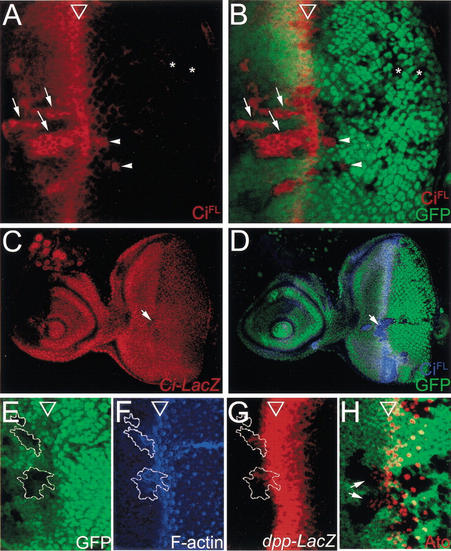
The Drosophila eye imaginal disc is an excellent model system for developmental study. Cells are undifferentiated and dividing randomly anterior to the MF, and cells posterior to the MF are differentiating into different types of cells. Thus, we can observe Nedd8 phenotypes in cells of different differentiation states in a single eye disc. The Hh pathway is the major signaling pathway in eye development, and the protein level of its effector Ci is tightly regulated in Drosophila (Ingham and McMahon 2001). We focused our studies on how Nedd8 regulates the CiFL level in the Hh pathway and the effects of Ci upregulation on eye development. We found that in the Nedd8 clones that located anterior to the MF, CiFL accumulated to a level identical to that in the MF cells that transduce the Hh signaling pathway (Fig. 2A,B, arrows). Accumulation of CiFL also existed in posterior mutant cells that located proximally (Fig. 2A,B, arrowheads) but not distally (Fig. 2A,B, asterisks) to the MF. CiFL accumulation in Nedd8 mutant cells was not caused by an increase in the ci transcription level, because expression of ci-lacZ that recapitulates endogenous ci expression (Jiang and Struhl 1998) remained constant in Nedd8 mutant cells (Fig. 2C,D), indicating that posttranscriptional defects resulted in CiFL accumulation.
Figure 2.
Nedd8 affects transduction of the Hh pathway in Drosophila eye development. (A–D) The CiFL protein, but not ci expression, is upregulated in the Nedd8 mutant cells (revealed by the lack of GFP expression, in green). (A,B) A late third-instar eye disc containing Nedd8 mutant clones and stained by anti-CiFL antibody (2A1) in red. The open arrowheads mark the position of the MF in this and following figures. In the mutant cells anterior to the MF, CiFL accumulation (arrows) reaches a level identical to that observed in the MF where Hh signaling is fully induced. In the posterior mutant cells, CiFL accumulation is persistent at a lower level abutting the MF (arrowheads) and not detected in more posterior cells (asterisks). Note that the anterior clones are obviously larger than the posterior clones, suggesting a survival defect in the Nedd8 mutant cells. (C,D) A late third-instar eye disc containing Nedd8 mutant clones and double stained for ci-LacZ expression in red (C) and CiFL in blue (D). (C) The ci-LacZ expression level is ubiquitous throughout the whole eye disc, and remains unchanged in the Nedd8 mutant clones (C, arrow), in which the CiFL protein accumulates (D, arrow). (E–H) Accumulated CiFL responds to Hh signaling to induce MF fate in Nedd8 mutant clones. (E–G) A late third-instar eye disc containing Nedd8 mutant clones was double stained for F-actin in blue (F) and dpp-LacZ in red (G). (E) The Nedd8 mutant clones are marked by the lack of GFP expression and outlined by a white line in E–G. (F) F-actin staining (blue) by phalloidin shows that the Nedd8 mutant cells constrict precociously. (G) Expression of dpp-lacZ (red) is induced in the mutant cells abutting the MF anteriorly. (H) In the Nedd8 mutant clones, as shown by lack of arm-lacZ expression (green), expression of the proneural protein Atonal (red) is induced (arrows).
Elevated CiFL levels caused anterior Nedd8 mutant cells to adopt MF fate precociously. Nedd8 mutant cells constricted in the apical surface, as revealed by the intensified phalloidin staining (Fig. 2F), and expressed the Hh-target gene, dpp, as detected by the expression of dpp-lacZ reporter gene (Fig. 2G; Blackman et al. 1991). Furthermore, the early photoreceptor marker, Atonal, was induced (Fig. 2H, arrows). These phenotypes were observed only in mutant cells abutting the MF anteriorly, suggesting that accumulated CiFL in Nedd8 mutant cells was able to respond to Hh signaling.
Nedd8-mediated CiFL processing is downstream of Smo signaling and PKA phosphorylation
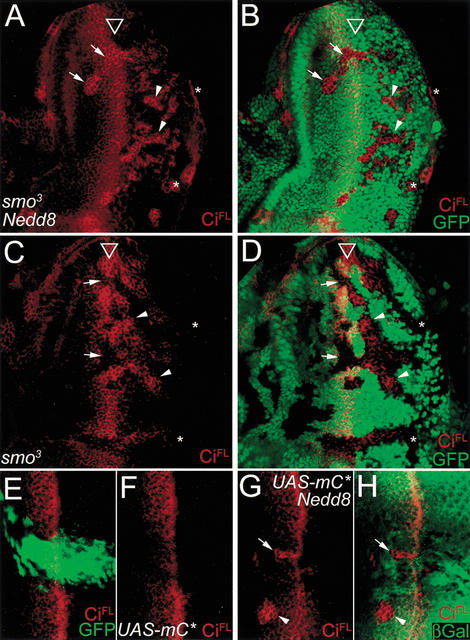
Although CiFL accumulation in Nedd8 cells could result from a defect in the machinery controlling CiFL protein processing, it was equally possible that CiFL accumulation could be caused by the activation of Hh signaling. For example, smo has been shown to be under posttranscriptional regulation by Hh signaling (Alcedo et al. 2000; Denef et al. 2000; Ingham et al. 2000). Therefore, Nedd8 activity could have affected the protein stability of Smo, leading to signal activation and CiFL accumulation. To test whether the effect of Nedd8 on CiFL processing is dependent on smo, we generated smo3 and Nedd8 double mutant clones in the eye disc. When the double mutant clones were located anterior to the MF, the level of CiFL accumulation was indistinguishable from that in the Nedd8 mutant clones (cf. arrows in Figs. 2A and 3A). As a control, anterior smo3 clones failed to enhance CiFL accumulation (Fig. 3C,D). This result indicated that Nedd8-mediated CiFL processing was independent of smo. (CiFL accumulation in posterior cells is described below.)
