Abstract
Pharmacogenetic approaches are widely expected to bring about a “revolution” in medicine. While the application of molecular genetic approaches to disease research will provide us with new opportunities for progressively more targeted and, hopefully, more effective treatments, these developments will be evolutionary in nature and will, for their realization, still require the painstaking process that discovering and developing a new drug entails. It is also quintessential for the realization of these promises that we support a more rational understanding and more realistic expectations in the public at large through dialogue and information.
Keywords: drug-response, genetics, pharmacogenetics, pharmacogenomics, pharmacology
Introduction
There can be no doubt that the advances of molecular biology and molecular genetics and genomics, and of the associated methods and technologies, have had major impact on our understanding of biology and drug action, and that these tools are quintessential and indispensable for future progress in biomedicine and health care. The interface between these methods and concepts, and the discovery, development, and use of new medicines are being recognized as new ‘disciplines’, or facets of biomedical science, termed pharmacogenetics and pharmacogenomics.
Definition of terms
There is widespread indiscriminate use of, and thus confusion about the terms ‘pharmacogenetics’ and ‘pharmacogenomics’. While no universally accepted definition exists, there is emerging consensus on the differential connotation of the two terms (see Table 1).
Table 1.
Terminology.
| Pharmacogenetics: |
| •differential effects of a drug – in vivo – in different patients, dependent on the presence of inherited gene variants |
| •assessed primarily by genetic (SNP) and genomic (expression) approaches |
| •a concept to provide more patient/disease-specific health care |
| •one drug – many genomes (i.e. different patients) |
| •focus: patient variability |
| Pharmacogenomics: |
| •differential effects of compounds – in vivo or in vitro – on gene expression, among the entirety of expressed genes |
| •assessed by expression profiling |
| •a tool for compound selection/drug discovery |
| •many ‘drugs’ (i.e. early stage compounds) – one genome (i.e. ‘normative’ genome [database, technology platform]) |
| •focus: compound variability |
Pharmacogenomics
Pharmacogenomics, and its close relative toxicogenomics are etymologically linked to ‘-genomics’, the study of the genome and of the entirety of expressed and nonexpressed genes. These two fields of study are concerned with a comprehensive, genome-wide assessment of the effects of certain interventions, mainly drugs or toxicants. Pharmacogenomics is concerned with the systematic assessment of how chemical compounds modify the overall expression pattern in certain tissues of interest. In contrast to pharmacogenetics, pharmacogenomics does not focus on differences from one person to the next with regard to the drug's effects, but rather focuses on differences among several drugs or compounds with regard to a ‘generic’ set of expressed or nonexpressed genes (most commonly using quantitative measures of expression) and their (possible) association with phenotype characteristics.
Pharmacogenetics
In contrast, the term ‘-genetics’ relates etymologically to the presence of individual properties as a consequence of having inherited them. Thus, the term pharmacogenetics describes the interactions between drug and individuals' characteristics (which may be related to inborn traits to a larger or lesser extent). Pharmacogenetics therefore is based on observations of clinical efficacy and/or the safety and tolerability profile of a drug in individuals – the phenotype – and tests the hypothesis that interindividual differences in the observed response may be associated with the presence or absence of individual-specific biological markers that may allow prediction of individual drug response. Such markers are most commonly polymorphisms at the level of the nuclear DNA, but conceivably also other types of nucleic acid-derived data, such as quantitative gene expression measurements, which serve as surrogates for the presence of underlying variants in the DNA.
Thus, although both pharmacogenetics and pharmacogenomics refer to the evaluation of drug effects using nucleic acid technology, the directionalities of their approaches are distinctly different: pharmacogenetics represents the study of differences among a number of individuals with regard to clinical response to a particular drug, whereas pharmacogenomics represents the study of differences among a number of compounds with regard to gene expression response in a single (normative) genome/expressome. Accordingly, the fields of intended use are distinct: the former will help in the clinical setting to find the best medicine for a patient, the latter in the setting of pharmaceutical research and development to find the best drug candidate from a given series of compounds under evaluation.
Pharmacogenomics: finding new medicines quicker and more efficiently
Once a screen (assay) has been set up in a drug discovery project, and lead compounds are identified, the major task becomes the identification of an optimized clinical candidate molecule among the many compounds synthesized by chemists. Conventionally, such compounds are screened in a number of animal or cell models for efficacy and toxicity, experiments that – while having the advantage of being conducted in the in vivo setting – commonly take significant amounts of time and depend entirely on the similarity between the experimental animal condition/setting and its human counterpart, i.e. the validity of the model.
