Abstract
Aims
To investigate in a large panel of 50 human liver samples the contribution of CYP2C9, CYP2D6, and CYP3A4 to the overall formation of the potent antioestrogen Z-4-hydroxy-tamoxifen, and how various genotypes affect its formation from tamoxifen.
Methods
The formation of Z-4-hydroxy-tamoxifen from 10 µm tamoxifen was studied in human liver microsomes (n = 50), characterized for CYP2B6, CYP2C9, CYP2D6 and CYP3A4 expression, and CYP2B6, CYP2C9 and CYP2D6 genotype. The effect of chemical and monoclonal antibody inhibitors, and the formation in supersomes expressing recombinant CYP isoforms was also investigated. Z-4-hydroxy-tamoxifen was quantified using LC-MS analysis.
Results
Z-4-hydroxy-tamoxifen was formed by supersomes expressing CYP2B6, CYP2C9, CYP2C19 and CYP2D6, but not CYP3A4. In agreement with these data, the mean formation of Z-4-hydroxy-tamoxifen was inhibited 49% by sulphaphenazole (P = 0.001), 38% by quinidine (P<0.05) and 13% by monoclonal antibody against CYP2B6 (MAB-2B6, P<0.05). Furthermore, Z-4-hydroxy-tamoxifen formation significantly correlated with both CYP2C9 expression (rs = 0.256, P<0.05) and CYP2D6 expression (rs = 0.309, P<0.05). Genotypes of CYP2D6, CYP2B6 and CYP2C9 had an effect on metabolite formation in such a way that samples with two nonfunctional CYP2D6, or two variant CYP2C9 or CYP2B6 alleles, showed lower enzyme activity compared with those with two functional or wild-type alleles, (5.0 vs 9.9 pmol mg−1 protein min−1, P = 0.046, 5.1 vs 9.9 pmol mg−1 protein min−1, P = 0.053, and 6.8 vs 9.4 pmol mg−1 protein min−1, P = 0.054, respectively). CYP2D6 and CYP2C9 contribute on average 45 and 46%, respectively, to the overall formation of Z-4-hydroxy-tamoxifen.
Conclusions
CYP2B6, CYP2C9 and CYP2D6 genotypes all affected Z-4-hydroxy-tamoxifen formation and can predict individual ability to catalyse this reaction.
Keywords: CYP2B6, CYP2C9, CYP2D6, genotype, Z-4-hydroxy-tamoxifen
Introduction
Tamoxifen is a triphenylethylene compound that possesses antioestrogenic activity. Although it is the drug of first choice for the treatment of oestrogen receptor positive breast cancer, 50% of these patients show no therapeutic response [1]. Furthermore, the administration of tamoxifen as a preventative measure for women with an increased risk of developing breast cancer [2] continues to be investigated. It is extensively metabolized in human liver with the major metabolites being N-desmethyl-tamoxifen, tamoxifen-N-oxide (tam-N-oxide) and Z-4-hydroxy-tamoxifen (Z-4-OH-tam) [3–5].
Compared with tamoxifen, Z-4-OH-tam is a potent antioestrogen, with a 33-fold higher affinity for oestrogen receptors in human breast cancer tissue [6]. Therefore, it is likely that Z-4-OH-tam could contribute substantially to its antioestrogen effects. However, the overall contribution will depend on the fraction of the dose converted to Z-4-OH-tam, such that variability in its formation, could be a factor in nonresponse to tamoxifen. Several studies have identified which cytochrome P450 (CYP) isoforms play a role in the formation of Z-4-OH-tam [7–9]. The most comprehensive of these reported that CYP2C9, CYP2D6 and CYP3A4, predominantly mediate this metabolic pathway in human liver, and the contributions of the individual isoforms to the overall formation are highly variable in this small sample size [9]. However, interindividual variability in the contribution of these isoforms has not been assessed in a large sample population characterized in terms of expression and/or genotype of these CYPs. Moreover, the role of CYP2B6 has not been investigated.
Since all of the above CYP isoforms display genetic polymorphisms, their expression and/or function varies substantially between individuals. Mutations of CYP2D6 and CYP2C19 lead to either decreased enzyme expression or loss of function [10, 11]. Both the CYP2B6*5 and *7 allelic variants lead to decreased CYP2B6 expression and reduced S-mephenytoin N-demethylase activity [12], with the functional significance of the other allelic variants still to be elucidated. With regard to CYP2C9, the CYP2C9*2 mutation results in an Arg144Cys amino acid substitution which alters the interaction between the cytochrome P450 and NADPH:cytochrome P450 oxidoreductase [13], and the CYP2C9*3 mutation results in a Ile359Leu amino acid substitution in the substrate-binding-site of CYP2C9 [14]. In vitro investigations have revealed that the extent of the functional effect of the *2 mutation is somewhat dependent on the substrate [15–18]. In contrast, the *3 mutation results in a reduced intrinsic clearance of tolbutamide and S-warfarin in vitro[16, 19], and has been associated with sensitivity to warfarin therapy in vivo[20]. Finally, the expression of CYP3A4 has been shown to be highly variable amongst individuals [21, 22]. Therefore, the interindividual variation in Z-4-OH-tam formation previously observed with a small sample size (n = 10 [9]), is not surprising, and indicates that elucidation of the contribution of the isoforms to overall formation in a larger population is worthwhile.
In addition, despite the identification of the roles of CYP2C9, CYP2C19 and CYP2D6 in Z-4-OH-tam formation to date, there has been no investigation of how various genotypes affect formation rates. Therefore, the aims of this study were to elucidate the role of CYP2B6 and the contribution of CYP2C9, CYP2D6 and CYP3A4 in the overall formation of Z-4-OH-tam from tamoxifen, and investigate the relationship between the formation rates and CYP2B6, CYP2C9, CYP2C19 and CYP2D6 genotypes in microsomes from a large panel of human livers. In addition, we report a new LC-MS method for the detection and quantification of the Z-4-OH-tam.