Figure 3.
The Nedd8 pathway on CiFL proteolytic processing is downstream of the Hh receptor Smo and PKA activity. (A–D) CiFL accumulation in Nedd8 mutant cells is independent of smo activity. (A,B) A late third-instar eye disc carrying smo and Nedd8 double mutant clones and stained for CiFL in red. CiFL accumulates in all double mutant clones (marked by the lack of GFP staining in B) regardless of their locations in the eye disc. (C,D) A late third-instar eye disc carrying smo clones and stained for CiFL in red. In the smo mutant cells marked by the lack of GFP expression (green in D), CiFL expression is missing in clones anterior to and in the MF (arrows). However, CiFL accumulation is detected in mutant cells in the posterior clones (arrowheads) except near the posterior margin (asterisks). (E–H) Nedd8 is required for PKA activity in promoting CiFL proteolytic processing. (E,F) A late third-instar eye disc expressing PKA catalytic domain (UAS-mC*) driven by eq-GAL4 in the equator region, as visualized by the coexpressed GFP (green in E). Constitutive activation of PKA suppresses CiFL accumulation in MF (red in F). (G,H) A late third-instar eye disc expressing UAS-mC* under the control of eq-GAL4 and containing Nedd8 mutant clones. CiFL accumulation (red) persists in the Nedd8 mutant clones even when PKA is constitutively activated in the mutant cells (arrows). The accumulated CiFL level is similar to that in the mutant cells without mC* expression (arrowheads). The Nedd8 mutant clones are marked by the lack of arm-lacZ expression (green in H), and the effect of misexpressed PKA catalytic subunit can be observed by the downregulated CiFL level in MF cells near the Nedd8 clone.
Ci protein processing is known to depend on the phosphorylation status of CiFL by PKA (Chen et al. 1998; Price and Kalderon 1999). The level of CiFL is downregulated when PKA is constitutively activated by the expression of its catalytic subunit (UAS-C*; Li et al. 1995). Therefore, we examined the functional relationship between PKA activity and Nedd8 modification. When the UAS-C* transgene was driven by eq-GAL4 for misexpression in the equator region of the eye disc, as visualized by the coexpressed GFP (green in Fig. 3E), the level of CiFL in the equator region was reduced (Fig. 3F), consistent with the observations that PKA phosphorylates Ci and promotes Ci proteolysis. We then generated Nedd8 mutant clones in the equator region where PKA is constitutively activated. In Nedd8 clones that overlapped the eq-GAL4 expression domain, CiFL accumulated to a high level (Fig. 3G, arrow), identical to the level in the Nedd8 clone located externally to the eq-GAL4 expression domain (Fig. 3G, arrowhead). These results indicated that CiFL downregulation by PKA activity requires Nedd8 activity, and the effect of the Nedd8 pathway on CiFL processing is unlikely to be mediated through modulation of PKA activity.
Nedd8 modifies Cul1 and modulates Cul1 stability in Drosophila
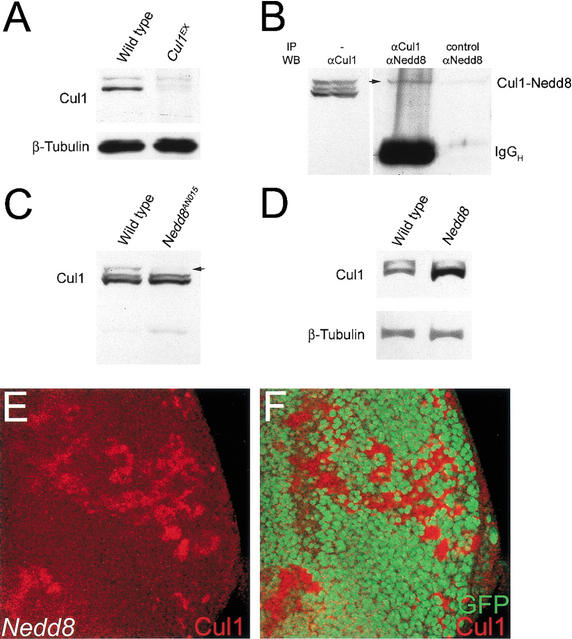
The genetic evidence described above suggested that Nedd8 could be directly required for CiFL proteolytic processing, consistent with the hypothesis that neddylation affects CiFL proteolysis through regulating SCFSlimb activity. Cullin proteins are the identified targets for Nedd8 modification. In the Drosophila genome, six Cullin proteins are identified, each corresponding to a mammalian homolog (Fig. 6I, below). Among them, Cul1 is involved in the formation of SCF complexes that function as E3 ligase. We generated a null Cul1 allele, Cul1EX (see Materials and Methods). In Cul1EX homozygous larvae in the first instar stage, the Cul1 signal detected by αCul1 antibodies was almost gone (Fig. 4A, right lane). The residual Cul1 protein in Cul1EX larvae was probably maternally contributed.
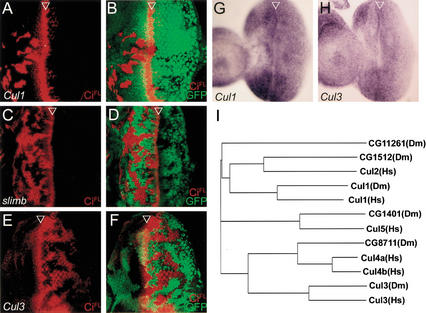
Figure 6.