Although such experiments will never be entirely replaced by expression profiling on either the nucleic acid (genomics) or the protein (proteomics) level, these techniques offer powerful advantages and complimentary information. First, efficacy and profile of induced changes can be assessed in a comprehensive fashion (within the limitations – primarily sensitivity and completeness of transcript representation) of the technology platform used. Second, these assessments of differential efficacy can be carried out much more expeditiously than in conventionally used, physiology-based animal models. Third, the complex pattern of expression changes revealed by such experiments may provide new insights into possible biological interactions between the actual drug target and other biomolecules, and thus reveal new elements, or branch-points of a biological pathway. Fourth, increasingly important, these tools serve to determine specificity of action among members of gene families that may be highly important for both efficacy and safety of a new drug. It must be borne in mind that any and all such experiments are limited by the coefficient of correlation with which the surrogate markers examined are linked to the desired in vivo physiological action of the compound.
As a subcategory of this approach, toxicogenomics is increasingly evolving as a powerful adjuvant to classic toxicological testing. As pertinent databases are being created from experiments with known toxicants, revealing expression patterns that may potentially be predictive of longer-term toxic liabilities of compounds, future drug discovery efforts should benefit by insights allowing earlier ‘killing’ of compounds likely to cause such complications.
It is imperative, however, to understand the probabilistic nature of such experiments: a promising profile on pharmacogenomic and toxicogenomic screens will enhance the likelihood of having selected an ultimately successful compound, and will achieve this goal quicker than conventional animal experimentation, but will do so only with a certain likelihood of success. The less reductionist approach of the animal experiment will still be needed. It is to be anticipated, however, that such approaches will constitute an important, time- and resource-saving first evaluation or screening step that will help to focus and reduce the number of animal experiments that will ultimately need to be conducted.
Pharmacogenetics: more targeted, more effective medicines for our patients
Genes and environment
It is common knowledge that today's pharmacopea – in as much as it represents enormous progress compared with what our physicians had only 15 or 20 years ago – is far from perfect. Many patients respond only partially, or fail to respond altogether, to the drugs they are given, and others suffer serious adverse events. If we accept, reasonably, that all common complex diseases – i.e. the health problems that are the main contributors to public and private health spending – are the results of complex, multifactorial interactions between inborn predispositions and susceptibilities on the one hand, and external, environmental factors on the other, then the problem of interindividual variance of response to medication is but one of the aspects of this complexity, and may, likewise, be assumed to have as much to do with external influences (e.g. noncompliance, wrong dose) as with inherent (i.e. inherited, genetically determined) ones.
Clearly, a better, more fundamental understanding of the nature of genetic predispositions to disease, and of pathology and of drug action on the molecular level, is essential for future progress in health care. Current progress in molecular biology and genetics has indeed provided us with some of the prerequisite tools to reach this more refined understanding.
Drugs, among all the ‘environmental factors’ that we are exposed to, may be particularly likely to ‘interact’ specifically and selectively with the genetic properties of a given individual, as their potency pitches them into a narrow ‘therapeutic window’, precariously balanced between potent potions and perilous poisons. We would predict that, based on a patient's innate, individual biological makeup – as it affects the interaction with a drug – one or the other of these properties may manifest itself; this phenomenon is covered by the term pharmacogenetics.
An attempt at a systematic classification
Several conceptually very different scenarios of such individual-specific drug response may be distinguished (see Table 2). They include, on the one hand, differential pharmacokinetics, due to interindividual differences in absorption, distribution, metabolism (with regard to both activation of prodrugs, inactivation of the active molecule, and generation of derivative molecules with biological activity), or excretion of the drug. In any of these cases, differential effects are observed due to the presence at the intended site of action either of inappropriate concentrations of the pharmaceutical agent, or of inappropriate metabolites, or of both. Pharmacogenetics, as it relates to pharmacokinetics, has of course been recognized as an entity ever since Archibald Garrod's seminal observations and his visionary interpretation as interindividual differences in detoxification of drugs. We have since come to understand the underlying genetic causes for many of the previously known differences in enzymatic activity, most prominently with regard to the P450 enzyme family, and these have been the subject of recent reviews [1, 2].
Table 2.
Pharmacogenetics systematic classification.
| Pharmacokinetics |
| Absorption |
| Metabolism |
| Activation of prodrugs |
| De-activation |
| Generation of biologically active metabolites |
| Distribution |
| Elimination |
| Pharmacodynamics |
| Causative drug action: related to molecular pathology |
| Palliative drug action: related to molecular physiology |
On the other hand, interindividual differences in a drug's effects may also be observed in the presence of appropriate concentrations of the intended compound at the intended site of action, i.e. be due to differential pharmacodynamics. Here, two conceptually quite different conceptual scenarios may be distinguished that relate to the two principal mechanisms by which drugs act: aetiology-specific and palliative.