Methods
Materials
Z-3-OH-tam (droloxifene, 3-OH-tam), Z-4-OH-tam, Z-tam-N-oxide and Z-tamoxifen were gifts from Klinge Pharma GmbH (Munich, Germany). Z-α-OH-tam was obtained from Toronto Research Chemicals Inc. (Toronto, ON, Canada). S-(+)-mephenytoin was a kind gift from W. Trager, Seattle, USA. Other materials were obtained from the following sources: acetonitrile from J.T. Baker (Deventer, Holland); β-nicotinamide adenine dinucleotide phosphate (β-NADPH, tetrasodium salt, reduced form), coumarin, furafylline, quinidine (free-base) and sulphaphenazole from Sigma (Deisenhofen, Germany); troleandomycin from ICN Biomedicals Inc. (Aurora, OH, USA); dimethyl sulfoxide and magnesium chloride from Merck (Darmstadt, Germany); and potassium dihydrogen phosphate from Fluka Chemie AG (Buchs, Switzerland). CYP1A2, CYP2A6, CYP2B6, CYP2C9.1, CYP2C9.2, CYP2C9.3, CYP2C19, CYP2D6, CYP2E1 and CYP3A4 supersomes, and human CYP2B6 monoclonal antibody were purchased from BD GENTEST™ (A BD Biosciences Company, Woburn, MA, USA). All other chemicals and solvents were of analytical grade.
Due to poor aqueous solubility of tamoxifen, stock solutions of 0.1, 0.25, 0.5 and 1 mm were prepared in 100% dimethyl sulfoxide. These stock solutions were added directly to the incubations, with the final concentration of dimethyl sulfoxide in incubations being 1%. All stock solutions of Z-4-OH-tam for calibration samples and quality controls and 3-OH-tam (internal standard) were dissolved in 100% acetonitrile.
Human liver microsomes
Human liver tissue was obtained from liver resections of tumour patients with primary or secondary liver tumours. The collection of tissue was done according to institutional guidelines and the patients' written consent. Microsomes from 50 randomly selected livers from this tissue bank were prepared and total protein content measured as previously described [12]. The mean±s.d. (range) age of the patients was 57.5±16.8 (4–77) years, 32 were male and 18 were female.
Formation of Z-4-OH-tam by expressed CYP450 isoforms
The in vitro formation of Z-4-OH-tam by supersomes (baculovirus insect cells) expressing recombinant CYP isoforms was investigated initially via incubation of 12.5 pmol CYP with tamoxifen (10 µm, 1% dimethyl sulfoxide), 2 mm NADPH, 33 mm magnesium chloride and 100 mm potassium phosphate buffer (pH 7.5, microsomal incubation buffer) in a total volume of 100 µl. Due to the inhibition of CYP2E1 by even small percentages of dimethyl sulfoxide (0.02%, personal communication with BD GENTEST™), metabolism of tamoxifen by this isoform was investigated using a tamoxifen saturated 100 mm potassium dihydrogen phosphate buffer pH 7.5 (∼5 µm tamoxifen). If initial incubations revealed quantifiable amounts of Z-4-OH-tam, additional incubations with tamoxifen (1, 2.5 and 5 µm, 1% dimethyl sulfoxide) were performed to obtain an estimate of intrinsic clearance of tamoxifen by that particular CYP isoform. Incubations were initiated by the addition of ice-cold supersomes and carried out in a water bath at 37° C for 30 min, and stopped by the addition of 40 µl of 100% acetonitrile. 3-OH-tam (internal standard, 10 µl of 5 µm) was added, and samples were centrifuged at 1500 g for 5 min. The resultant supernatant was removed and 10 µl injected onto the LC-MS system.
Formation of Z-4-OH-tam by human liver microsomes
The in vitro formation of Z-4-OH-tam was investigated in human liver microsomes (n = 50) via incubation of tamoxifen (10 µm, 1% dimethyl sulfoxide) for 30 min with 10 µg microsomal protein, in a final incubation volume of 100 µl with identical incubation constituents to those described previously. Conditions of incubations and sample processing did not alter from above. The formation of Z-4-OH-tam was observed to be linear with microsomal protein concentrations between 0.01 and 0.1 mg ml−1.
Inhibition studies
Inhibition of Z-4-OH-tam formation was studied in microsomes from 10 of the human liver samples, selected in terms of CYP2D6 expression. Furafylline (25 µm), coumarin (100 µm), sulphaphenazole (5 µm), quinidine (1 µm), troleandomycin (10 µm) were used to study the inhibition of CYP1A2, CYP2A6, CYP2C9, CYP2D6, and CYP3A4, respectively [23, 24], and S-mephenytoin (100 µm) to study the inhibition of CYP2C19. In addition, lack of specificity of orphenadrine, a CYP2B6 chemical inhibitor [25], led to the use of a monoclonal antibody raised against human CYP2B6 (MAB-2B6, 10 µl·100 µg−1 microsomal protein, BD GENTEST™). Tamoxifen (10 µm, 1% dimethyl sulfoxide) and the inhibitors were incubated in a total volume of 100 µl with identical incubation constituents as described previously. Conditions of incubations were identical, except that furafylline and troleandomycin required 15 min preincubation at 37° C prior to the addition of tamoxifen, and MAB-2B6 required 15 min preincubation on ice prior to the addition of all other incubation constituents. All chemical inhibitor stocks, excluding coumarin which was soluble in water, were made in 2% dimethyl sulfoxide (final concentration, including addition from tamoxifen, 1.5%), and incubations containing equivalent amounts of dimethyl sulfoxide and water were used as controls.