SCFSlimb complex and a Cul3-based degradation machinery to control the CiFL stability in the anterior and posterior cells of the eye discs, respectively. (A–D) The Cul1-based, SCFSlimb complex functions in the anterior cells to promote CiFL proteolytic processing. In the Cul1EX (A) and slimb1 (C) mutant clones located anterior to the MF, CiFL accumulation is detected (red), whereas in the posterior clones, no accumulation can be observed. The mutant clones of Cul1EX (B) and slimb1 (D) are marked by the lack of GFP expression. (E,F) In the Cul3gft2 mutant clones, marked by the lack of GFP expression (green), only posterior clones accumulate CiFL (red). (G,H) The ubiquitous expression patterns of Cul1 (G) and Cul3 (H) in the eye discs as revealed by in situ hybridization with antisense probes of Cul1 and Cul3, respectively. In the control experiments, the sense probes gave no signals (data not shown). (I) The phylogenetic tree of the Drosophila Cullin family (denoted by Dm) to their human counterparts (Hs).
Figure 4.
Nedd8 modifies Cul1 and controls its stability. (A) Western blot by αCul1 antibodies. Cell extracts were prepared from first-instar larvae of wild-type (left lane) and Cul1EX mutant (right lane). The levels of Cul1 proteins of 90 kD are greatly reduced in Cul1EX cell extract. (B) Conjugation of Nedd8 to Cul1. Western blot of cell extract prepared from the eye discs and brain lobes of third-instar larvae by αCul1 antibodies (left lane). Immunocomplex of Cul1 from precipitation by αCul1 antibodies is blotted by αNedd8 antibodies (middle lane). An arrow indicates the putative Nedd8-modified Cul1 form. In the control experiment without the addition of αCul1 antibodies in precipitation, no Nedd8 positive signal can be detected (right lane). (C) Immunoblotting of cell extract prepared from first-instar larvae of wild-type (left lane) and Nedd8AN015 (right lane) by αCul1 antibodies. The Nedd8-modified form of Cul1 is missing in Nedd8 larvae (right lane, arrow). (D) In the absence of Nedd8 modification, the Cul1 level is increased. Western blot analysis of cell extract from third-instar eye discs and brain lobes by αCul1 antibodies indicates that the Cul1 protein is accumulated in Nedd8AN015/Nedd8EP(2)2063 (right lane). (E,F) A late third-instar eye disc containing Nedd8 mutant clones and stained with αCul1 antibodies (red). Cul1 accumulates in the Nedd8 mutant clones (red in E) that are marked by the lack of GFP expression (green in F).
We further investigated whether Nedd8 conjugates to Cul1 in Drosophila. Cell extracts of eye-antennal discs and brain lobes isolated from wild-type third instar larvae were immunoprecipitated by αCul1 antibodies and immunoblotted by αNedd8 antibodies (Fig. 4B, middle lane). The Nedd8-positive signal, indicated by the arrow in Figure 4B, suggests that Cul1 was modified by Nedd8 in Drosophila. Consistently, the Nedd8-modified Cul1 signal disappeared in cell extracts prepared from the first instar Nedd8 mutant larvae (Fig. 4C, arrow).
In addition, we found that unmodified Cul1 accumulated in the Nedd8 hypomorphic mutants Nedd8EP(2)2063/AN015 (Fig. 4D). This effect of Cul1 accumulation was also observed in Nedd8 null mutant clones in third-instar eye discs (Fig. 4E,F). These data suggested that Nedd8 modification of Cul1 might modulate Cul1 stability.
Cul1 functions in the same pathway as Nedd8 in modulating protein stability
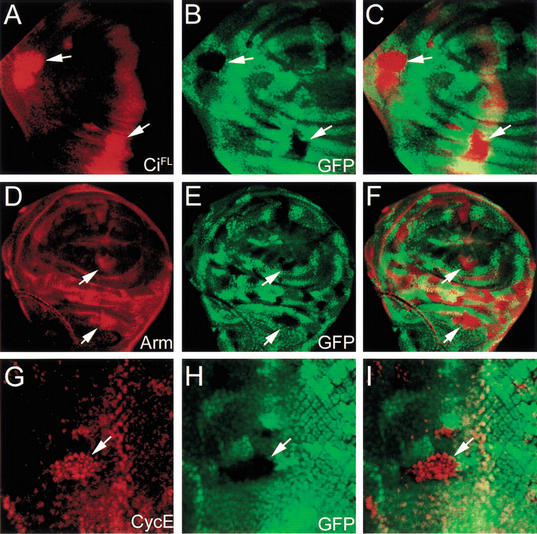
To test whether Nedd8 activity in controlling protein stability is mediated through Cul1, we examined whether depleting Cul1 would exhibit the same phenotypes as depleting Nedd8. Cul1EX clones were generated in developing wing discs, and the protein levels of CiFL and Arm were examined. Accumulations of CiFL and Arm were found in Cul1EX mutant cells (Fig. 5A–C and D–F, respectively), identical to the phenotypes observed in Nedd8 mutant clones. In the eye disc, CycE accumulated in Cul1 mutant cells (Fig. 5 G–I). These results suggest that Nedd8 and Cul1 function together for controlling the protein stability of Ci and Arm in the wing disc and CycE in the eye disc.
Figure 5.
Accumulation of CiFL, Arm, and CycE in Cul1 mutant cells. Cul1EX mutant clones are marked by the lack of GFP expression in wing discs (green in B,E) and eye disc (green in H). (A–C) CiFL (red in A,C) accumulates in the Cul1EX mutant clones (indicated by arrows) in the anterior compartment of the wing discs. (D–F) In Cul1EX mutant cells, Arm is upregulated (red in D,F) in cytosol (arrows) of wing disc cells. (G–I) Enhancement of CycE protein level (red in G,I) is observed in Cul1EX mutant clones (arrows) in the eye disc.