The former relates to drugs that work by targeting, and mitigating or correcting the actual cause of the disease or one of its aetiologically contributing elements. In contrast, palliative drugs modulate disease-phenotype-relevant (but not disease-cause-relevant) pathways that are not dysfunctional but can be used to counterbalance the effect of a disease-causing, dysfunctional pathway. These drugs do not directly address the underlying cause or aetiological contribution.
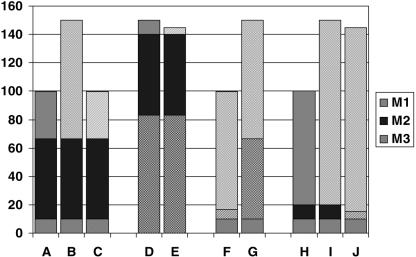
There is general agreement today that any of the major clinical diagnoses, such as diabetes or cancer, are comprised of a number of aetiologically (i.e. at the molecular level) more or less distinct subcategories. In the case of an aetiologically acting drug this implies that it will only be appropriate in a fraction of the patients that carry the clinical diagnosis; namely in those in whom the dominant molecular aetiology, or at least one of the contributing aetiological factors matches the mechanism of the drug given. A schematic (Figure 1) is enclosed to help clarify these somewhat complex concepts, in which a hypothetical case of a complex disease is depicted where excessive function of one of the trait-controlling pathways causes symptomatic disease – assume, e.g. the trait is blood pressure, and the associated disorder is hypertension (for the case of a defective function of a pathway, an analogous schematic could be constructed, and again for a deviant function). Since a causative treatment will only work if the mechanism it addresses is indeed contributing to the patient's disease (Figure 1a,b,c), such a treatment may be ineffective if that mechanism is not operative (Figure 1d,e). Thus, unrecognized and undiagnosed disease heterogeneity at the molecular level provides an important explanation for differential drug response and likely represents a substantial fraction of what we today somewhat indiscriminately subsume under the term ‘pharmacogenetics’.
Figure 1.
a: normal physiology: 3 molecular mechanisms (M1, M2, M3) contribute to a trait; b: diseased physiology D1: derailment (cause/contribution) of molecular mechanism 1 (M1); c: diseased physiology D1: causal treatment T1 (aimed at M1); d: diseased physiology D3: derailment (cause/contribution) of molecular mechanism 3 (M3); e: diseased physiology D3, treatment T1: treatment does not address cause; f: diseased physiology D1, palliative treatment T2 (aimed at M2); g: diseased physiology D1, palliative treatment T2; T2-refractroy gene variant in M2; h: normal physiology variant: differential contribution of M1 and M2 to normal trait; i: diseased physiology D1-variant: derailment of mechanism M1; j: diseased physiology D1-variant: treatment with T2.
On the other hand, in the case of a drug that works palliatively, molecular variations in the structure of the drug's biological target that affect the target's interaction with a drug, as well as interindividual differences in the activity of the targeted pathways (and thus in the relative disease-counterbalancing effect of inhibiting or enhancing them) provide a second, conceptually different explanation for differential drug response based on pharmacodynamics. Thus, a palliative treatment (Figure 1f) may not be effective either if the target molecule represents a variant that does not respond to the treatment (Figure 1g), or if the particular mechanism targeted by the palliative drug is not phenotype-relevant in the patient in question, due to a genetic variant or other reasons (Figure 1h,i,j). Here we are faced with disease-aetiology-unrelated, interindividual variability as the root cause for differential drug response.
Pharmacogenetics as a consequence of ‘subclinical’ differential diagnosis
An increasingly sophisticated and precise diagnosis of disease, arising from a deeper, more differentiated understanding of pathology at the molecular level, that will subdivide today's clinical diagnoses into molecular subtypes, will foster medical advances which, if considered from the viewpoint of today's clinical diagnosis, will appear as ‘pharmacogenetic’ phenomena. However, the sequence of events commonly expected as characteristic for a ‘pharmacogenetic scenario’ – namely, exposing patients to the drug, recognizing a differential (i.e. [quasi-]bimodal-) response pattern, discovering a marker predicting this response, and creating a diagnostic product to be comarketed with the drug henceforth – is likely to be reversed. Rather, we will search for a new drug specifically, and a priori, based on a new diagnosis (i.e. a newly found ability to diagnose a molecular subentity of a previously more encompassing, broader, and less precise clinical disease definition). Thus, pharmacogenetics will not be so much about finding the ‘right medicine for the right patient’, but about finding the ‘right medicine for the disease(−subtype)’, as we have aspired to do all along throughout the history of medical progress. This is, in fact, good news: the conventional ‘pharmacogenetic scenario’ will invariably present major challenges from both a regulatory and a business development and marketing standpoint, as it confronts development teams with a critical change in the drug's profile at a very late point during the development process. In addition, the timely development of an approvable diagnostic in this situation is difficult at best, and its marketing as an ‘add-on’ to the drug a less than attractive proposition to diagnostic businesses. Thus, the ‘practice’ of pharmacogenetics will, in many instances, be marked by progress along the very same path that has been one of the main avenues of medical progress for the last several hundred years: differential diagnosis.