Liver content of CYP isoforms
The content of CYP2B6, CYP2C9, CYP2D6 and CYP3A4 in the human liver microsomes was determined by quantitative Western blots. Microsomal proteins (10–25 µg) were separated on a 10% SDS-polyacrylamide gel, transferred to nitrocellulose and probed with specific primary antibodies: MAB-2B6 and MB-3A4 (BD GENTEST™), rabbit antihuman cytochrome P450 2C9 (Research Diagnostics Inc., Flanders, NJ), and antihuman CYP2D6 monoclonal antibody 114 [26]. Recombinant CYP450(s) expressed in either baculovirus infected insect cells, or lymphoblastoid cells (both from BD GENTEST™), or immuno-isolated CYP2D6 [27], were analysed to determine specificity of the antibodies used and for immuno-quantification. Secondary antibodies were obtained as standard reagents from common commercial sources. The ECL™ (enhanced chemiluminescence) kit (Amersham Pharmacia Biotech, UK Ltd, Buckinghamshire, UK) was used for detection. Band intensities were densitometrically analysed, following photography with a CCD camera (Raytest, Straubenhardt, Germany), with FUJIFILM Luminescent Image Analyser LAS-1000 and Image Reader LAS-1000 for Windows® Version 1.0 software (FUJI Photo Film (Europe) GmbH, Düsseldorf, Germany).
CYP2B6, CYP2C9 and CYP2D6 genotyping
Genomic DNA was isolated from blood samples of the same patients using standard methods. Genotyping for CYP2B6*2–7 was performed by allele-specific polymerase chain reaction (PCR) as previously described [12]. Genotyping for CYP2C9*2 and *3 alleles was performed by an allele-specific PCR assay previously described [16] utilizing shorter forward primers with the following sequences: CYP2C9*2 5′ TACAAATACAATGAAAATATC 3′, and CYP2C9*3 5′ TGCACGAGGTCCAGAGATGC 3′. PCR products of 689 and 165 bp were obtained for the CYP2C9*2 and *3 assay, respectively, with restriction fragments of 521+168 bp and 135+30 bp. Genotyping for CYP2D6*2,*2×2,*3,*5,*7,*8,*9 and *10 alleles was performed by allele-specific PCRs as previously described [28, 29]. Genotyping for CYP2D6*4 and *6 was performed using predeveloped TaqMan assay reagents (PDAR) (Applied Biosystems, Weiterstadt, Germany) according to the manufacturer's instructions, with a total volume of 25 µl.
Assessment of the influence of 1% dimethyl sulfoxide on CYP supersome activity
Inhibition by dimethyl sulfoxide of Z-4-OH-tam formation by expressed recombinant CYP450(s) was assessed by comparison of incubations with a semisaturated solution of tamoxifen in 100 mm potassium phosphate buffer pH 7.5 (final concentration ∼5 µm tamoxifen), with (n = 2) and without (n = 2) 1% dimethyl sulfoxide. Incubation constituents were identical to those above, except that 50 µl of tamoxifen saturated buffer was the substrate in a total volume of 100 µl, and 100% dimethyl sulfoxide was added to obtain a final concentration of 1% for assessment of inhibition. Incubation conditions and sample processing did not alter from above.
Analysis of tamoxifen metabolites
Chromatographic separation of Z-4-OH-tam from other tamoxifen metabolites and tamoxifen, and quantification was performed using a LUNA C8 analytical column (150×2 mm I.D., 5 µm particle size; Phenomenex, Hösbach, Germany) and a HP1100 LC-system (Agilent Technologies, Waldbronn, Germany) with a binary pump. The mobile phases were: (A) 1% acetic acid in water and (B) 1% acetic acid in acetonitrile. Gradient runs were programmed as follows: 30% B at a flow-rate of 0.5 ml min−1 for 5 min, increase to 50% B in 5 min, increase to 90% B in 2 min, 90% B for 4 min, increase flow-rate to 0.8 ml min−1 in 0.2 min, 90% B at a flow-rate of 0.8 ml min−1 for 2.8 min, decrease to 30% B in 1 min, 30% B at a flow-rate of 0.8 ml min−1 for 2.8 min, then re-equilibration with 30% B at a flow-rate of 0.5 ml min−1 for 2.8 min, until the next sample was injected. A column switching valve was used that switched the liquid flow through the detector from 1.8 to 14 min after injection.
A HP1100 single quadrupole mass spectrometer (Agilent Technologies) equipped with an electrospray source was used. Electrospray parameters were: spray voltage, 3000 V; fragmentor, 80 V; gas temperature, 350° C; nebuliser pressure, 30 p.s.i.; drying gas, 10 l min−1. Positive ionization with selected ion monitoring mode (SIM) was used for all analytes, the multiplier gain was set on one. α-OH-tam, E(Cis)-4-OH-tam, Z-4-OH-tam, 3-OH-tam and tam-N-oxide were monitored at m/z 388. N-didesmethyl-tam was monitored at m/z 344, and N-desmethyl-tam was monitored at m/z 358.
A calibration curve was constructed for Z-4-OH-tam, with six final concentrations ranging from 3.9 to 2000 nm. Low, medium and high quality control (QC) samples were also prepared, with final concentrations of 12, 100 and 750 nmα-OH-tam, tam-N-oxide, N-didesmethyl-tam and N-desmethyl-tam were also present in the calibration and QC samples at identical concentrations to monitor chromatographic interference. Calibration and QC samples were prepared identically to the microsomal incubations with heat inactivated human liver microsomal protein (excluding the addition of tamoxifen) and placed in a water bath at 37° C for 30 min. Following this, 40 µl of 100% acetonitrile was added and sample preparation as described above was performed.
The robustness of the method was assessed by assaying 15 QC samples (five each of low, middle and high concentrations) on a single assay day to determine the intraday accuracy and precision. Inter-day accuracy and precision were determined by analysis of three QC samples (one each of low, middle and high concentrations) on five different assay days.
Peak areas of Z-4-OH-tam were converted into peak area ratios using the peak area of the internal standard, and linear regression analysis of peak area ratios against peak amount ratios provided an estimate of slope and coefficient of determination (r2). Peak areas and subsequent calculation of calibration curves and amounts of Z-4-OH-tam formed was performed with HP Chemstation (98) software (Agilent Technologies).