The SCFSlimb complex controls CiFL processing exclusively in cells anterior to the MF of the eye disc
We then tested whether the Cul1-based SCF complex is involved in controlling CiFL stability in the eye disc. When located anterior to the MF, Cul1EX cells accumulated high levels of CiFL (Fig. 6A,B), a phenotype similar to Nedd8 mutant cells. However, Cul1EX cells that located posterior to the MF only expressed a basal level of CiFL, suggesting that the Cul1-based SCF complex may not control Ci stability in the posterior cells.
In addition to the Nedd8-Cul1 core component, the SCF complex also includes a substrate-specific F-box protein. To investigate whether SCF activity in CiFL processing is limited to the anterior cells of the eye disc, we examined the mutant phenotype of slimb that is required for CiFL proteolytic processing in tissues such as wing and leg discs (Jiang and Struhl 1998). When slimb1 mutant clones were generated in eye discs, high levels of CiFL accumulation were detected exclusively in clones located anterior to the MF. No accumulation of CiFL could be detected in posterior slimb1 clones (Fig. 6C,D). Suppression of CiFL accumulation in the posterior cells was not due to possible residual activity present in hypomorphic slimb1, because we observed identical results of CiFL accumulation in the strong hypomorphic allele slimb2 and the null allele slimbP (data not shown).
In summary, our results strongly suggested that in vivo, the Nedd8-modified, Cul1-based SCFSlimb complex controls CiFL proteolysis in anterior cells. Following the sweep of the MF, CiFL stability in the posterior cells is controlled by an SCFSlimb-independent mechanism.
Ci downregulation in the posterior cells of the eye disc requires Smo signaling and Nedd8 modification activity
The finding that CiFL accumulated in posterior smo3 clones (Fig. 3C,D, arrowheads; Dominguez 1999) indicated that Smo signaling contributes to the downregulation of CiFL in the posterior cells of the eye disc. This effect is in contrast to the smo role in the MF, where smo is required for CiFL activation (Fig. 3C,D, arrows). CiFL accumulation was also observed in the posterior Nedd8 mutant clones located proximally to the MF (Fig. 2A,B, arrowheads). In the smo3 Nedd8 double mutant clones, the level of CiFL was further enhanced (Fig. 3A,B, arrowheads), even in clones located distally to the MF (Fig. 3A,B, asterisks), whereas no CiFL accumulation was detected in Nedd8 (Fig. 2A,B, asterisks) or smo3 clones (Fig. 3C,D, asterisks), suggesting that Nedd8 and Smo function partially redundantly to downregulate Ci stability in the posterior cells of the eye disc.
CiFL degradation in the posterior cells of the eye disc is mediated by a Cul3-dependent mechanism
The involvement of Nedd8 in controlling CiFL levels in the posterior cells of the eye disc suggests that Cullin proteins other than Cul1 may be involved in the posterior mechanism to control Ci stability. Among the mammalian Cullin family, Cul3 shares with the Cul1-based SCF complex the substrate CycE (Singer et al. 1999). To test whether Cul3 affects CiFL degradation in the eye disc, we analyzed the available Drosophila Cul3 mutants (see Materials and Methods). We found that CiFL accumulated in Cul3 mutant clones located posterior to the MF (Fig. 6E,F), with a higher level in nondifferentiating cells that surround differentiating photoreceptor clusters. In contrast, no CiFL accumulation was detected in anterior Cul3 mutant clones, indicating that Cul3 controls CiFL protein stability only in the posterior cells of the eye disc. Ci accumulation in posterior Cul3 mutant cells was controlled at the posttranscriptional level because ci expression was normal, as revealed by in situ hybridization (data not shown). These results showed that the CiFL degradation machinery in the posterior cells of the eye disc requires a Cul-3-mediated degradation mechanism. Ci accumulation was also detected in Cul3 mutant cells located in the A/P boundary of the wing disc (data not shown). The level of Arm in Cul3 mutant clones in wing discs and the level of CycE in Cul3 mutant clones in eye discs remained constant (data not shown), suggesting that Cul3 activity was specific to Ci.
In contrast to the Cul1-based SCFSlimb complex that controls CiFL processing only in the anterior cells of the eye disc, the Cul3-mediated Ci degradation mechanism is specific to the posterior cells. These specific activities in controlling Ci protein stability were not caused by differential gene expression of Cul1 and Cul3 in the eye disc. We detected ubiquitous mRNA expression patterns of both Cul1 (Fig. 6G) and Cul3 (Fig. 6H), and ubiquitous Cul1 protein expression all along the eye disc (Fig. 4E), suggesting that control of specificity is mediated by mechanisms other than regulation of Cul1 and Cul3 expression.
CiFL degradation in the posterior cells of the eye disc is constitutive and PKA-independent
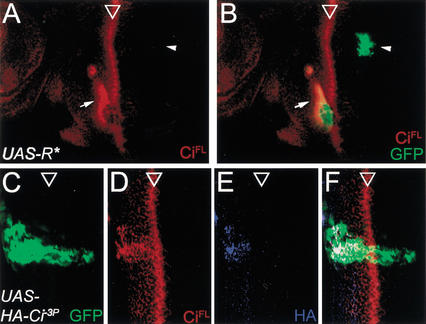
PKA phosphorylation promotes CiFL processing, and plays a role in the Hh signaling pathway for Ci activation. We therefore examined the requirement of PKA in CiFL degradation in the posterior cells of the eye disc. We generated PKA-deficient cells by overexpressing the regulatory subunit of PKA (UAS-R*) that functions to sequester the catalytic subunit and, therefore, to inhibit PKA activity (Li et al. 1995). The UAS-R* transgene was expressed in random clones of cells labeled with GFP (see Materials and Methods). Consistent with the requirement of PKA activity in CiFL processing, CiFL accumulated when PKA activity was inhibited in GFP-positive cells located anterior to the MF (Fig. 7A,B, arrows). In contrast, no CiFL accumulation could be detected in GFP-positive cells located posterior to the MF (Fig. 7A,B, arrowheads), indicating that CiFL downregulation is not regulated by PKA activity.