The sequence of events in this case would likely involve, first, the development of an in vitro diagnostic test as a stand-alone product that may even be marketed on its own merits, allowing the physician to establish an accurate, state-of-the-art diagnosis of the molecular subtype of the patient's disease. Sometimes such a diagnostic may prove helpful even in the absence of specific therapy by guiding the choice of existing medicines and/or of nondrug treatment modalities, such as specific changes in diet or lifestyle. Availability of such a diagnostic – as part of the more sophisticated understanding of disease – will undoubtedly foster and stimulate the search for new, more specific drugs; and once such drugs are found, availability of the specific diagnostic will be important for carrying out the appropriate clinical trials. This will allow a prospectively planned, much more systematic approach towards clinical and business development, with a commensurate greater chance of actual realization and success.
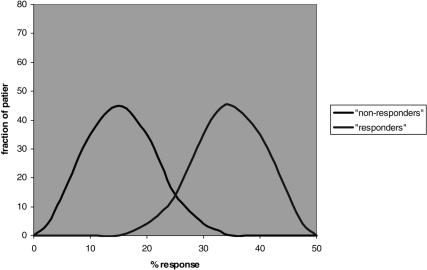
In practice, some extent of guesswork will remain, due to the nature of common complex disease. First, all diagnostic approaches will ultimately only provide a measure of probability, not of certainty: thus, although the variances of patient response among patients who do or do not carry the drug-specific subdiagnosis will be smaller, there will still be a distribution of differential responses; thus, although by-and-large the drug will work better in the ‘responder’ group, there will be some who respond less ore not at all in that group, and conversely, not everyone belonging to the ‘non-responder’ group will completely fail to respond, depending ultimately on the relative magnitude with which the particular mechanism contributes to the disease. Thus, it is important to bear in mind that even in the case of fairly obvious bimodality individual patients will still fall into a distribution pattern of responses, and all predictions as to responder- or non-responder status will be of a probabilistic nature (Figure 2). In addition, based on our current understanding of the polygenic and heterogeneous nature of these disorders, we will – even in an ideal world where we would know about all possible susceptibility gene variants for a given disease and have treatments for them – only be able to exclude, in any one patient, those that do not appear to contribute to the disease, and therefore rule out certain treatments. We will, however, most likely find ourselves left with a small number – two to four, perhaps – of potentially disease-contributing gene-variants whose relative contribution to the disease will be very difficult, if not impossible, to rank in an individual patient. Likely then, trial and error, and this great intangible quantity, physician experience will still play an important role, albeit on a more limited and subselective basis.
Figure 2.
Hypothetical example of bimodal distribution according to marker that indicates ‘non-responder’ or ‘responder’ status. Note that in both cases a distribution is present, with overlaps, thus, the categorization into ‘responders’ or ‘nonresponders’ based on the marker must be understood to convey only the probability to belong to one or the other group.
Today, the most frequently cited example for this category of ‘pharmacogenetics’ is trastuzamab (HERCEPTIN®), a humanized monoclonal antibody directed against the her-2-oncogene. This breast cancer treatment is prescribed based on the level of her-2-oncogene expression in the patient's tumour tissue. Differential diagnosis at the molecular level not only provides an added level of diagnostic sophistication, but also actually represents the prerequisite for choosing the appropriate therapy. Because tastuzamab specifically inhibits a ‘gain-of-function’ variant of the oncogene, it is ineffective in the 2/3 of patients who do not ‘over-express’ the drug's target, whereas it significantly improves survival in the 1/3 of patients that constitute the ‘subentity’ of the broader diagnosis ‘breast cancer’ in whom the gene is expressed [3]. (Some have argued against this being an example of ‘pharmacogenetics’, because the parameter for patient stratification (i.e. for differential diagnosis) is the somatic gene expression level rather than a particular ‘genotype’ data [4]. This is a difficult argument to follow, since in the case of a treatment-effect-modifying germ-line mutation it would obviously not be the nuclear gene variant per se, but also its specific impact on either structure/function or on expression of the respective gene/gene product that would represent the actual physiological corollary underlying the differential drug action. Conversely, an a priori observed expression difference is highly likely to reflect a – potentially as yet undiscovered – sequence variant. Indeed, as pointed out below, there are a number of examples where the connection between genotypic variant and altered expression has already been demonstrated [5, 6]).