Data analysis
The rates of Z-4-OH-tam formation were expressed as pmol mg−1 protein min−1 or pmol pmol−1 P450 min−1. Estimates of intrinsic clearance (µl min−1 pmol−1 P450) by a particular CYP isoform were obtained from the slope of the linear regression of the formation rates against the initial tamoxifen concentrations (Prism®, Version 3.0, GraphPad Software Inc., San Diego, CA, USA). The influence of 1% dimethyl sulfoxide on the activity of the expressed recombinant CYP(s) was assessed by the percentage of the ratio of the amount of Z-4-OH-tam formed by supersomes in the presence of dimethyl sulfoxide to the amount formed without dimethyl sulfoxide. Inhibition data were expressed as a percentage of the corresponding controls, and a two-tailed Wilcoxon signed rank test was used to determine if medians were significantly different from controls. In addition, Spearman rank correlations were used to determine if there was a relationship between: 1) the expression of CYP isoforms in the human liver microsomes and the formation rate of Z-4-OH-tam and 2) the expression of CYP isoforms in human liver microsomes and the inhibition by either chemical inhibitors or monoclonal antibodies. Mann–Whitney U-tests were performed to determine if the median rate of Z-4-OH-tam formation was significantly different between: samples with zero and those with two functional CYP2D6 alleles; and samples with two variant and those with two wild-type CYP2C9 or CYP2B6 alleles. For multiple analysis of CYP2B6, CYP2C9 and CYP2D6 expression between CYP2B6, CYP2C9 and CYP2D6 genotype groups, respectively, a Kruskal–Wallis test followed by Dunn's multiple comparison test was used. All data are tabulated as mean±s.d., with 95% confidence intervals (CI). A P value of 0.05 or less was considered significant.
Results
H.p.l.c. separation and validation
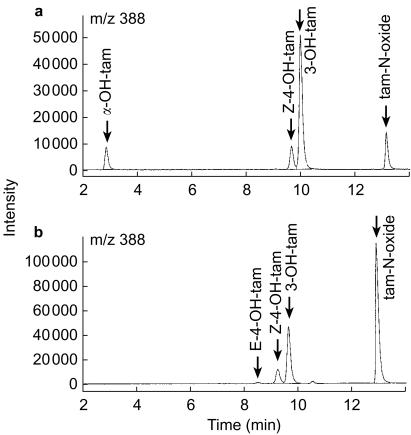
Chromatograms resulting from the analysis of an incubated intermediate concentration QC sample (100 nm of α-4-OH-tam, Z-4-OH-tam, tam-N-oxide and 500 nm 3-OH-tam) and a 10-µm tamoxifen incubation with CYP2C19 supersomes are shown in Figure 1a and b, respectively. The incubations containing human liver microsomes and supersomes with expressed CYP isoforms resulted in peaks with identical retention time to the various metabolites in the QC sample. Under the h.p.l.c. conditions described, the retention times for E-4-OH-tam, Z-4-OH-tam, and 3-OH-tam were 9.3, 9.8, and 10.1 min, respectively.
Figure 1.
Representative chromatograms from the analysis of samples containing 100 nmα-OH-tam, Z-4-OH-tam and tam-N-oxide and 500 nm 3-OH-tam (internal standard) (a) and incubated CYP2C19 supersomes with 10 µm tamoxifen and 500 nm 3-OH-tam (b).
Calibration curves were linear over the concentration range used for Z-4-OH-tam with a mean r2 value of 0.999 (n = 5). The mean (±s.d.) slopes of the calibration curves over five independent assay days for Z-4-OH-tam were 0.890 (±0.034). All intra- and interassay bias and precision values were below ±12%. The limit of quantification for Z-4-OH-tam was 0.39 pmol per sample.
Formation of Z-4-OH-tam by CYP supersomes
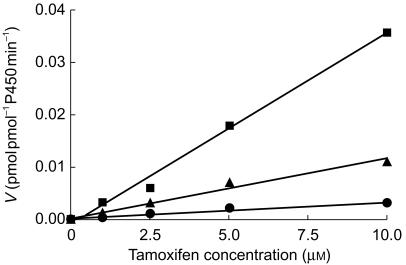
Z-4-OH-tam was formed by expressed recombinant wild-type CYP2B6, CYP2C9, CYP2C19 and CYP2D6. No formation by expressed recombinant wild-type CYP1A2, CYP2A6, CYP2E1 or CYP3A4 was observed. The derived intrinsic clearance values following further experiments with several initial tamoxifen concentrations are shown in Table 1. In addition, the intrinsic clearance of tamoxifen to Z-4-OH-tam by expressed CYP2C9.2 and CYP2C9.3 variant proteins was 48 and 89% lower, respectively, in comparison with expressed CYP2C9.1 (0.0011 and 0.0003 compared with 0.0036 µl min−1 pmol−1 P450, Figure 2).
Table 1.
Estimates of intrinsic clearance (µl min−1 pmol−1 CYP, r2) of tamoxifen to Z-4-OH-tam by supersomes expressing wild-type recombinant CYP450 isoform protein.
| Expressed CYP450 isoform | Intrinsic clearance (r2) |
|---|---|
| CYP2B6 | 2.91×10−2 (0.998) |
| CYP2C9 | 3.71×10−2 (0.992) |
| CYP2C19 | 3.93×10−2 (0.999) |
| CYP2D6 | 4.20×10−2 (0.991) |
Figure 2.
Metabolism of tamoxifen (1–10 µm, 1% dimethyl sulfoxide, n = 1) to Z-4-OH-tam in supersomes with expressed CYP2C9.1 (▪), CYP2C9.2 (▴) and CYP2C9.3 (•) proteins. The line represents that of best fit from linear regression with an estimate of in vitro intrinsic clearance provided by the slope.
Influence of 1% dimethyl sulfoxide on CYP supersome activity
Dimethyl sulfoxide (1%) inhibited the formation of Z-4-OH-tam by expressed recombinant wild-type CYP2B6, CYP2C19, CYP2D6 protein by ∼15, 14, and 11%, respectively.
Formation of Z-4-OH-tam by human liver microsomes
There was considerable variability in the rates of the formation of Z-4-OH-tam between the 50 samples, ranging from 1.7 to 27.8 pmol mg−1 protein min−1 with a mean of 8.7 pmol mg−1 protein min−1.