Figure 7.
Regulation of CiFL degradation in posterior cells of the eye disc. (A,B) PKA activity is not required for CiFL degradation in the posterior cells. To inhibit PKA activity, the dominant negative form of PKA regulatory domain is expressed in random clones marked by GFP expression (see Materials and Methods). The protein level of CiFL (red) is upregulated in anterior (arrow) but not posterior clones (arrowhead). (C–F) CiFL degradation is independent of PKA phosphorylation. The PKA phosphorylation site-mutated form of Ci, Ci−3P, is expressed by eq-GAL4 in the equator, as shown by the coexpressed GFP (C). (D) Misexpressed Ci−3P is refractory to PKA-mediated proteolytic processing in the anterior cells, as shown by antibody staining for CiFL. However, no Ci−3P expression can be detected in the posterior cells. (E) The Ci−3P is fused with an HA tag in the N terminus, and no signal can be detected with an antibody against HA in the posterior cells, suggesting that both the full-length and the short form of Ci are degraded completely. (F) The merged image of C,D,E.
PKA-independent CiFL downregulation in posterior cells of the eye disc could cause either proteolytic processing of CiFL to the short form, Ci75, or complete Ci degradation. To address this question, we expressed the UAS-HACi−3P transgene in which an HA tag was fused to the N terminus of Ci and the three PKA phosphorylation sites required for Ci processing were mutated (Wang et al. 1999). Expression of the phosphorylation-defective form of CiFL was driven by eq-GAL4 in the equator region of the eye disc and monitored by the coexpressed GFP (Fig. 7C). While the αCiFL antibody detected a high level of CiFL in the anterior equator of the eye disc where PKA phosphorylation is required for CiFL processing, expression of CiFL was completely diminished in the posterior cells in which the GFP signal was strongly detected (Fig. 7D). The αHA antibody that could recognize the full-length Ci (Ci155) and the short form of Ci (Ci75) also failed to detect any signal in the posterior region (Fig. 7E), suggesting that instead of processing to Ci75, CiFL was completely degraded. We also tested another Ci mutant in which, in addition to the phosphorylation sites, the proteolytic processing site of Ci was also mutated (Wang et al. 1999). Expression of the uncleavable and unphosphorylated CiFL (UAS-HACiU−3P) by eq-GAL4 in the equator region revealed no detectable Ci level in the posterior equator region (data not shown), similar to the result obtained from expression of UAS-HACi−3P. This result suggested that proteolytic processing of CiFL to the short form Ci75 is not a prerequisite for complete degradation in the posterior cells, in contrast to the proteolytic processing of the phosphorylated CiFL to the short form Ci75 in the anterior cells. To sum up, our results suggested that in the posterior cells of the eye disc, CiFL is degraded constitutively, and this degradation process is independent of PKA phosphorylation.
Discussion
Nedd8-modified Cul1 is required for the function of different SCF complexes in vivo
In this work, we first isolated and characterized Drosophila Nedd8 and Cul1 mutants. Immunostaining of the developing discs showed that Nedd8 and Cul1 are required for controlling the protein stabilities of Arm, CiFL, and CycE (Figs. 1,5). In Nedd8 mutant clones, the presence of Cul1, although at a higher level (Fig. 4E), is unable to downregulate the protein stability. In addition, we showed that in Drosophila Nedd8 modifies Cul1 in biochemical assays (Fig. 4B,C). These results indicate that neddylation of Cul1 is essential for Cul1 activity in vivo.
Accumulation of Arm and CiFL has also been reported for slimb mutants (Jiang and Struhl 1998). The identified F-box protein Ago has been shown to control CycE degradation in vivo (Moberg et al. 2001). These results are consistent with the idea that individual SCF targets specific substrates for protein degradation via a recognition process mediated by a specific F-box protein, such as Slimb for Arm and Ci, and Ago for CycE in flies. Since Nedd8 affects substrate stability that depends on either Slimb or Ago, we infer that Nedd8 modification regulates SCF activity, but not substrate specificity.
Nedd8 modification regulates both activity and stability of Cul1
We found that in biochemical assay, the unmodified Cul1 accumulated with the reduction of Nedd8 activity (Fig. 4D). In Nedd8 mutant cells in the developing eye disc, Cul1 also accumulated to a high level (Fig. 4E,F), raising the possibility that neddylation activates Cul1 and at the same time turns the modified Cul1 into a labile state for protein degradation. In this way, Cul1 activity could be tightly controlled in signaling pathways during developmental processes. Although the mechanism by which Nedd8-modified Cul1 is targeted for protein degradation is not clear, it has been shown that some unknown proteins with Nedd8 conjugation are targeted for proteasome degradation by the Nedd8-interacting protein NUB1 (Kamitani et al. 2001).
Possible roles of Nedd8 modification and SCFSlimb in Ci proteolytic processing
In anterior cells of developing discs, we show that CiFL proteolytic processing requires the activity of the Nedd8-modified, Cul1-based SCFSlimb complex. This CiFL proteolytic processing is inhibited by Smo signaling and promoted by PKA phosphorylation on CiFL. The mechanism by which CiFL is proteolyzed from CiFL to Ci75 is not clear. We propose that Nedd8 modifies and activates SCFSlimb for Ci ubiquitination and then proteolysis, because of Cul1 modification by Nedd8 and CiFL accumulation in Nedd8, Cul1, and slimb mutants. Consistently, proteolysis of CiFL depends on 26S proteasome activity (Ingham 1998). However, ubiquitinated Ci is not detected in cells treated with 26S proteasome inhibitors (Chen et al. 1999). We could not exclude the possibility that Nedd8 directly modifies Ci through the SCF complex, because it has been shown that Nedd8 might direct its conjugates to proteasome-dependent protein degradation through NUB1 (Kamitani et al. 2001). In this case, SCF could function as the E3 ligase for Nedd8 modification on its substrate (Kamura et al. 1999a).