Another example, although still hypothetical, of how proper molecular diagnosis of relevant pathomechanisms will significantly influence drug efficacy, is in the evolving class of anti-AIDS/HIV drugs that target the CCR5 cell-surface receptor [7–9]. These drugs are predicted to be ineffective in the rare patients who carry the delta-32 variant, but who nevertheless have contracted AIDS or test HIV-positive (most likely due to infection with an SI-virus phenotype that utilizes CXCR4) [10, 11].
It should be noted that the pharmacogenetically relevant molecular variant need not affect the primary drug target, but may equally well be located in another molecule belonging to the system or pathway in question, both up- or downstream in the biological cascade with respect to the primary drug target.
Pharmacogenetic effects of palliative drugs due to structural target diversity
The alternative scenario, where differential drug response and/or safety occurs with regard to a ‘palliative’ drug is likely to pose, as discussed, considerably greater difficulty in planning and executing a clinical development program because, presumably, it will be more difficult to anticipate or predict differential responses a priori. When such a differential response occurs, it will also potentially be more difficult to find the relevant marker(s), unless it happens to be among the ‘obvious’ candidate genes implicated in the disease physiopathology or the treatment's mode of action. Although screening for molecular variants of these genes, and testing for their possible associations with differential drug response is a logical first step, if unsuccessful, it may be necessary to embark on an unbiased genome-wide screen, using single nucleotide polymorphisms (SNPs) as molecular flagpoles. Despite recent progress in high-throughput genotyping, the obstacles that will have to be overcome on the technical, data-analysis, and cost levels are formidable. They will limit the deployment of such programs, at least for the foreseeable future, to select cases in which there are very solid indications for doing so, based on clinical data showing a near-categorical (e.g. bimodal) distribution of treatment outcomes. Even then, we may expect to encounter for every success – that will be owed to a favourably strong linkage-disequilibrium across considerable genomic distance in the relevant chromosomal region – as many or more failures, in cases where the culpable gene variant cannot be found due to the higher recombination rate or other characteristics of the stretch of genome that it is located on.
Several of the more persuasive examples we have accumulated to date for such palliative-drug-related pharmacogenetic effects have been observed in the field of asthma. The treatment of asthma relies on an array of drugs aimed at modulating different ‘generic’ pathways, thus mediating bronchodilation or anti-inflammatory effects. Pharmacogenetic effects have been demonstrated in situations where these pathways do not respond as expected. Thus, molecular variants of the β2-adrenoceptor have been shown associated with differential treatment response to β2-agonists [13, 14]. Individuals carrying one or two copies of a variant allele that contains a glycine in place of arginine in position 16 were found to have a 3- and 5-fold reduced response to the agonist, respectively. This was shown in both in vitro [15, 16] and in vivo [16] studies to correlate with an enhanced rate of agonist-induced receptor down-regulation, but no difference in gene transcriptional or translational activity, or agonist binding. In contrast, a second polymorphism affecting position 19 of the β upstream peptide has been shown to affect translation (but not transcription) of the receptor itself, with a 50% decrease in receptor numbers associated with the variant allele – which happens to be in strong linkage disequilibrium with a variant allele position 16 in the receptor. The simultaneous presence of both mutations would thus be predicted to result in low expression and enhanced down-regulation of an otherwise functionally normal receptor, depriving patients carrying such alleles of the benefits of effective bronchodilation as a ‘palliative’ (i.e. noncausal) counter-measure to their pathological airway hyper-reactivity. (In the schematic depicted in Figure 1, the common type of β-receptor response would be represented by situation f, the variant by situation g). Importantly, there is no evidence that any of the allelic variants encountered are associated with the prevalence or incidence, and thus potentially the aetiology of the underlying disease [17, 18].
Similarly, inhibition of leukotriene synthesis proved clinically ineffective in a small fraction of patients who carried only non-wild-type alleles of the 5-lipoxygenase promoter region [12]. These allelic variants had previously been shown to be associated with decreased transcriptional activity of the gene [5]. It stands to reason – consistent with the clinical observations – that in the presence of already reduced 5-lipoxygenase activity pharmacological inhibition may be less effective (corresponding to situations h–j in Figure 1). Of note, again, there is no evidence for a primary, disease-causing or -contributing role of 5-lipoxygenase variants; all of them were observed at equal frequencies in affected and nonaffected individuals [5].