Inhibition studies
Quinidine inhibition in those samples with no CYP2D6 expression (n = 3) were excluded from the statistical analysis. Formation of Z-4-OH-tam was significantly inhibited by sulphaphenazole, quinidine and MAB-2B6, with mean (±s.d., CI)% inhibition compared with control of 49 (±19, 36–62)% (P = 0.001), 38 (±14, 25–52)% (P<0.05), and 13 (±11, 5–21)% (P<0.005), respectively.
Expression of CYP isoforms
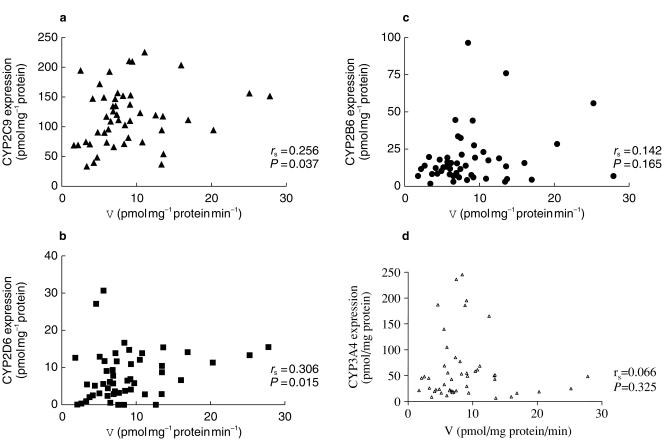
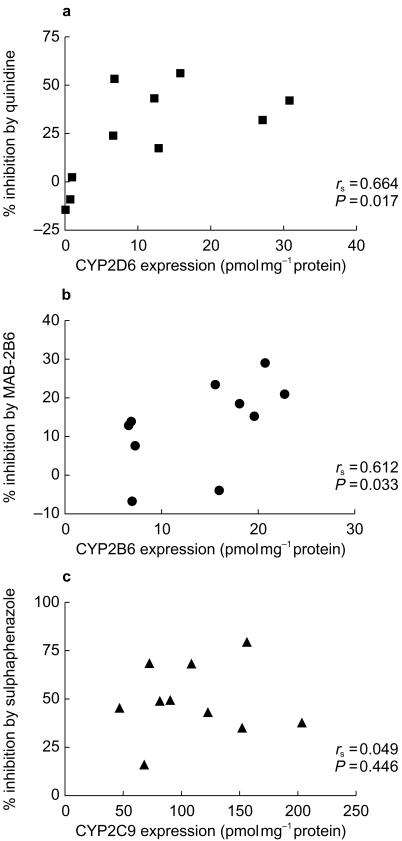
The expression of various CYP isoforms in the human liver microsomes (n = 50) determined by quantitative Western blots is shown in Table 2. The expression of both CYP2C9 and CYP2D6 in the livers correlated with Z-4-OH-tam formation rate (rs = 0.256, P<0.05, rs = 0.309, P<0.05, Figure 4a and b). No significant relationship was observed between Z-4-OH-tam formation and the expression of CYP2B6 or CYP3A4 (Figure 4c,d). Significant correlations between the percentage inhibition by quinidine of Z-4-OH-tam formation and the expression of CYP2D6 (rs = 0.684, P<0.05, Figure 3a), and the percentage inhibition by MAB-2B6 and the expression of CYP2B6 (rs = 0.612, P<0.05, Figure 3b). There was no correlation between the percentage inhibition by sulphaphenazole of Z-4-OH-tam formation and the expression of CYP2C9 (rs = 0.049, P>0.05, Figure 3c).
Table 2.
Expression of CYP isoforms in human liver microsomes (n = 49–50).
| CYP expression (pmol mg−1 protein) | ||
|---|---|---|
| mean ± s.d. | range | |
| CYP2B6 | 18.2±18.1 | 1.30–95.9 |
| CYP2C9 | 116±48.1 | 32.6–224 |
| CYP2D6 | 7.99±6.59 | 0–30.8 |
| CYP3A4 | 64.1±68.7 | 6.60–295 |
Figure 4.
Spearman rank correlation between CYP2C9 (a), CYP2D6 (b), CYP2B6 (c) and CYP3A4 (d) expression (pmol mg−1 protein) and formation rate of Z-4-OH-tamoxifen (V, pmol mg−1 protein min−1) in human liver microsomes (n = 50).
Figure 3.
Spearman rank correlation between percentage inhibition of Z-4-OH-tam formation from 10 µm tamoxifen compared with control by quinidine (a), MAB-2B6 (b) and sulphaphenazole (c) and CYP2D6, CYP2B6 and CYP2C9 expression, respectively (n = 10).
CYP2B6, CYP2C9 and CYP2D6 genotyping
CYP2B6 genotyping revealed the presence of CYP2B6*2, *4, *5, *6 and *7 allelic variants. Ten samples had two variant CYP2B6 alleles, 21 had one variant and one wild-type allele, and 18 had two wild-type alleles (Table 3a). CYP2D6 functional alleles were CYP2D6*1, *2, *9, *10, or *41. CYP2D6 genotyping revealed four poor metabolizers (zero functional alleles), 18 extensive metabolizers with one functional allele and 26 extensive metabolizers with two functional alleles (Table 3b). The CYP2C9 genotyping assay revealed four samples with zero wild-type alleles (CYP2C9*2/*2 or *2/*3), 19 with one wild-type allele (CYP2C9*1/*2 or *1/*3) and 26 with two wild-type alleles (CYP2C9*1/*1) (Table 3C).
Table 3.