In the Hh signaling pathway, it is not clear how Smo signaling prevents CiFL from proteolysis. According to our double mutant analysis (Fig. 3), Nedd8 could be downstream or parallel to Smo and PKA signaling. Thus, it is possible that Hh signaling prevents CiFL from proteolysis through downregulating the level of Nedd8-modified Cul1. However, we failed to detect a change in the level of Nedd8-modified Cul1 in cell extracts prepared from the eye discs with ectopic Hh expression (data not shown). We infer that Hh may affect CiFL proteolysis through a Nedd8-independent mechanism.
A distinct protein degradation mechanism in controlling Ci stability in posterior cells of the eye disc
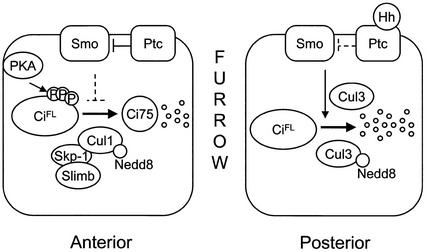
We propose two modes of Ci downregulation in Drosophila eye development (Fig. 8). In the undifferentiated cells anterior to the MF, Ci is phosphorylated by PKA constantly and processed by an SCFSlimb-dependent mechanism to generate the repressor form of Ci75. Upon binding to Hh, cells in the MF transduce Smo signaling to prevent this proteolytic processing. Thus, the transcriptional activator CiFL is preserved for activation of downstream genes in the MF.
Figure 8.
A proposed model for Ci degradation in Drosophila eye development (see Discussion for details).
In the posterior cells that are undergoing differentiation, a novel mechanism controls Ci degradation. Our mutant analyses suggest that this mechanism is comprised of Smo signaling, Nedd8 modification, and Cul3 activity. The effect of Smo signaling in promoting Ci degradation in the posterior cells is in contrast to its effect on the anterior cells, in which Smo signaling prohibits CiFL processing. In addition to Smo signaling, Nedd8 modification activity also participates in this posterior Ci degradation. Further Cul1 mutant analysis suggests that Cullin proteins other than Cul1 are likely involved in this posterior degradation mechanism. This hypothesis has led to the identification of Cul3 as one candidate functioning in Ci degradation. More surprisingly, Cul3 activity is very restricted; Cul3 controls Ci degradation in the posterior, but not anterior, cells of the eye disc. CiFL accumulation may have an impact on proper differentiation of the posterior cells. In Cul3 mutants, cone cell differentiation is affected (C.-Y. Ou and C.-T. Chien, unpubl.), probably due to the accumulation of CiFL.
Furthermore, the Ci degradation process is also distinct in posterior cells; Ci degradation is independent of PKA phosphorylation and proteolytic processing to the short form Ci-75 (Fig. 7). Based on these results, we propose that Smo signaling, acting in concert with the Nedd8 pathway, activates a Cul3-based ubiquitin ligase to degrade Ci in a PKA-independent mechanism in posterior cells of the eye disc.
It is not clear how Nedd8 modifies Cul3 in flies. We observed strong genetic interaction between Nedd8 and Cul3 during eye and antennal development (Y.-F. Lin and C.-T. Chien, unpubl.), suggesting that Nedd8 may also regulate Cul3. However, depletion of Nedd8 activity only affects posterior cells abutting the MF (Fig. 2A), in contrast to depletion of Cul3 activity, which increases the CiFL level in all posterior clones (Fig. 6E), indicating that some Cul3 activity is Nedd8-independent. It is possible that a basal Cul3 activity for Ci degradation is further enhanced by Nedd8 modification near the MF in which accumulated Ci may require efficient degradation for cells to enter proper differentiation.
Switch of Ci degradation mechanisms
Different protein-protein interactions may result in a switch between two Ci degradation mechanisms in eye discs. Ci is known to interact with Cos2, Fu, and Su(fu) to comprise a protein complex (for review, see Ingham and McMahon 2001) that promotes Ci degradation. Cos2, a motor-like protein with a kinesin motif, is required for tethering Ci in the cytosolic compartment and Ci proteolytic processing in the Drosophila developing wing. Similarly, Fu, a serine/threonine kinase, is also required for Ci processing. However, in Su(fu) mutants, levels of both long and short forms of Ci are reduced (Ohlmeyer and Kalderon 1998; Lefers et al. 2001), suggesting that Su(fu) plays an additional role in Ci protein stability. Interestingly, the role of Su(fu) in controlling Ci stability seems modulated by Hh signaling. Our results indicate that, in contrast to the effect of Hh signaling in the anterior cells, Hh signaling downregulates the Ci level in the posterior cells of the eye disc. It is possible that the Ci protein complex is modulated by the sweep of the MF, and this change requires Hh signaling to expose Ci to the Cul3-based protein degradation machinery. Alternatively, additional factors may be activated by the sweeping of the MF and be required for Hh signaling to induce Cul3 activity that leads to constitutive Ci degradation.
Similar to Ci, CycE is degraded by two different mechanisms in mammalian cells. The Cul1-based SCF complex recognizes the phosphorylated form of Cdk2-bound CycE for ubiquitination (Dealy et al. 1999; Skowyra et al. 1999; Koepp et al. 2001; Strohmaier et al. 2001; Yeh et al. 2001), and Cul3 targets unbound CycE for ubiquitination, which is independent of protein phosphorylation (Singer et al. 1999). β-catenin is also degraded by two different mechanisms in mammalian cells (Polakis 2001). One mechanism involves the SCFβTrCP complex (βTrCP is the mammalian homolog of Slimb) that recognizes phosphorylated β-catenin. The other mechanism involves the Ebi complex comprised of Ebi, Skp1, SIP, and Siah-1, which targets β-catenin in a phosphorylation-independent manner.