Pharmacogenetic stratification allows not only recognition of responders and nonresponders with regard to the intended treatment effect, but also with regard to undesirable responses, i.e. the occurrence of adverse effects. An example for this scenario is provided by the well-documented ‘pharmacogenetic’ association between molecular sequence variants of the 12S rRNA, a mitochondrion-encoded gene, and aminoglycoside-induced ototoxicity [19]. Intriguingly, the mutation that is associated with susceptibility to ototoxicity renders the sequence of the human 12S rRNA similar to that of the bacterial 12S rRNA gene, and thus effectively turns the human 12S rRNA into the (bacterial) target for aminoglycoside drug action – presumably mimicking the structure of the bacterial binding site of the drug [20]. As in the other examples, presence of the 12S rRNA mutation per se has no primary, drug-treatment-independent effect.
Analogously, within one species such ‘molecular mimicry’ may occur: adverse events may arise if the selectivity of a drug is lost because a gene that belongs to the same gene-family as the primary target, loses its ‘identity’ vis-à-vis the drug and attains, based on its structural similarity with the principal target, similar affinity to the drug. Depending on the biological role of the ‘imposter’ molecule, adverse events may occur. Although we currently have no clear actual examples for this, it is certainly imaginable for classes of receptors and enzymes.
Different classes of markers
Pharmacogenetic phenomena, as pointed out previously, need not be restricted to the observation of a direct association between allelic sequence variation and phenotype, but may extend to a broad variety of indirect manifestations of underlying, but often un-recognized, sequence variation. Thus, differential methylation of the promoter-region of O6-methylguanine-DNA-methylase has recently been reported to be associated with differential efficacy of chemotherapy with alkylating agents. If methylation is present, expression of the enzyme that rapidly reverses alkylation and induces drug-resistance is inhibited, and therapeutic efficacy is greatly enhanced [21].
Complexity is to be expected
In the real world, it is likely that not only one of the scenarios depicted, but a combination of several ones may affect how well a patient responds to a given treatment, or how likely it is that he or she will suffer an adverse event. Thus, a fast-metabolizing patient with poor-responder pharmacodynamics may be particularly unlikely to gain any benefit from taking the drug in question, while a slow-metabolizing status may counterbalance in another patient the same pharmacodynamics, whereas a third patient, being a slow metabolizer and displaying normal pharmacodynamics, may be more likely to suffer adverse events. In all of them, both the pharmacokinetic and pharmacodynamics properties may result from the interaction of several of the mechanisms described above. In addition, we know of course that coadministration of other drugs, or even the consumption of certain foods, may affect and further complicate the picture for any given treatment.
Pharmacogenetic testing for drug efficacy vs safety
In principle, pharmacogenetic approaches may be useful both to raise efficacy and to avoid adverse events, by stratifying patient eligibility for a drug according to appropriate markers. In both cases, clinical decisions and recommendations must be supported by data that have undergone rigorous biostatistical scrutiny. Based on the substantially different prerequisites for and opportunities to acquiring such data, and to applying them to clinical decision-making, we expect the use of pharmacogenetics for enhanced efficacy to be considerably more common than for the avoidance of adverse events.
The likelihood that adequate data on efficacy in a subgroup may be generated is reasonably high, given the fact that unless the drug is viable in a sizeable number of patients, it will probably not be developed for lack of a viable business case, or at least only in the protected environment of orphan regulations. Implementation of pharmacogenetic testing to stratify for efficacy, provided that safety in the nonresponder group is not an issue, will primarily be a matter of physician preference and sophistication, and potentially of third-party payer directives, but would appear less likely to become a matter of regulatory mandate. Indeed, an argument can be made against depriving those who carry the ‘nonresponder’ genotype of eligibility for the drug, but who individually, of course, may respond to the drug with a certain, albeit lower probability. From a regulatory aspect, use of pharmacogenetics for efficacy, if adequate safety data exist, appears largely unproblematic – the worst-case scenario (a genotypically inappropriate patient receiving the drug) resulting in treatment without expected beneficial effect, but with no increased odds to suffer adverse consequences, i.e. much of what one would expect under conventional paradigms.