Rates of formation (V, pmol mg−1 protein min−1) of Z-4-OH-tam from 10 µm tamoxifen (1% dimethyl sulfoxide) and CYP expression (pmol mg−1 protein) in human liver microsomes from patients with varying CYP2B6 (a), CYP2D6 (b) and CYP2C9 (c) genotypes. Data are mean±s.d. (CI).
| a | ||
|---|---|---|
| CYP2B6 variant alleles | V (pmol mg−1 protein min−1) | CYP2B6 expression |
| zero (n = 18) | 9.44±5.47 (6.72–12.2) | 26.3±18.6 (17.1–35.6)a,b |
| one (n = 20) | 8.83±6.08 (6.06–11.6) | 15.4±19.2 (6.67–24.1) |
| two (n = 10) | 6.82±3.76 (4.13–9.51) | 8.77±5.88 (4.56–13.0) |
| b | ||
|---|---|---|
| CYP2D6 functional alleles | V (pmol mg−1 protein min−1) | CYP2D6 expression |
| two (n = 26) | 9.93±5.43 (7.74–12.1)c | 10.4±7.08 (7.51–13.4)d |
| one (n = 18) | 8.10±5.48 (5.38–10.8) | 6.45±4.82 (4.05–8.84)e |
| zero (n = 4) | 5.04±3.17 (−0.002–10.1) | 0.155±0.161 (−0.1–0.4) |
| c | CYP2C9 variant alleles | V (pmol mg−1 protein min−1) | CYP2C9 expression | ||
|---|---|---|---|---|---|
| zero: CYP2C9*1/*1 (n = 26) | 9.98±6.67 (7.29–12.7) | 123±52.0 (102–144) | |||
| one: CYP2C9*1/*2 or *1/*3 (n = 19) | 7.81±2.69 (6.51–9.10) | 111±39.6 (91.8–130) | |||
| two: CYP2C9*2/*2 or *2/*3 (n = 4) | 5.12±3.93 (−1.13–11.4) | 94.9±66.1 (−10.3–200) | |||
P<0.05 vs one variant allele
P<0.05 vs two variant alleles
P<0.05 vs zero functional alleles
P<0.001 vs zero functional alleles
P<0.05 vs zero functional alleles.
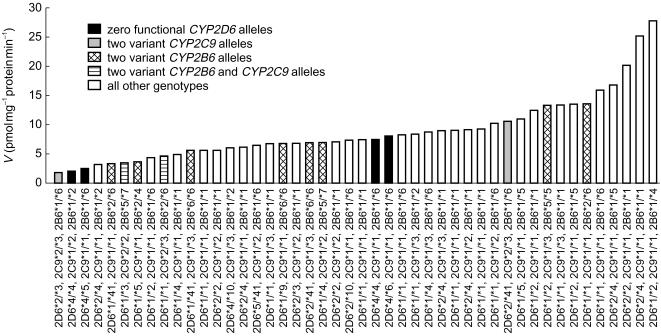
The mean expression of CYP2B6 and CYP2D6 in the livers was dependent on the number of wild-type CYP2B6 and CYP2D6 functional alleles, respectively (Table 3A and B). No significant differences in CYP2C9 expression levels were observed in livers with different CYP2C9 genotypes (Table 3C). Combining genotype for all three CYP isoforms, it was observed that two samples carried both two variant CYP2B6 alleles and two variant CYP2C9 alleles however, none had both zero functional CYP2D6 alleles and two variant CYP2C9 alleles (Figure 5).
Figure 5.
Formation rates of Z-4-OH-tam (V, pmol mg−1 protein min−1) from 10 µm tamoxifen (1% dimethyl sulfoxide) in human liver microsomes with various CYP2B6, CYP2C9 and CYP2D6 genotypes (n = 46).
A significantly higher rate of formation of Z-4-OH-tam was observed in samples with two CYP2D6 functional alleles when compared with those with no functional alleles (P = 0.046), with a trend towards significance when compared with those with one functional allele (P = 0.068, Table 3b, Figure 5). Furthermore, although there were no statistical differences between the rates of formation in the CYP2B6 and CYP2C9 genotype groups, lower formation rates were observed in samples with two variant alleles when compared with homozygous wild-type (P = 0.054 and P = 0.053, respectively, Table 3a and c, Figure 5).
Discussion
The potential importance of the formation of Z-4-OH-tam with regards to both therapy and use as a preventative agent is obvious, considering it is at least a 30-fold more potent antioestrogen compared to tamoxifen itself. This study elucidated the contribution of particular CYP isoforms to the interindividual variability in formation of this active metabolite. Amongst the 50 livers studied a 16-fold range in the formation rate was observed which is markedly higher than the 2-fold (n = 3 [7]) and 4-fold (n = 12 [8], n = 10 [9]) variability previously reported. Furthermore, the mean formation rate observed was lower in comparison to two of the previous studies (2.5-fold, n = 12 [8], 3.5-fold, n = 3 [7]), but higher in comparison to the other (∼6-fold, n = 10 [9]). Unfortunately, no information regarding CYP expression or genotype (with the exception of CYP2D6 genotype from Crewe and colleagues [9]), was available from these studies for comparison with the liver samples used in the present study. Therefore, it is likely that these differences in both the mean and range of formation rates could be due to differences in enzyme expression.
This explanation has merit since combined data from the present study indicate predominant roles for CYP2D6 and CYP2C9 in the formation of Z-4-OH-tam, with a minor contribution from CYP2B6. Comparisons between metabolite formation in samples with no CYP2D6 functional alleles and all other samples indicate that CYP2D6 contributes on average 45% to the overall formation of Z-4-OH-tam. This is in agreement with the significant inhibition observed by quinidine in the subset of samples, and the relationship observed between CYP2D6 expression and percentage inhibition with quinidine. In addition, CYP2C9 contributes on average 46% to the overall formation, as indicated by the percentage reduction in Z-4-OH-tam formed when incubated with sulphaphenazole, although this inhibition did not correlate with expression of CYP2C9. The latter is expected since Western blot analysis does not distinguish between the wild-type and variant forms of CYP2C9. Furthermore, it was observed that the contribution of CYP2C9 to the overall formation increased in those samples with no functional CYP2D6 alleles. Therefore, it is concluded that interindividual differences exist in the contribution of CYP2D6 and CYP2C9 to the overall formation of Z-4-OH-tam. These data are mostly in agreement with the report of Crewe and coworkers [9], who showed that the inhibition of metabolite formation by sulphaphenazole, quinidine and ketoconazole was highly variable in the subset of 10 samples.