Degradation of the Gli proteins in vertebrates
In vertebrates, the three Ci-related proteins Gli1, Gli2, and Gli3 transduce Hh signaling in different developmental processes (for review, see Ingham and McMahon 2001). Ectopic expression of the Gli proteins in Drosophila showed that Gli2 and Gli3, but not Gli1, are proteolyzed to generate repressor forms. Although the proteolytic cleavage of Gli3 is under the regulation of Hh signaling, Gli2 proteolysis is independent of Hh (Aza-Blanc et al. 2000). Consistently, proteolytic processing of Gli3, but not Gli1, has been observed in mouse embryos (Dai et al. 1999). In cultured cells, Gli3 processing is dependent on Hh signaling and PKA activity, in contrast to Gli1 and Gli2 (Dai et al. 1999; Ruiz i Altaba 1999). Apparently, the Gli proteins are controlled by different protein downregulation mechanisms. It will be interesting to investigate whether Nedd8, Cul1, Cul3, and perhaps other Cullins are differentially involved in protein degradation of the Gli proteins.
Materials and methods
Genetics
The Drosophila Nedd8 alleles, Nedd8AN015 and Nedd8AN024 (Fig. 1A), were recovered in an EMS screen for lethal mutations that failed to complement Df(2L)TW3 (36F7–9, 37B2–7). The insertion site of EP(2)2063 is located 91 bp upstream of the translation start site of Nedd8. Imprecise excision of the EP element generated a null allele Nedd8172 that included a deletion of the N-terminal half of the Nedd8 protein (aa 1–45), and a weak allele Nedd8203 that included a small insertion of 45 bp in the EP insertion site.
The Drosophila lin-19 allele (from Bloomington Stock Center) contains a P-element insertion in the first exon, 990 bp upstream of the start ATG codon. lin-19 homozygotes died in the third-instar larval stage. Excision of the P-element rescued the larval lethality, suggesting that the P-element insertion caused the lethality. Immunoblotting lin-19 cell extracts for Cul1, and complementation test with a Cul1 null allele, Cul1EX, suggested that lin-19 is a hypomorphic allele of Cul1. We performed imprecise excision of the P-element to isolate a null allele Cul1EX, which includes a deletion of aa 1–90 of the Cul1 protein. When homozygous, or in trans to the deficiency Df(2R)CA53 (43E7–18;44B6) that uncovers Cul1, Cul1EX caused lethality in the second-instar larval stage, suggesting that Cul1EX is a null allele. Cul306430 is a P-element insertion line, and Cul3gft2 is an EMS allele (both from Bloomington Stock Center), and both function like genetic null and exhibit identical phenotypes described. To identify homozygotes of Nedd8, Cul1, or Cul3, the green balancer CyO Kr-GAL4 UAS-GFP was used.
Other fly stocks used were f36a, M(2L)24F f+30b (Cifuentes and Garcia-Bellido 1997), ey-FLP, cl2L3 (Newsome et al. 2000), ubi-nlsGFP (gift from S. Luschnig), arm-LacZ FRT40A (gift from J. Treisman), ci-LacZ (Eaton and Kornberg 1990), dpp-LacZ (Blackman et al. 1991), smo3 (Chen and Struhl 1996; van den Heuvel and Ingham 1996), Act<y+<Gal4 UAS-GFPS65T (Ito et al. 1997), eq-GAL4 (gift from H. Sun), UAS-mC*, UAS-R* (Li et al. 1995), UAS-HACi−3P (Wang et al. 1999), slimb1, slimb2, and slimbP1493 (Jiang and Struhl 1998). The following flies were used to generate Nedd8 clones: hs-FLP122; Nedd8AN015 FRT40A/cl2L3 P[ubi-nlsGFP] FRT40A (Fig. 1G–L), w ey-FLP; Nedd8AN015 FRT40A/cl2L3 P[arm-LacZ] FRT40A (Figs. 1M–O, 2H), and w ey-FLP; Nedd8AN015 FRT40A/cl2L3 P[ubi-nlsGFP] FRT40A (Figs. 2A–G and 4E,F). We used w ey-FLP; smo3 FRT40A/cl2L3 P[ubi-nlsGFP] FRT40A flies for generation of smo clones, and w ey-FLP; smo3 Nedd8AN015 FRT40A/cl2L3 P[ubi-nlsGFP] FRT40A for Nedd8 and smo double mutant clones. To generate Nedd8 clones in mC* expression domain in eye discs, UAS-mC* Nedd8AN015 FRT40A/cl2L3 P[ubi-nlsGFP] FRT40; eq-Gal4 UAS-GFP UAS-Flp flies were used. To generate clones expressing R*, hs-Flp, Act<y+<Gal4 UAS-GFPS65T/UAS-R* flies were used. To generate Cul1 and Cul3 clones, we used w ey-FLP (or hs-FLP); FRT42 Cul1EX/FRT42 P[ubi-nlsGFP] and w ey-FLP; Cul3gft2 FRT40A/cl2L3 P[ubi-nlsGFP] FRT40A flies, respectively.
Immunohistochemistry, Western blotting, and immunoprecipitation
The imaginal discs were fixed and stained as described previously (Chen and Chien 1999). The primary antibodies used were rat αCi (1:1; Motzny and Holmgren 1995), rabbit αAto (1:1000; Jarman et al. 1994), rabbit αβ-galactosidase (1:1000; Cappel), rat αElav (1:125; Developmental Studies Hybridoma Bank), mouse αArm N27A1 (1:100; Peifer et al. 1994), mouse αCycE (1:3; Moberg et al. 2001), mouse αHA (1:100; Roche), and rabbit αMyc (9E10, 1:250; Santa Cruz).