The utility and clinical application of pharmacogenetic approaches towards improving safety, in particular with regard to serious adverse events, will meet with considerably greater hurdles and is therefore less likely expected to become reality. A number of reasons are cited for this: first, in the event of serious adverse events associated with the use of a widely prescribed medicine, withdrawal of the drug from the market is usually based almost entirely on anecdotal evidence from a rather small number of cases – in accordance with the Hippocratic mandate ‘primum non nocere’. If the sample size is insufficient to statistically demonstrate a significant association between drug exposure and event, it will most certainly be insufficient to allow meaningful testing for genotype-phenotype correlations; this becomes progressively more difficult as many markers are tested and the number of degrees of freedom applicable to any analysis continues to rise. Therefore, the fraction of attributable risk shown to be associated with a given at-risk (combination of) genotype(s) would have to be very substantial for regulators to accept such data. Indeed, the low prior probability of the event will, by definition, result in an expected equally low positive (or negative) predictive value. Second, the very nature of safety issues raises the hurdles substantially because in this situation the worst-case scenario – administration of the drug to the ‘wrong’ patient – will result in higher odds to harm to the patient. Therefore, it is likely that the practical application of pharmacogenetics towards limiting adverse events will be restricted to diseases with dire prognosis, where a high medical need exists, where the drug in question offers unique potential advantages (usually bearing the characteristics of a ‘life-saving’ drug), and where the tolerance even for relatively severe side-effects is a-priori substantial, and accepted in favour of the drug's beneficial effects. This applies primarily to areas like oncology or HIV/AIDS. In most other indications, the sobering biostatistical and regulatory considerations discussed represent barriers that are unlikely to be overcome easily; and the proposed, conceptually highly attractive, routine deployment of pharmacogenetics as a generalized drug surveillance practice following the introduction of a new pharmaceutical agent [22] faces these as well as formidable economic hurdles.
Ethical-societal aspects of pharmacogenetics
No discussion about the use of genetic/genomic approaches to health care can be complete without considering their impact on the ethical, societal, and legal level. Arguments have been advanced that genotype determinations for pharmacogenetic characterization, in contrast to ‘genetic’ testing for primary disease risk assessment, are less likely to raise potentially sensitive issues with regard to patient confidentiality, the misuse of genotyping data or other nucleic-acid-derived information, and the possibility of stigmatization. While this is certainly true when pharmacogenetic testing is compared to predictive genotyping for highly penetrant Mendelian disorders, in it is not apparent why in common complex disorders issues surrounding predictors of primary disease risk would be any more or less sensitive than those pertaining to predictors of likely treatment success/failure. Indeed, two lines of reasoning may actually indicate an increased potential for ethical issues and complex confrontations among the various stakeholders to arise from pharmacogenetic data.
First, while access to genotyping and other nucleic acid-derived data related to disease susceptibility can be strictly limited, the very nature of pharmacogenetic data calls for a rather more liberal position regarding use: if this information is to serve its intended purpose, i.e. improving the patients chance for successful treatment, then it is essential that it is shared among at least a somewhat wider circle of participants in the health care process. Thus, the prescription for a drug that is limited to a group of patients with a particular genotype will inevitably disclose the receiving patient's genotype to anyone of a large number of individuals involved in the patients care at the medical and administrative level. The only way to limit this quasi-public disclosure of this patient's genotype data would be if he or she were to sacrifice the benefits of the indicated treatment for the sake of data confidentiality.
Second, patients profiled to carry a high disease probability along with a high likelihood for treatment response may be viewed, from the standpoint of, e.g. insurance risk, as quite comparable with patients displaying the opposite profile, i.e. a low risk to develop the disease, but a high likelihood not to respond to medical treatment, if the disease indeed occurs. For any given disease risk, then, patients less likely to respond to treatment would be seen as a more unfavourable insurance risk, particularly if non-responder status is associated with chronic, costly illness rather than with early mortality, the first case having much more far-reaching economic consequences. The pharmacogenetic profile may thus, under certain circumstances, even become a more important (financial) risk-assessment parameter than primary disease susceptibility, and would be expected – in as much as it represents but one stone in the complex-disease mosaic – to be treated with similar weight, or lack thereof, as other genetic and environmental risk factors.
Practically speaking, the critical issue is not only, and perhaps not even predominantly, the sensitive nature of the information, and how it is, if at all, disseminated and disclosed, but how and to what end it is used. Obviously, generation and acquisition of personal medical information must always be contingent on the individual's free choice and consent, as must be all application of such data for specific purposes. Beyond this, however, there is today an urgent need for the requisite dialogue and discourse among all stakeholders within society to develop and endorse a set of criteria by which the use of genetic, indeed of all personal medical information should occur. It will be critically important that society as a whole endorses, in an act of solidarity with those destined to develop a certain disease, guidelines that support the beneficial and legitimate use of the data in the patient's interest while at the same time prohibiting their use in ways that may harm the individual, personally, financially, or otherwise. As long as we trust our political decision processes to reflect societal consensus, and as long as such consensus reflects the principles of justice and equality, the resulting set of principles should assert such proper use of medical information. Indeed, both aspects – data protection and patient/subject protection, are seminal components of the mandates included in the WHO's ‘Proposed International Guidelines on Ethical Issues in Medical Genetics and Genetic Services’ [23] which mandate autonomy, beneficence, no maleficence, and justice.