The major discrepancy between our data and that of Crewe and colleagues [9] is the role of CYP3A4. Possible explanations are the use of different organic solvents to dissolve tamoxifen, whose effect on the activities of CYP isoforms has been highlighted previously [30–32], or differences in the expression systems used for studying formation by recombinant CYP3A4 (baculovirus vs lymphoblastoid cells). However, in the present study only formation of Z-4-OH-tam by CYP3A4 supersomes was below the limit of quantification, regardless of the presence of dimethyl sulfoxide. In addition, the formation of Z-4-OH-tam was linear from 1 to 10 µm tamoxifen from expressed recombinant CYP2C9, CYP2C19 and CYP2D6. Therefore the difference in substrate concentration used in this study (10 µm) compared with Crewe and colleagues (1 µm) [9] would not change observations on the relative contributions of each CYP isoform. Another consideration is that the present study used troleandomycin as a CYP3A4 specific inhibitor compared with the use of ketoconazole by Crewe et al.[9] which at the 2 µm concentration has been shown to not only inhibit CYP3A4 activity, but also that of CYP2B6 (44%) and of CYP2C9 (17%) [25]. Moreover, there was no relationship between Z-4-OH-tam formation rates and expression of CYP3A4, and no inhibition of enzyme activity by troleandomycin. Consequently, although our data indicate that CYP3A4 does not contribute to the formation of Z-4-OH-tam, previous evidence to the contrary indicates that a minor role cannot be excluded.
Since CYP2D6 and CYP2C9 are the major contributors in the formation of Z-4-OH-tam the influence of their genotypes was investigated by comparisons between the genotypic groups. A significant difference between the formation rates in samples with two vs no functional CYP2D6 alleles is not surprising considering that CYP2D6 expression increased with the number of functional alleles. This is the first study to demonstrate that CYP2D6 genotype is a determinant of an individual's ability to form Z-4-OH-tam.
A similar trend was observed between the genotypes of CYP2C9. Previous in vitro studies have shown that the CYP2C9*2 and *3 mutations lead to functional changes in CYP2C9 variant proteins [15, 16, 18], rather than to altered expression levels, with the extent of these changes dependent on the substrate. This study has shown for the first time that expressed recombinant CYP2C9.2 and CYP2C9.3 variant proteins in supersomes have decreased intrinsic clearances of tamoxifen to Z-4-OH-tam when compared with wild-type CYP2C9.1. In addition, these data imply that this reduction in intrinsic clearance would be larger in patients with the CYP2C9*3/*3 genotype when compared with patients of CYP2C9*2/*2 and CYP2C9*2/*3 genotypes. Moreover, the largest influence of genotype is expected for patients who are both CYP2D6 poor metabolizers and carry a CYP2C9*3 mutation, which is predicted to be one patient in 1000. Although investigations with a population of patients with CYP2C9*3/*3 genotype are required, the data indicate that CYP2C9 genotype may also influence the ability of an individual to form Z-4-OH-tam.
The in vitro intrinsic clearance and inhibition data demonstrate that CYP2B6 plays a minor role in Z-4-OH-tam formation. Nonetheless, CYP2B6 genotype was also observed to have an influence on formation rates. The functional and clinical significance of the allelic variants remains largely unknown. However, this could be elucidated further by studying Z-4-OH-tam formation by recombinant CYP2B6 variant proteins.
In conclusion, this study has shown a significant association between CYP2D6 genotype and Z-4-OH-tam formation in microsomes from a large number of human liver samples. In addition, a relationship between tamoxifen Z-4-hydroxylation and CYP2B6 and CYP2C9 genotype was observed, in that the majority of samples with low Z-4-OH-tam formation carried allelic variants of at least one of the three enzymes. These data are of potential clinical significance since CYP2B6, CYP2C9 and CYP2D6 genotype all contribute to the variability in the amount of the potent antioestrogen Z-4-OH-tam formed from tamoxifen, which could be a factor affecting the efficacy of the drug in patients.
Acknowledgments
We thank S. Brod, B. Klumpp and I. Liebermann for their excellent technical assistance. This study was supported by The Robert Bosch Foundation, Stuttgart, Germany, and grant number 01 GG 9846 from The German Federal Ministry of Research and Education, Berlin, Germany. Dr J. Coller is a recipient of a C.J. Martin Post-Doctoral Fellowship from the National Health and Medical Research Council of Australia.