The Western blotting and immunoprecipitation were performed as in standard protocol. The primary antibodies used were rabbit αCul1 (1:250; Zymed) and rabbit αNedd8 (1:500; Alexis). For immunoprecipitation of Cul1, 200-μL cell extracts from eye imaginal discs and brains of 50 third-instar larvae were added with 5 μg of rabbit αCul1 antibodies. In the control experiment, the rabbit αCul1 antibodies were not added.
Acknowledgments
We thank H.Y. Sun, A. Garcíbia-Bellido, B. Dickson, J. Treisman, J. Jiang, R. Holmgren, T. Kornberg, S. Luschnig, H. Richardson, J.-C. Hsu, T.-B. Chou, and the Bloomington Stock Center for fly stocks and reagents. We especially thank J. Jiang for discussions and comments on the manuscript. We are grateful to J.-L. Chao, C.-C. Chang, Y.-C. Tsai, J.-M. Bai, Y.-H. Chou, S.-D. Yeh, R.-Y. Lin, and all lab members for advice and technical support. This work was supported by grants from Academia Sinica and the National Science Council of Taiwan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ctchien@ccvax.sinica.edu.tw; FAX 886-2-2782-6085.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1011402.
References
- Alcedo J, Zou Y, Noll M. Posttranscriptional regulation of Smoothened is part of a self-correcting mechanism in the Hedgehog signaling system. Mol Cell. 2000;6:457–465. doi: 10.1016/s1097-2765(00)00044-7. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Lin HY, Ruiz i Altaba A, Kornberg TB. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development. 2000;127:4293–4301. doi: 10.1242/dev.127.19.4293. [DOI] [PubMed] [Google Scholar]
- Blackman RK, Sanicola M, Raftery LA, Gillevet T, Gelbart WM. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development. 1991;111:657–666. doi: 10.1242/dev.111.3.657. [DOI] [PubMed] [Google Scholar]
- Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- Chen CK, Chien CT. Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc Natl Acad Sci. 1999;96:5055–5060. doi: 10.1073/pnas.96.9.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for Patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gallaher N, Goodman RH, Smolik SM. Protein kinase A directly regulates the activity and proteolysis of cubitus interruptus. Proc Natl Acad Sci. 1998;95:2349–2354. doi: 10.1073/pnas.95.5.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes FJ, Garcia-Bellido A. Proximo-distal specification in the wing disc of Drosophila by the nubbin gene. Proc Natl Acad Sci. 1997;94:11405–11410. doi: 10.1073/pnas.94.21.11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- Dealy MJ, Nguyen KV, Lo J, Gstaiger M, Krek W, Elson D, Arbeit J, Kipreos ET, Johnson RS. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet. 1999;23:245–248. doi: 10.1038/13886. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Dominguez M. Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development. 1999;126:2345–2353. doi: 10.1242/dev.126.11.2345. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Hafen E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes & Dev. 1997;11:3254–3264. doi: 10.1101/gad.11.23.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Kornberg TB. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes & Dev. 1990;4:1068–1077. doi: 10.1101/gad.4.6.1068. [DOI] [PubMed] [Google Scholar]
- Greenwood S, Struhl G. Progression of the morphogenetic furrow in the Drosophila eye: The roles of Hedgehog, Decapentaplegic and the Raf pathway. Development. 1999;126:5795–5808. doi: 10.1242/dev.126.24.5795. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Wolff T, Rubin GM. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75:913–926. doi: 10.1016/0092-8674(93)90535-x. [DOI] [PubMed] [Google Scholar]
- Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- Ingham PW. Transducing Hedgehog: The story so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes & Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Nystedt S, Nakano Y, Brown W, Stark D, van den Heuvel M, Taylor AM. Patched represses the Hedgehog signalling pathway by promoting modification of the Smoothened protein. Curr Biol. 2000;10:1315–1318. doi: 10.1016/s0960-9822(00)00755-7. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F- box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Kamitani TF, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;3:3. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes & Dev. 1999a;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999b;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Lefers MA, Wang QT, Holmgren RA. Genetic dissection of the Drosophila Cubitus interruptus signaling complex. Dev Biol. 2001;236:411–420. doi: 10.1006/dbio.2001.0345. [DOI] [PubMed] [Google Scholar]
- Li W, Ohlmeyer JT, Lane ME, Kalderon D. Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell. 1995;80:553–562. doi: 10.1016/0092-8674(95)90509-x. [DOI] [PubMed] [Google Scholar]
- Ma C, Zhou Y, Beachy PA, Moses K. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell. 1993;75:927–938. doi: 10.1016/0092-8674(93)90536-y. [DOI] [PubMed] [Google Scholar]
- Maniatis T. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways. Genes & Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCFskp2 toward p27kip1. Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- Motzny CK, Holmgren R. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes & Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Saeki M, Katayama S, Aida N, Toh EA, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Sweeton D, Casey M, Wieschaus E. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. More than one way to skin a catenin. Cell. 2001;105:563–566. doi: 10.1016/s0092-8674(01)00379-8. [DOI] [PubMed] [Google Scholar]
- Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Price MA, Kalderon D. Proteolysis of Cubitus interruptus in Drosophila requires phosphorylation by protein kinase A. Development. 1999;126:4331–4339. doi: 10.1242/dev.126.19.4331. [DOI] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. Nedd8 modification of Cul-1 activates SCFβTrCP-dependent ubiquitination of IκBα. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: Implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes & Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes & Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 1998;251:688–692. doi: 10.1006/bbrc.1998.9532. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes & Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes & Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: New wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yeh KH, Kondo T, Zheng J, Tsvetkov LM, Blair J, Zhang H. The F-box protein SKP2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin-dependent degradation of cyclin E. Biochem Biophys Res Commun. 2001;281:884–890. doi: 10.1006/bbrc.2001.4442. [DOI] [PubMed] [Google Scholar]