In summary, pharmacogenetics, in the different scenarios included in this term, will represent an important new avenue towards understanding disease pathology and drug action, and will offer new opportunities of stratifying patients to achieve optimal treatment success. As such, it represents a logical, consequent step in the history of medicine – evolution, rather than revolution. Its implementation will take time, and will not apply to all diseases and all treatments equally. If society finds ways to sanction the proper use of this information, thus allowing and protecting its unencumbered use for the patient's benefit, important progress in health care will be made.
References
- 1.Dickins M, Tucker G. Drug disposition: to phenotype or genotype. Int J Pharm Med. 2001;15:70–73. also see: http://www.imm.ki.se/CYPalleles/
- 2.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapies. Science. 1999;206:487–491. doi: 10.1126/science.286.5439.487. also see: http://www.sciencemag.org/feature/data/1044449.shl/ [DOI] [PubMed]
- 3.Baselga J, Tripathy D, Mendelsohn J, et al. Phase II study of weekly intravenous recombinant humanized anti-p185 (HER2) monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- 4.Haseltine WA. Not quite pharmacogenomics (letter; comment) Nat-Biotechnol. 1998;16:1295. doi: 10.1038/4244. [DOI] [PubMed] [Google Scholar]
- 5.In KH, Asano K, Beier D, et al. Naturally occurring mutations in the human 5-lipoxygenase gene promoter that modify transcription factor binding and reporter gene transcription. J Clin Invest. 1997;99:1130–1137. doi: 10.1172/JCI119241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGraw DW, Forbes SL, Kramer LA, Liggett SB. Polymorphisms of the 5′ leader cistron of the human beta2-adrenergic receptor regulate receptor expression. J Clin Invest. 1998;102:1927–1932. doi: 10.1172/JCI4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Paxton WA, Wolinsky SM, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nature Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 8.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 9.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien TR, Winkler C, Dean M, et al. HIV-1 infection in a man homozygous for CCR5 32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. [DOI] [PubMed] [Google Scholar]
- 11.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzious C. Seroco Study Group. HIV-1 infection in an individual homozygous for CCR5 32. Lancet. 1997;349:1219–1220. [PubMed] [Google Scholar]
- 12.Drazen JM, Yandava CN, Dube L, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat-Genet. 1999;22:168–170. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- 13.Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100:3184–3188. doi: 10.1172/JCI119874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan S, Hall IP, Dewar J, Dow E, Lipworth B. Association between beta 2-adrenoceptor polymorphism and susceptibility to bronchodilator desensitisation in moderately severe stable asthmatics. Lancet. 1997;350:995–999. doi: 10.1016/S0140-6736(97)03211-X. [DOI] [PubMed] [Google Scholar]
- 15.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 16.Green SA, Turki J, Bejarano P, Hall IP, Liggett SB. Influence of beta 2-adrenergic receptor genotypes on signal transduction in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 1995;13(1):25–33. doi: 10.1165/ajrcmb.13.1.7598936. [DOI] [PubMed] [Google Scholar]
- 17.Reihsaus E, Innis M, MacIntyre N, Liggett SB. Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am J Respir Cell Mol Biol. 1993;8:334–349. doi: 10.1165/ajrcmb/8.3.334. [DOI] [PubMed] [Google Scholar]
- 18.Dewar JC, Wheatley AP, Venn A, Morrison JFJ, Britton J, Hall IP. β2-adrenoceptor polymorphisms are in linkage disequilibrium, but are not associated with asthma in an adult population. Clin Exp Allergy. 1998;28:442–448. doi: 10.1046/j.1365-2222.1998.00245.x. [DOI] [PubMed] [Google Scholar]
- 19.Fischel-Ghodsian N. Genetic factors in aminoglycoside toxicity. Ann NY Acad Sci. 1999;884:99–109. doi: 10.1111/j.1749-6632.1999.tb08639.x. [DOI] [PubMed] [Google Scholar]
- 20.Hutchin T, Cortopassi G. Proposed molecular and cellular mechanism for aminoglycoside ototoxicity. Antimicrob Agents Chemother. 1994;38:2517–2520. doi: 10.1128/aac.38.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene mgmt and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 22.Roses A. Pharmacogentics and future drug development and delivery. Lancet. 2000;355:1358–1361. doi: 10.1016/S0140-6736(00)02126-7. [DOI] [PubMed] [Google Scholar]
- 23.Proposed international guidelines on ethical issues in medical genetics and genetic services. http://www.who.int/ncd/hgn/hgnethic.htm. [PubMed]