References
- 1.Osborne CK, Fuqua AW. Mechanisms of tamoxifen resistance. Breast Cancer Res Treat. 1994;32:49–55. doi: 10.1007/BF00666205. [DOI] [PubMed] [Google Scholar]
- 2.Fisher B, Constantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 3.Foster AB, Griggs LJ, Jarman M, Van Maanen JMS, Schulten H-R. Metabolism of tamoxifen by rat liver microsomes: formation of the N-oxide, a new metabolite. Biochem Pharmacol. 1978;29:1977–1979. doi: 10.1016/0006-2952(80)90116-1. [DOI] [PubMed] [Google Scholar]
- 4.Ruenitz PC, Bagley JR, Pape CW. Some chemical and biochemical aspects of liver microsomal metabolism of tamoxifen. Drug Metab Dispos. 1984;12:478–483. [PubMed] [Google Scholar]
- 5.McCague R, Seago A. Aspects of metabolism of tamoxifen by rat liver microsomes. Identification of a new metabolite: E-1-[4-(2-dimethylaminoethoxy)-phenyl]-1, 2-diphenyl-1-buten-3-ol N-oxide. Biochem Pharmacol. 1986;35:827–834. doi: 10.1016/0006-2952(86)90251-0. [DOI] [PubMed] [Google Scholar]
- 6.Fabian C, Tilzer L, Sternson L. Comparative binding affinities of tamoxifen, 4-hydroxytamoxifen, and desmethyltamoxifen for estrogen receptors isolated from human breast carcinoma: correlation with blood levels in patients with metastatic breast cancer. Biopharm Drug Dispos. 1981;2:381–390. doi: 10.1002/bdd.2510020407. [DOI] [PubMed] [Google Scholar]
- 7.Mani C, Gelboin HV, Park SS, Pearce R, Parkinson A, Kupfer D. Metabolism of the antimammary cancer antiestrogenic agent tamoxifen. I. Cytochrome P-450-catalyzed N-demethylation and 4-hydroxylation. Drug Metab Dispos. 1993;21:645–656. [PubMed] [Google Scholar]
- 8.Dehal SS, Kupfer D. CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res. 1997;57:3402–3406. [PubMed] [Google Scholar]
- 9.Crewe HK, Ellis SW, Lennard MS, Tucker GT. Variable contribution of cytochromes P4502D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol. 1997;53:171–178. doi: 10.1016/s0006-2952(96)00650-8. [DOI] [PubMed] [Google Scholar]
- 10.Daly AK, Brockmöller J, Broly F, et al. Nomenclature for human CYP2D6 alleles. Pharmacogenetics. 1996;6:193–201. doi: 10.1097/00008571-199606000-00001. [DOI] [PubMed] [Google Scholar]
- 11.Xie H-G, Stein CM, Kim RB, Wilkinson GR, Flockhart DA, Wood AJ. Allelic, genotypic and phenotypic distributions of S-mephenytoin 4′-hydroxylase (CYP2C19) in healthy Caucasian populations of European descent throughout the world. Pharmacogenetics. 1999;9:539–549. [PubMed] [Google Scholar]
- 12.Lang T, Klein K, Fischer J, et al. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics. 2001;11:399–415. doi: 10.1097/00008571-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Crespi CL, Miller VP. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH. cytochrome P450 oxidoreductase. Pharmacogenetics. 1997;7:203–210. doi: 10.1097/00008571-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 15.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan-Klose TH, Ghanayem BI, Bell DA, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Miners JO, Birkett DJ. Cytochrome P4502C9. An enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–538. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasar U, Tybring G, Hidestrand M, et al. Role of CYP2C9 polymorphism in losarton oxidation. Drug Metab Dispos. 2001;29:1051–1056. [PubMed] [Google Scholar]
- 19.Haining RL, Hunter AP, Veronses ME, Trager WF, Rettie AE. Allelic variants of human cytochrome P450 2C9: Baculovirus-mediated expression, purification, structural characterization, substrate stereoselectivity, and prochiral selectivity of the wild-type and I359L mutant forms. Arch Biochem Biophys. 1996;333:447–458. doi: 10.1006/abbi.1996.0414. [DOI] [PubMed] [Google Scholar]
- 20.Steward DJ, Haining RL, Henne KR, et al. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics. 1997;7:361–367. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Forrester LM, Henderson CJ, Glancey MJ, et al. Relative expression of cytochrome P450 isoenzymes in human liver and association with the metabolism of drugs and xenobiotics. Biochem J. 1992;281:359–368. doi: 10.1042/bj2810359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkatakrishnan K, von Moltke LL, Court MH, Harmatz JS, Crespi CL, Greenblatt DJ. Comparison between cytochrome P450 (CYP) content and relative activity approaches to scaling from cDNA-expressed CYPs to human liver microsomes: Ratios of accessory proteins as sources of discrepancies between the approaches. Drug Metab Dispos. 2000;28:1493–1504. [PubMed] [Google Scholar]
- 23.Newton DJ, Wang RW, Lu AYH. Cytochrome P450 inhibitors: Evaluation of specificities in the in vitro metabolism of therapeutic agents by human liver microsomes. Drug Metab Dispos. 1995;23:154–158. [PubMed] [Google Scholar]
- 24.Eagling VA, Tija JF, Back DJ. Differential selectivity of cytochrome P450 inhibitors against probe substrates in human and rat liver microsomes. Br J Clin Pharmacol. 1998;45:107–114. doi: 10.1046/j.1365-2125.1998.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sai Y, Dai R, Yang TJ, et al. Assessment of specificity of eight chemical inhibitors using cDNA-expressed cytochromes P450. Xenobiotica. 2000;30:327–343. doi: 10.1080/004982500237541. [DOI] [PubMed] [Google Scholar]
- 26.Zanger UM, Hauri HP, Loeper J, Homberg JC, Meyer UA. Antibodies against human cytochrome P-450db1 in autoimmune hepatitis type II. Proc Natl Acad Sci USA. 1988;85:8256–8260. doi: 10.1073/pnas.85.21.8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanger UM, Fischer J, Raimundo S, et al. Comprehensive analysis of the genetic factors determining expression and function of hepatic CYP2D6. Pharmacogenetics. 2001;11:573–585. doi: 10.1097/00008571-200110000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Stüven T, Griese EU, Kroemer HK, Eichelbaum M, Zanger UM. Rapid detection of CYP2D6 null alleles by long distance- and multiplex-polymerase chain reaction. Pharmacogenetics. 1996;6:417–421. doi: 10.1097/00008571-199610000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Griese EU, Zanger UM, Brudermanns U, et al. Assessment of the predictive power of genotypes for the in-vivo catalytic function of CYP2D6 in a German population. Pharmacogenetics. 1998;8:15–26. doi: 10.1097/00008571-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 30.Hickman D, Wang J-P, Unadkat JD. Evaluation of the selectivity of in vitro probes and suitability of organic solvents for the measurement of human cytochrome P450 monooxygenase activities. Drug Metab Dispos. 1998;26:207–215. [PubMed] [Google Scholar]
- 31.Chauret N, Gauthier A, Nicoll-Griffith DA. Effect of common organic solvents on in vitro cytochrome P450-mediated metabolic activities in human liver microsomes. Drug Metab Dispos. 1998;26:1–4. [PubMed] [Google Scholar]
- 32.Bubsy WF, Ackermann JM, Crespi CL. Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of cDNA-expressed human cytochromes P-450. Drug Metab Dispos. 1999;27:246–249. [PubMed] [Google Scholar]